Abstract
Campylobacter species are fastidious to culture, and the ability to directly quantify biomass in microbiologically complex substrates using real-time quantitative (RTQ) PCR may enhance our understanding of their biology and facilitate the development of efficacious mitigation strategies. This study reports the use of nested RTQ-PCR to directly quantify Campylobacter jejuni and Campylobacter lanienae in cattle feces. For C. jejuni, the single-copy mapA gene was selected. For C. lanienae, the three-copy 16S rRNA gene was targeted. RTQ-PCR primers were tested alone or they were nested with species-specific primers, and amplification products were detected using the intercalating dye SYBR Green. Nesting did not increase the specificity or sensitivity of C. jejuni quantification, and the limit of quantification was 19 to 25 genome copies (≈3 × 103 CFU/g of feces). In contrast, nested RTQ-PCR was necessary to confer specificity on C. lanienae by targeting the 16S rRNA gene. The limit of quantification was 1.8 genome copies (≈250 CFU/g of feces), and there was no discernible difference between the two C. lanienae secondary primer sets evaluated. Detection and quantification of C. jejuni in naturally infested cattle feces by RTQ-PCR were comparable to the results of culture-based methods. In contrast, culturing did not detect C. lanienae in 6 of 10 fecal samples positive for the bacterium and substantially underestimated cell densities relative to nested RTQ-PCR. The results of this study illustrate that RTQ-PCR can be used to directly quantify campylobacters, including very fastidious species, in a microbiologically and chemically complex substrate. Furthermore, targeting of a multicopy universal gene provided highly sensitive quantification of C. lanienae, but nested RTQ-PCR was necessary to confer specificity. This method will facilitate subsequent studies to elucidate the impact of this group of bacteria within the gastrointestinal tracts of livestock and studies of the factors that influence colonization success and shedding.
Campylobacter species are recognized as one the most frequent causes of acute diarrheal disease in humans in North America (Centers for Disease Control and Prevention-U.S. Department of Agriculture-Food and Drug Administration Collaborating Sites Foodborne Disease Active Survey Network [Foodnet; http://www.cd.gov/foodnet/annuals.htm]). Alberta, Canada, possesses a very large beef cattle population (≈5 million head) centered in the Southern region of the province, but relatively limited research has investigated the prevalence of Campylobacter species associated with beef cattle. The prevalence of Campylobacter infections in humans in this region is considerably higher than the national average (Health Canada website [http://dsol-smed.hc-sc.gc.ca/dsol-smed/ndis/index_e.html]), and cattle in this region shed a variety of Campylobacter species (8). In a PCR-based survey of 380 feedlot steers, 83% of the animals were observed to be shedding campylobacters, and the most prevalent taxa were Campylobacter lanienae (49%) and Campylobacter jejuni (38%) (9).
Our research is focusing on the elucidation of factors affecting the colonization of the gastrointestinal (GI) tracts of cattle by campylobacters and their release in feces. The ability to rapidly and accurately quantify the biomasses of different Campylobacter species will greatly facilitate such studies. The quantification of campylobacters by plating methods can provide erroneous results. For example, the motility of Campylobacter species, their association with specific tissues (e.g., mucosa), their occupation of various niches within the GI tract, the differential selectivity of isolation media, and the logistical limitations of plating methods (i.e., samples must be processed rapidly, and it is a very labor-intensive process) can all limit the utility of culture-based enumeration methods. Real-time quantitative (RTQ) PCR has been used to quantify the biomass of C. jejuni in pure culture (24, 35). Furthermore, Sails et al. (28) used RTQ-PCR to quantify C. jejuni DNA in foods. However, they used enrichment prior to the application of RTQ-PCR. The ability to quantify Campylobacter species directly in microbiologically complex habitats such as feces without relying on an enrichment step may provide a more accurate measure of the biomass present in such substrates.
Many Campylobacter species, including species commonly associated with cattle, are very fastidious, and media used to selectively isolate C. jejuni inhibit the growth of these species (8). Furthermore, direct RTQ-PCR is attractive for logistical reasons (e.g., time effectiveness), and it may provide more accurate information on the spatial distribution of campylobacters in niches, such as within the GI tract. The overall objective of the present study was to develop a direct RTQ-PCR method for quantifying campylobacters commonly shed in cattle feces. SYBR Green is an intercalating dye that can be used for RTQ-PCR because it fluoresces 50 to 100 times more brightly in the presence of double-stranded DNA (i.e., that resulting from the extension step of each cycle) than unbound dye. The primary advantage of using SYBR Green over molecular probes (e.g., Taqman) is cost, and it requires less optimization. The salient disadvantages of using SYBR Green are related to nonspecific binding to any double-stranded nucleic acid. Nested PCR (nPCR) is often used to increase the sensitivity of detection of bacteria in naturally contaminated substrates (8, 10, 23, 31, 34), and the application of nested RTQ-PCR to increase accuracy has been reported (5, 22). However, the application of nested RTQ-PCR has not been widely adopted. Critical and strict evaluation of newly developed assays is a prerequisite to obtain reliable data, and the following specific objectives were considered imperative for the development of an economical yet accurate RTQ-PCR method using SYBR Green: (i) to develop primers for RTQ-PCR of C. jejuni (targeting the mapA gene) and C. lanienae (targeting the 16S rRNA gene), (ii) to measure the sensitivity and specificity of nested RTQ-PCR for quantifying C. jejuni and C. lanienae in inoculated cattle feces, and (iii) to assess the accuracy of RTQ-PCR for detecting and quantifying C. jejuni and C. lanienae in naturally infested cattle feces.
MATERIALS AND METHODS
Bacterial strains.
C. jejuni ATCC 49943 and C. lanienae L52 were used for preparing the quantification standards for RTQ-PCR and as inocula in the fecal inoculation experiment; the strain of C. lanienae was originally isolated from cattle feces at the Agriculture and Agri-Food Canada Research Centre, Lethbridge, Canada (8). The strain of C. jejuni was cultured on Brucella agar (Difco, Detroit, Mich.) at 37°C in anaerobic gas jars (Oxoid, Nepean, Ontario, Canada). Microaerophilic conditions were generated with the CampyPak Plus microaerophilic system (BBL, Cockeysville, Md.) with palladium catalyst (Becton Dickinson, Sparks, Md.). C. lanienae was cultured on Karmali agar (Oxoid) (12) at 40°C under the same microaerophilic conditions. The two media were not amended with antibiotics.
C. lanienae 16S rRNA gene copy number.
To determine the copy number of the C. lanienae 16S rRNA gene, Southern hybridization was conducted. The genomic DNAs of C. lanienae L52 and C. jejuni ATCC 49943 were restricted with PstI, SacI, SalI, SmaI (Invitrogen Canada Inc., Burlington, Ontario, Canada), and NcoI (Promega, Madison, Wis.) according to the manufacturers' instructions. The digests were electrophoresed in 0.8% agarose in Tris-acetate-EDTA buffer for ≈4 h at 85 V. The DNA was transferred to a Hybond N+ nylon membrane (Amersham Biosciences Corp., Piscataway, N.J.) overnight by capillary transfer using established procedures. The probe used was prepared by PCR amplification of the 16S rRNA gene of C. lanienae L52 with the primers UNI27F and UNI519R (16). The PCR products were purified using the Qiaquick PCR purification kit (Qiagen Inc., Mississauga, Ontario, Canada). The probe was labeled with 32P using the Prime-It RmT random primer-labeling kit (Stratagene, Cedar Creek, Tex.), and hybridization was conducted according to the method of Sambrook and Russell (30). The blot was visualized on Biomax MS film (Kodak Canada Inc., Vancouver, British Columbia, Canada).
Genome size of C. lanienae.
Pulsed-field gel electrophoresis (PFGE) was conducted according to the basic protocol of Ribot et al. (25). C. lanienae L52 and Campylobacter fetus subsp. fetus ATCC 25936 were included, and the restriction enzymes SmaI and SalI were used (29). Plugs were loaded into individual wells of 1% SeaKem gold agarose (Mandel Scientific, Guelph, Ontario, Canada), and electrophoresis was conducted using a CHEF-DRII (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada). The conditions consisted of an initial switch time of 6.75 s and a final switch time of 38.35 s (the gradient of 6 V/cm included an angle of 120°). The gels were electrophoresed for ≈18 h in 0.5× Tris-borate-EDTA. The gels were stained in ethidium bromide, restriction fragments were visualized under UV light, and the fragments were sized relative to a MidRange II PFG marker (New England Biolabs) using Gel-Pro Analyzer software (MediaCybernetics, Carlsbad, Calif.).
Quantification standard.
C. jejuni and C. lanienae cells were scraped from the surfaces of the Brucella and Karmali agars, respectively, after 48 h. DNA was extracted from the harvested cell mass using a DNeasy kit (Qiagen Inc.) according to the manufacturer's protocol. The DNA was measured fluorimetrically using a Hoefer DyNA Quant 200 apparatus (Amersham Biosciences Corp.); calf thymus DNA (Calbiochem, San Diego, Calif.) was used as a standard. The numbers of genome copies of C. jejuni (based on a genome size of 1.6 Mbp) and C. lanienae (based on a genome size of 808 kbp) in 1 ng of DNA were 5.649 × 105 and 1.147 × 106, respectively. Genomic DNA standards for both bacteria were diluted in a 10-fold dilution series in 10 mM Tris (pH 8.5); standard DNA was thawed and frozen a maximum of two times. The procedure was conducted on three separate occasions for each bacterium.
Primer design.
The primers used (Sigma Genosys, Mississauga, Ontario, Canada) are presented in Table 1. For C. jejuni and C. lanienae, the mapA and 16S rRNA genes, respectively, were targeted. Primary amplification of the C. jejuni mapA gene produces a 589-bp amplicon (3). For C. lanienae, primary amplification produces a 920-bp amplicon (8). Both primary primer sets have been shown to be specific for C. jejuni and C. lanienae found in cattle feces (8).
TABLE 1.
Primer sequences for the amplification of Campylobacter species from cattle feces
| PCR target genea | Primer | Tab (°C) | Sequence (5′ to 3′)c | Size (bp) | Reference |
|---|---|---|---|---|---|
| C. jejuni mapA | Primary | ||||
| MDmapA1Upper | 58 | CTATTTTATTTTTGAGTGCTTGTG | 589 | Denis et al. (3) | |
| MDmapA2Lower | GCTTTATTTGCCATTTGTTTTATTA | Denis et al. (3) | |||
| C. jejuni mapA | Nested | ||||
| QCjmapANF | 58 | GGTTTTGAAGCAAAGATTAAAGG | 94 | New primera,d | |
| QCjmapANR | AAGCAATACCAGTGTCTAAAGTGC | New primera,d | |||
| C. lanienae 16S rRNA | Primary | ||||
| CLAN76F | 54 | GTAAGAGCTTGCTCTTATGAG | 920 | Logan et al. (19) | |
| CLANL521021R | TCGTATCTCTACAAGGTTCTTA | Inglis and Kalischuk (8) modified CLAN1021R from Logan et al. (19) | |||
| C. lanienae 16S rRNA | Nested 1 | ||||
| QClanN1F | 58 | ACACGGTCCAGACTCCTACG | 103 | New primerd | |
| QClanN1R | ACGCTCCGAAAAGTGTCATC | New primerd | |||
| C. lanienae 16S rRNA | Nested 2 | ||||
| QClanN2F | 58 | AGGCGATCTGCTGGAACTTA | 89 | New primerd | |
| QClanN2R | GTTTAGGGCGTGGACTACCA | New primerd | |||
| C. jejuni, C. coli, C. hyoilei 16S rRNA | Primary | ||||
| MD16S1Upper | 58 | ATCTAATGGCTTAACCATTAAAC | 857 | CCCJ609F from Linton et al. (18) modified by Denis et al. (3) | |
| MD16S2Lower | GGACGGTAACTAGTTTAGTATT | CCCJ1442R from Linton et al. (18) modified by Denis et al. (3) |
Primer developed using Primer3 software.
Ta, annealing temperature.
Boldfaced nucleotides indicate regions of the primer that were modified.
Net Primer (Primer Biosoft International, Palo Alto, Calif.) was used to check the design.
When SYBR Green is used, RTQ-PCR primers should be 18 to 25 nucleotides long, they should have similar melting temperatures (≈58 to 60°C), they should have no more than two Gs or Cs among the last 5 nucleotides at the 3′ end, and the G/C content should be ≈50 to 60% (Plant-Microbe Genomics Facility at The Ohio State University website [http://www.biosci.ohio-state.edu/∼pmgf/procedures_rt-pcr.pdf]). Furthermore, the amplicon should be between 75 and 150 bases long and must contain no or minimal hairpin structure. For C. jejuni, the nested RTQ-PCR primer sets were designed with Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) and provided a 94-bp amplicon (Table 1); a search of the National Center for Biotechnology Information database (for short, nearly exact sequences) indicated that the QCjmapANF (24 of 24 bases) and QcjmapANR (23 of 23 bases) primers were predicted to be specific for C. jejuni AL139077 and X80135. No other bacteria were predicted. For C. lanienae, two nested RTQ-PCR primer sets were designed using the 16S rRNA gene. Primers were designed manually from sequences of C. lanienae AY288304 and AF043425 aligned with those of closely related Campylobacter species (C. fetus subsp. fetus AJ306568 and AJ306569, Campylobacter fetus subsp. venerealis AF482990, Campylobacter hyointestinalis M65010 and AF219235, and Campylobacter hyointestinalis subsp. lawsonii AF097688). All prospective primers were then checked for hairpin structure, dimers, cross dimers, palindromes, repeats, and runs using Net Primer software (Premier Biosoft International, Palo Alto, Calif.). Given the requirements placed on RTQ-PCR primers, it was not possible to design appropriate primers that were specific for C. lanienae using the 16S rRNA gene. A search of the National Center for Biotechnology Information database indicated that none of the nested primers designed for C. lanienae were exclusive for the bacterium. Other species of Campylobacter (i.e., Campylobacter coli, Campylobacter concisus, Campylobacter curvus, Campylobacter helveticus, C. jejuni, Campylobacter lari, Campylobacter mucosalis, Campylobacter rectus, Campylobacter sputorum, and Campylobacter upsaliensis) possessed 16S ribosomal DNA sequences identical to the primers QClanN1F, QClanN1R, and QClanN2F. QClanN2R was predicted to be nonspecific for Campylobacter species.
To test the specificities of RTQ-PCR primers, conventional PCR was conducted against the following reference strains: Arcobacter butzleri LMG 10243 and LMG 10828, Arcobacter cryaerophilus LMG 10209, Arcobacter skirrowii LMG 6621 and LMG 10238, C. coli ATCC 49941, C. fetus subsp. fetus ATCC 25936, C. hyointestinalis subsp. hyointestinalis ATCC 35217, C. hyointestinalis subsp. lawsonii NCTC 12901, C. jejuni ATCC 49943, C. lanienae L52 and NCTC 13004, C. lari ATCC 35221, C. sputorum subsp. fecalis LMG 8532, Campylobacter sputorum subsp. sputorum LMG 8535, and C. upsaliensis LCDC 5424. To obtain biomass, isolates were grown on Campylobacter blood-free selective agar base (modified Campylobacter charcoal differential agar [CCDA]) (Oxoid) without the antibiotic supplement or on Brucella agar without antibiotics. The cultures were maintained microaerophilically at 37°C for 48 h, and DNA was extracted from harvested cells as described above. The conditions for amplification were 1 cycle at 95°C for 15 min, followed by 25 cycles of 30 s at 94°C, 90 s at the appropriate annealing temperature (Table 1), and 60 s at 72°C, ending with an extension cycle of 10 min at 72°C. The PCR mixtures contained a total volume of 20 μl consisting of 1× reaction buffer, 0.2 mM deoxynucleoside triphosphates, 2 mM MgCl2, 0.5 μM each primer, 0.2 μg of bovine serum albumin (Promega), and 1 U of HotStar Taq polymerase (Qiagen Inc.). Each PCR was performed with a total of 2 μl of a 10−3 dilution of genomic DNA (≈100 pg/μl). All PCR products (10 μl) were electrophoresed in a 2% Tris-borate-EDTA-agarose gel (Invitrogen Corp.), visualized by being stained with ethidium bromide, and viewed under UV light. A 100-bp ladder (Promega) was used to size the products.
Primary PCR.
For nested RTQ-PCR, it is imperative that amplification in the primary step terminate within the exponential phase. The appropriate number of amplification cycles per copy of the C. jejuni mapA gene was calculated based on the following equation (5): n = log10 [Sn/Smax]/log10 (1 + e1), where n is the number of cycles, Sn is the maximum permitted product in the primary PCR step (typically 1/10 of the primer concentration in moles per liter), Smax is the maximum target sequence concentration to be tested (we used a maximum concentration of 105 copies) in moles per liter, and e1 is the primer PCR step efficiency, which is assumed to be 1 if it is unknown. We used a primer concentration of 0.5 μM, and therefore, the equation was solved as follows: Sn = 0.1 × 5 × 10−7 M = 5 × 10−8 M; Smax = 105 copies/6.022 × 1023 copies/mol = 1.66 × 10−19 mol/2.0 × 10−5 liter = 8.30 × 10−15 M. Solving for n gives n = log10 [5 × 10−8 M/8.3 × 10−15 M]/log10 (1 + 1) and n = log10 (6.024 × 106)/log10 (2) = 6.80/0.30 = 22.7 cycles.
For C. lanienae, 22 cycles were used in primary PCR. However, in preliminary experimentation, we observed that C. jejuni typically occurred in feces at densities of <105 copies (data not presented), and 25 cycles was chosen for primary amplification of the bacterium. The conditions used for primary amplification were 1 cycle at 95°C for 15 min, followed by 22 (C. lanienae) or 25 (C. jejuni) cycles of 30 s at 94°C, 90 s at the appropriate annealing temperature (Table 1), and 60 s at 72°C, ending with an extension cycle of 10 min at 72°C. The reaction mixture described above was used.
RTQ-PCR and melt curve analysis.
PCR was conducted using an iCycler iQ (Bio-Rad Laboratories Ltd). Reaction mixtures consisted of a total volume of 20 μl containing 1× QuantiTect SYBR Green PCR Master Mix (Qiagen Inc.), 0.5 μM each primer (Table 1), and 1 nM fluorescein (Bio-Rad Laboratories Ltd.). Each PCR was performed with a total of 2 μl of template DNA. The conditions for amplification were 1 cycle at 95°C for 15 min, followed by 40 cycles of 15 s at 94°C, 30 s at 58°C, and 30 s at 72°C for data acquisition. Melt curve analysis was conducted over a range of 55 to 95°C, with increments set at 0.5°C for 10 s (80 cycles); the melting point can be used as a measure of the specificity of amplification. In each run, standard samples were included to establish a standard curve. In all instances, each sample was run twice (i.e., two subsamples) per run.
The data were analyzed using iCycler iQ software (version 3.0; Bio-Rad Laboratories Ltd.). The user-defined “PCR Base Line Subtracted” (i.e., a set of baseline cycles is applied globally to all samples) and “Threshold Cycle Calculation” options were used to obtain the number of threshold cycles per well. Samples that produced no product or that produced an amplicon with an anomalous melting point were entered as missing. The linear equation for the standard curve (i.e., containing known quantities of DNA) was then used to interpolate the numbers of copies present in the unknown samples. The correlation coefficients for the standards averaged 0.97 (range, 0.92 to 0.99).
Inoculated cattle feces. (i) Inoculum preparation.
Cells of C. jejuni and C. lanienae were obtained as described above. Cells scraped from the media were suspended in sterile Brucella broth (Difco) for C. jejuni and in sterile Columbia broth for C. lanienae. For C. jejuni and C. lanienae, the turbidities (A600) of the suspensions were adjusted to 0.5 and 0.7, respectively. Tenfold dilutions were prepared in Brucella broth for C. jejuni and in Columbia broth for C. lanienae (i.e., density treatments); Brucella broth caused mortality of C. lanienae cells. To estimate cell densities in each of the suspensions, dilution spread plate counts were made on Brucella agar for C. jejuni or Karmali agar for C. lanienae. The cultures were maintained under microaerophilic conditions as described above. In all instances, C. jejuni cultures were maintained at 37°C, whereas C. lanienae cultures were incubated at 40°C. The colonies were enumerated at the dilution yielding 20 to 200 CFU per dish.
To quantify the relationship between CFU counts and actual Campylobacter cells present in inocula, a separate experiment was conducted. Inocula of C. jejuni and C. lanienae were prepared, and CFU were enumerated on Brucella and Karmali agars as described above. Simultaneously, the numbers of cells in the inocula were determined directly using a Petroff-Hausser counting chamber at ×400 magnification with differential interference contrast. To facilitate counting, the cell suspension was diluted two times in Columbia broth containing 40% glycerol. Given the motility of campylobacters, it was not possible to accurately count all of the cells within a 0.04-mm2 “large” square. Therefore, counts were restricted to five individual “small” squares (2.5 × 10−3 mm2) within each of five large squares. Three independent replicates were conducted, and differences in cell density as determined by direct counting relative to plating were calculated for each.
(ii) Inoculation of feces.
Fresh cattle feces were collected from cattle at the Lethbridge Research Centre. Fecal samples that were determined to be negative for Campylobacter DNA or that produced a weak amplicon using the Campylobacter genus 16S rRNA gene primers were selected (8). Feces that were free of Campylobacter DNA were infrequently obtained, and this necessitated the use of contaminated feces, albeit feces containing small amounts of campylobacters. Subsamples of feces were stored at −20°C until they were required, and samples were thawed once.
Feces were inoculated with C. jejuni or C. lanienae at a level of 2 ml per 18 g (wet weight) of feces for each density treatment; sterile Brucella and Columbia broths were used as controls for C. jejuni and C. lanienae, respectively. Immediately after the addition of inocula, the feces were thoroughly mixed with a metal spatula. The experiment was conducted on three separate occasions (replicates). For each replicate, two separate samples were removed from each density treatment (subsamples) for dilution plating and RTQ-PCR.
(iii) Dilution plating.
To enumerate campylobacters in cattle feces, 2.5 g of feces from each sample was placed in 22.5 ml of phosphate-buffered saline (consisting of 130 mM sodium chloride and 10 mM sodium phosphate buffer, pH 7.2) in a 50-ml Falcon tube (DiaMed, Missassauga, Ontario, Canada), and the suspension was vortexed at the maximum setting for 1 min. The suspension was diluted in a 10-fold dilution series, and 100 μl was spread in duplicate onto Campylobacter blood-free selective agar base (CCDA; Oxoid) containing selective supplement SR115E (Oxoid) for C. jejuni and on Karmali agar (Oxoid) with selective supplement CM935 (Oxoid) for C. lanienae. The cultures were incubated microaerophilically at 37 and 40°C as described above. The colonies were enumerated at the dilution yielding 20 to 200 CFU after 48 h, and the number of CFU per gram of feces (fresh weight) was calculated. In no instance were colonies of C. lanienae recovered from feces. For C. jejuni, a mean value (in CFU per gram) for the two cultures per subsample and density treatment per replicate was used to calculate the overall mean and standard error of the mean. Arbitrarily selected colonies were streaked for purity and stored in brucella broth amended with 30% glycerol at −20°C until they were processed. To identify the isolates, colony PCR was applied (8).
(iv) DNA extraction and RTQ-PCR.
The QIAamp DNA stool minikit (Qiagen) was used to extract DNA from 200 ± 5 mg of feces from each sample according to the manufacturer's protocol for isolation of DNA from stools for pathogen detection. To determine whether sufficient PCR inhibitors had been removed to allow amplification, an internal control was used (8). The internal control was constructed by deleting a fragment of the C. jejuni ATCC 49943 16S rRNA gene, and it was designed to amplify under the same PCR conditions described for the Campylobacter genus primer set; PCR amplification yielded a 475-bp product instead of the 816-bp product. Internal-control DNA was added to feces prior to extraction at a level of 10 μl per 200 mg of feces. The presence of either a genus-specific or internal-control amplicon, either of which indicated adequate removal of PCR inhibitors, was assessed with a nonnested Campylobacter genus-specific primer set (8). The DNA was stored at −20°C until it was processed.
Nested and nonnested RTQ-PCRs were conducted as described above. The numbers of copies of C. jejuni and C. lanienae genomic DNAs in 2 μl of template were determined relative to a DNA standard. Each sample was run in duplicate, and a mean value was obtained. In instances where one of the duplicate samples was negative, it was entered as a missing value (i.e., the one positive value was used).
(v) Statistical analysis.
For all primer set combinations, the relationship between log CFU per gram or log cells per gram (from dilution plating and Petroff-Hausser counts) and log genome copies (from RTQ-PCR) was determined using regression analysis (32); the resultant equations were used to convert log genome copies to log CFU per gram in the uninoculated beef cattle experiments described below. Analysis of covariance using linear (for C. jejuni) and quadratic polynomial (for C. lanineae) relationships, with log CFU per gram used as the covariant, was conducted to compare slopes and y intercepts for nested and nonnested RTQ-PCRs using the mixed procedure of SAS (32). For C. jejuni, the curves did not cross the y axis, and to prevent extrapolation, the smallest CFU per gram value at which an RTQ-PCR amplicon was detected was subtracted from all other values prior to analysis. To determine whether linear slopes differed significantly from 1, the regression procedure of SAS was applied (32).
Uninoculated beef cattle feces. (i) Feces acquisition.
Feces were extracted per rectum from 20 Angus cross cattle on 4 February 2003 at the Lethbridge Research Centre. Immediately following collection, the feces were placed on ice and transported to the laboratory for isolation of campylobacters by dilution plating and for allocation to tubes for DNA extraction (ca. 1 h). The feces from each animal were divided into two aliquots (A and B).
(ii) Dilution plating.
Feces from aliquot A were diluted in buffer, and 100 μl from each aliquot was spread on CCDA and Karmali agars containing antibiotic supplements in duplicate; the cultures were incubated microaerophilically at 37 and 40°C as described above for the inoculated cattle feces experiment. At 48- and 72-h intervals, the CFU of Campylobacter species were enumerated based on colony morphology and microscopic appearance. If possible, a minimum of 10 arbitrarily selected colonies deemed to be campylobacters were transferred to the same medium from which they were isolated, and streaked for purity. The isolates were then subjected to colony PCR using C. coli-C. jejuni, C. fetus-C. lanienae, and C. hyointestinalis multiplex primer sets (8). In most instances, groupings based on colony and cell morphologies were found to be monotaxic (based on PCR identification), and a mean value (in CFU per gram) for the duplicate cultures for each Campylobacter species per animal was calculated. Representative strains were stored in brucella broth amended with 30% glycerol at −20 and −80°C.
(iii) DNA extraction and PCR.
Genomic DNA was extracted from both fecal aliquots seeded with internal-control DNA, and conventional PCR using the nonnested primer set for Campylobacter genus DNA was applied (8). Conventional nPCR for C. jejuni and C. lanienae (Table 1) was then conducted on DNA from both aliquots. Nonnested conventional PCR for C. jejuni-C. coli-Campylobacter hyoilei (Table 1) was also carried out, but only on DNA extracted from fecal aliquot A; this primer set also detects C. lanienae in cattle feces. In addition, nested and nonnested RTQ-PCRs for C. jejuni and C. lanienae (i.e., both primer sets) were conducted in duplicate on DNA extracted from fecal aliquot A as described previously. The mean number of genome copies in 2 μl of template was determined relative to a standard curve, and values for log CFU per gram or log cells per gram were obtained from log genome copy values using the corresponding equation applied to feces inoculated with either C. jejuni or C. lanienae.
RESULTS
C. lanienae 16S rRNA gene copy number.
Southern analysis indicated that the C. lanienae L52 genome contained three copies of the 16S rRNA gene. This corresponded to the banding patterns observed for C. jejuni ATCC 49943, which possesses three copies of the gene.
Genome size of C. lanienae.
Following PFGE, the sum of the bands of C. lanienae L52 genomic DNA cut with SmaI was estimated to be 0.808 Mb compared to 0.729 Mb for DNA restricted with SalI (Table 2). The banding pattern obtained with SmaI was most distinct, and a genome size of 808 kbp was subsequently used to calculate the number of genome copies in 1 ng of DNA.
TABLE 2.
Molecular sizes of the restriction enzyme fragments of C. lanienae L52 and C. fetus ATCC 25936
| Fragment No. | Restriction enzyme fragment size (kbp)
|
|||
|---|---|---|---|---|
| Clan-SmaI | Clan-SalI | Cfet-SmaI | Cfet-SalI | |
| 1 | 260 | 243 | 324 | 245 |
| 2 | 220 | 219 | 253 | 213 |
| 3 | 166 | 167 | 187 | 186 |
| 4 | 88 | 58 | 116 | 155 |
| 5 | 74 | 42 | 74 | 136 |
| 6 | 57 | 116 | ||
| 7 | 47 | 106 | ||
| 8 | 96 | |||
| 9 | 66 | |||
| 10 | 37 | |||
| Total size | 808 | 729 | 1,058 | 1,356 |
Primer design.
The C. jejuni RTQ-PCR primers QCjmapANF and QCjmapANR, targeting the mapA gene, were specific to C. jejuni by conventional PCR (i.e., none of the other Campylobacter species were amplified) (data not shown). In contrast, the C. lanienae RTQ-PCR nested primer set 1 (QClanN1F and QClanN1R) amplified all strains tested with the exception of A. skirrowii. Similarly, the C. lanienae RTQ-PCR nested primer set 2 (QClanN2F and QClanN2R) amplified all of the Campylobacter strains tested, but it did not amplify any of the Arcobacter strains.
Inoculated cattle feces. (i) C. jejuni.
No C. jejuni cells were isolated from control feces. From feces inoculated at target densities of 102, 103, 104, 105, and 106 CFU/g and 2.49 ± 0.11, 3.38 ± 0.09, 4.28 ± 0.01, 5.28 ± 0.03, and 6.21 ± 0.05 log CFU/g were isolated on CCDA, respectively. The cell density, as estimated by plating, was similar to that obtained by direct counting using the Petroff-Hausser chamber. A differential of 1.3-fold (range, 1.0 to 1.8-fold) using the counter chamber relative to plating on CCDA was observed. The population size based on plating (in CFU per gram) was used for comparative analyses with RTQ-PCR data.
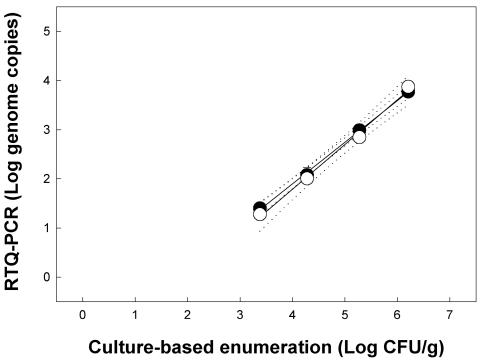
No PCR products were obtained from control feces by using nested RTQ-PCR for C. jejuni, and the results of the three independent replicates were indistinguishable. An amplicon was detected in only one of six samples from feces inoculated at a target density of 102 CFU/g. For feces inoculated at target densities of 103, 104, 105, and 106 CFU/g, 1.40 ± 0.26, 2.08 ± 0.15, 2.99 ± 0.03, and 3.78 ± 0.04 log genome copies, respectively, were observed (Fig. 1). The regression equation to describe the linear relationship between log genome copies and log CFU per gram was y = −1.49 + 0.85x, and the coefficient of correlation (r2) for this relationship was 0.99; the slope of the linear relationship differed from 1 (F = 21.6; df = 1, 11; P < 0.001). The limit of quantification for C. jejuni was 25 genome copies (antilog 1.40 copies), representing a density of ≈3 × 103 CFU/g of feces (Fig. 1). The melting point of the amplicon was 76.5 to 77.0°C.
FIG. 1.
Detection of C. jejuni in inoculated cattle feces by targeting the single-copy mapA gene with nested (solid symbols) and nonnested (open symbols) RTQ-PCRs. The log linear equations to describe the relationships are as follows: nested, y = −1.49 + 0.85x, r2 = 0.99; nonnested, y = −1.85 + 0.91x, r2 = 0.99. The error bars representing standard errors of the mean (n = 3) are within the symbols. The dotted lines represent 95% confidence intervals.
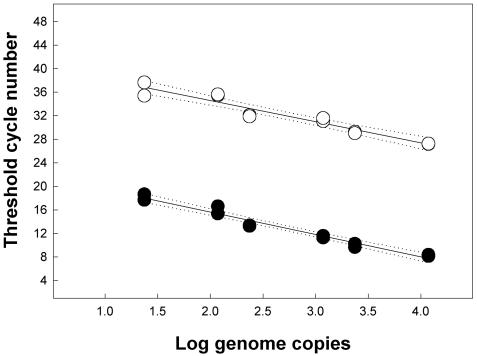
Nonnested RTQ-PCR of C. jejuni produced a relationship between log genome copies and log CFU/g very similar to that with nested RTQ-PCR (Fig. 1). The equation describing the log relationship was y = −1.85 + 0.91x, with an r2 of 0.99. The slope did not differ from 1 (F = 3.7; df = 1, 10; P = 0.084), and the limit of quantification was 19 genome copies (antilog 1.28 copies) (Fig. 1). In contrast to nested RTQ-PCR, the threshold cycles obtained with nonnested RTQ-PCR were substantially higher (by ≈19 cycles) (Fig. 2). There were no differences (F = 2.88; df = 1, 21; P = 0.10) in either the slope or y intercept (F = 1.5; df = 1, 22; P = 0.23) between the nested and nonnested RTQ-PCRs for C. jejuni by targeting the single-copy mapA gene.
FIG. 2.
Threshold values for C. jejuni in inoculated cattle feces by targeting the single-copy mapA gene for nested (solid symbols) and nonnested (open symbols) RTQ-PCRs. The linear equations to describe the relationships are as follows: nested, y = 23.22 − 3.80x, r2 = 0.97; nonnested, y = 41.74 − 3.58x, r2 = 0.92. The dotted lines represent 95% confidence intervals.
(ii) C. lanienae.
No Campylobacter cells were isolated from control feces or from feces inoculated with cells of C. lanienae, consistent with the fastidious nature of the bacterium. From inocula, densities of C. lanienae isolated on Karmali agar were 1.33 ± 0.25, 2.07 ± 0.22, 3.17 ± 0.18, 4.16 ± 0.19, 4.99 ± 0.22, and 6.00 ± 0.26 log CFU/g for the respective density treatments. Colonies of C. lanienae were very small and inconspicuous on Karmali agar, and considerable care was required for their enumeration. Relative to C. jejuni, more variability in CFU numbers was observed between the duplicate plates. A substantially higher density of C. lanienae cells was observed in inocula when the Petroff-Hausser chamber was used than was indicated by plating on Karmali agar. The differential was 188.8 to 1 (range, 122.6- to 306.5-fold). As a result, direct cell counts were used to determine the minimum level of quantification using RTQ-PCR.
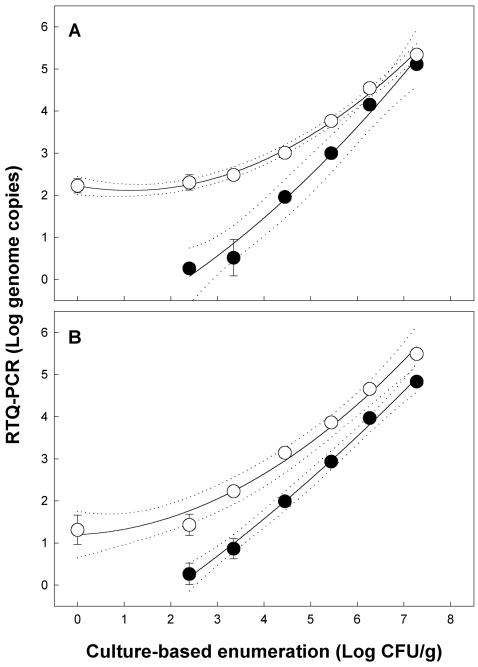
Relative to C. jejuni, more variation was observed for the RTQ-PCR results among the three replicates conducted with C. lanienae. In no instance was a PCR product obtained from control feces by using nested RTQ-PCR for C. lanienae. With nested RTQ-PCR primer set 1, 0.26 ± 0.08, 0.51 ± 0.44, 1.96 ± 0.09, 3.00 ± 0.13, 4.16 ± 0.06, and 5.12 ± 0.09 log genome copies were observed at a target density of 102, 103, 104, 105, 106, and 107 cells/g, respectively (Fig. 3A). Quadratic polynomial regression was applied, and the equation used to describe the relationship between log genome copies and log CFU per gram was y = −1.44 + 0.49x + 0.059x2 (r2 = 0.989); linear regression indicated that the slope did not differ from 1 (F = 0.90; df = 1, 15; P = 0.35). The limit of quantification was 1.8 to 1.9 genome copies (antilog 0.26 to 0.27), representing a density of ≈250 CFU/g of feces. The melting points of amplicons ranged from 84.0 to 85.0°C. With nested RTQ-PCR primer set 2, 0.27 ± 0.25, 0.87 ± 0.24, 1.99 ± 0.08, 2.94 ± 0.08, 3.97 ± 0.07, and 4.83 ± 0.13 log genome copies were observed at target levels of 102, 103, 104, 105, 106, and 107 cells/g, respectively (Fig. 3B). The quadratic polynomial regression equation to describe the relationship was y = −1.58 + 0.66x + 0.032x2 (r2 = 0.996). A limit of quantification very similar to that of primer set 1 was observed, and the linear slope of the relationship did not differ from 1 (F = 0.01; df = 1, 15; P = 0.92). The melting point of the primer set 2 amplicon was 81.5 to 82.5°C.
FIG. 3.
Quantification of C. lanienae in inoculated cattle feces by targeting the three-copy 16S rRNA gene with nested (solid symbols) and nonnested (open symbols) RTQ-PCRs. (A) Primer set 1, where the log quadratic polynomial equations to describe the relationships are as follows: nested, y = 2.23 − 0.20x + 0.087x2, r2 = 0.989; nonnested, y = −1.44 + 0.49x + 0.059x2, r2 = 0.996. (B) Primer set 2, where the log quadratic polynomial equations to describe the relationships are as follows: nested, y = −1.58 + 0.66x + 0.032x2, r2 = 0.997; nonnested, y = 1.20 + 0.053x + 0.077x2, r2 = 0.986. The error bars associated with the symbols represent standard errors of the mean (n = 3), and bars that are not visible are within the symbols. The dotted lines represent 95% confidence intervals.
In all instances a PCR product was obtained from control feces using nonnested RTQ-PCR for C. lanienae, and substantially different quadratic relationships were observed between log genome copies and log cells per gram for primer sets 1 and 2 relative to those obtained with nested RTQ-PCR (Fig. 3). The equations that described the relationship between log genome copies and log CFU per gram were y = 2.23 to 0.20x + 0.087x2 (r2 = 0.996) and y = 1.20 + 0.053x + 0.077x2 (r2 = 0.986) for nonnested primer sets 1 and 2, respectively. The melting points for all amplicons obtained with primer sets 1 and 2 were the same as noted previously using nested RTQ-PCR. For both nonnested primer sets 1 and 2, the slope of the quadratic regression lines differed from zero (F = 12.9 to 43.7; df = 1, 31 to 35; P ≤ 0.001). There were no differences between the slopes in the nested and nonnested methods for the corresponding primer sets (F = 0.8 to 2.2; df = 1, 30 to 34; P ≥ 0.15). However, the two methods produced substantially different y intercepts (F = 21.9 to 121.8; df = 1, 31 to 35; P < 0.001).
Uninoculated beef feces.
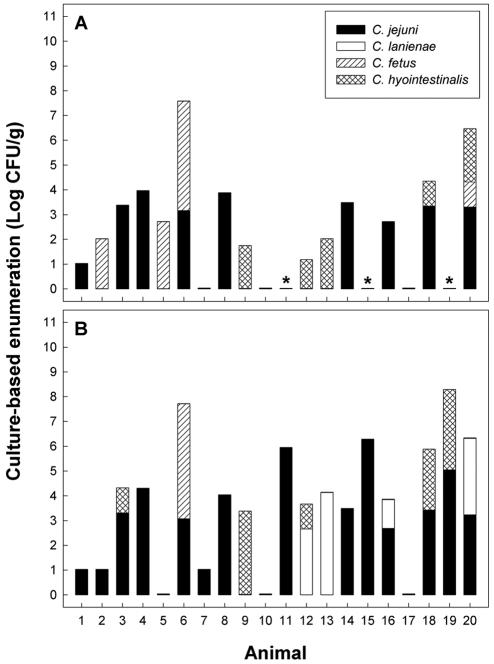
Considerable experience and diligence were frequently required to detect Campylobacter colonies on the two test media. A total of 326 colonies were recovered and characterized by colony PCR. The four taxa of Campylobacter occurring in cattle feces were C. fetus, C. hyointestinalis, C. jejuni, and C. lanienae (Fig. 4). C. fetus, C. hyointestinalis, and C. jejuni were isolated on both CCDA at 37°C and Karmali agar at 40°C, but C. lanienae was isolated only on Karmali agar at 40°C.
FIG. 4.
Populations of Campylobacter species in uninoculated cattle feces as determined by culture-based enumeration. (A) CCDA containing selective supplement SR115E at 37°C. (B) Karmali agar with selective supplement CM935 at 40°C. Identifications were made using colony PCR (8), and the asterisks indicate missing data.
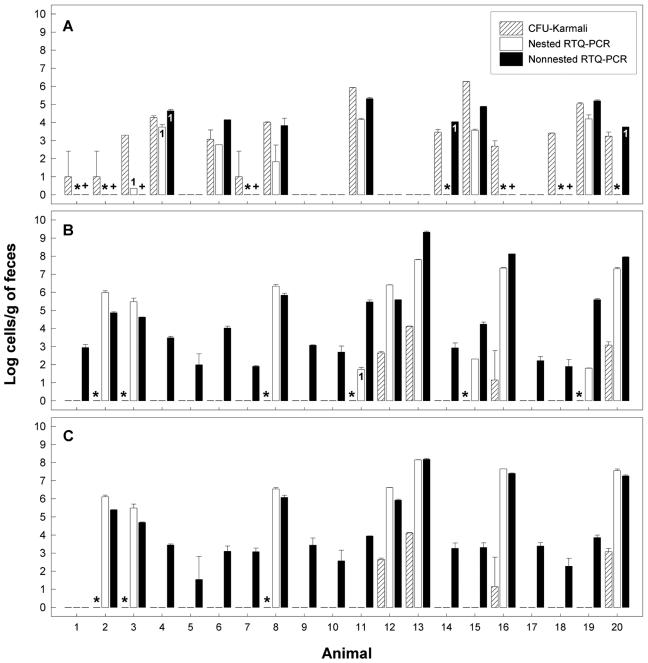
Of the 20 animals tested, C. jejuni was detected in 12 (60%) and 14 (70%) of the fecal samples using CCDA and Karmali agar, respectively (Fig. 4). Using nested RTQ-PCR, C. jejuni was detected in 7 of the 20 (35%) samples, and no amplicon was observed in the 4 samples in which populations exceeded 2.7 log CFU/g by plating (Fig. 5A). Estimates of C. jejuni populations were consistently smaller using nested RTQ-PCR. However, three of seven samples that were positive for C. jejuni were within 1 order of magnitude and an additional sample was within 2 orders of magnitude of the population size estimated by plating. C. jejuni was detected in eight samples by nonnested RTQ-PCR (Fig. 5A). Interestingly, two samples that were deemed negative for C. jejuni by nested RTQ-PCR (samples 14 and 20) were determined to contain large numbers of C. jejuni by nonnested RTQ-PCR. However, only one of the two subsamples of each of these samples was positive. Of the eight samples positive for C. jejuni by nonnested RTQ-PCR, six were within 1 order of magnitude and two were within 2 orders of magnitude of population sizes determined by plating. In no instance did conventional nPCR (i.e., nPCR followed by visualization and relative scoring of amplicon intensities) indicate the presence of C. jejuni in cattle feces, contrary to nested or nonnested RTQ-PCR (Table 3). However, feces from animal 6 were positive for C. jejuni by RTQ-PCR but not by conventional nPCR using the C. jejuni and C. jejuni-C. coli-C. hyoilei primer sets. Furthermore, C. jejuni was detected by conventional nPCR in fecal aliquot B but not in fecal aliquot A in two instances (i.e., animals 14 and 18). There were numerous cases in which C. fetus and/or C. hyointestinalis was detected in cattle feces by plating and the sample was deemed negative for C. jejuni or C. lanienae by RTQ-PCR.
FIG. 5.
Quantification comparison of C. jejuni and C. lanienae in uninoculated beef cattle feces (20 animals) among culture-based isolation on Karmali agar containing selective supplement CM935 at 40°C (CFU-Karmali), nested RTQ-PCR, and nonnested RTQ-PCR. (A) C. jejuni by targeting the mapA gene. (B) C. lanienae by targeting the 16S rRNA gene with primer set 1. (C) C. lanienae by targeting the 16S rRNA gene with primer set 2. With the exception of bars labeled 1 (n = 1), the error bars represent standard deviations (n = 2). *, detection disparity between culture-based isolation and nested RTQ-PCR; +, detection disparity between culture-based isolation and nonnested RTQ-PCR.
TABLE 3.
Results of nested and nonnested conventional PCRs and RTQ-PCR for campylobacters in cattle feces
| Animal no. | PCR results
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nested
|
Nonnested
|
||||||||
| Q-Cj Aa | C-Cj Ab | C-Cj Bc | Q-Cl Ad | C-Cl Ae | C-Cl Bf | C-Cjch Ag | Q-Cj Ah | Q-Cl Ai | |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 0 | 4 |
| 3 | 1 | 0 | 0 | 2 | 3 | 3 | 2 | 0 | 4 |
| 4 | 4 | 3 | 4 | 0 | 0 | 0 | 1 | 4 | 4 |
| 5 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 4 |
| 6 | 3j | 0j | 0j | 0 | 0 | 0 | 0 | 4 | 4 |
| 7 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| 8 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 4 |
| 9 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 4 |
| 10 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 4 |
| 11 | 4 | 4 | 3 | 0 | 1 | 0 | 4 | 4 | 4 |
| 12 | 0 | 0 | 0 | 4 | 3 | 4 | 4 | 0 | 4 |
| 13 | 0 | 0 | 0 | 4 | 4 | 4 | 4 | 0 | 4 |
| 14 | 0l | 0k | 3k | 0 | 0 | 0 | 0 | 4l | 4 |
| 15 | 4 | 4 | 3 | 0 | 0 | 0 | 2 | 4 | 4 |
| 16 | 0 | 0 | 0 | 4 | 4 | 4 | 4 | 0 | 4 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| 18 | 0 | 0k | 2k | 0 | 0 | 0 | 0 | 0 | 4 |
| 19 | 4 | 4 | 2 | 0 | 1 | 0 | 4 | 4 | 4 |
| 20 | 0l | 0 | 0 | 4 | 4 | 4 | 4 | 4l | 4 |
Nested RTQ-PCR for C. jejuni from fecal aliquot A (1, ≤1; 2, >1 and ≤2; 3, >2 and ≤3; and 4, >3 log CFU/g).
Conventional nested PCR for C. jejuni from fecal aliquot A (scale of 1 to 4 based on amplicon intensity).
Conventional nested PCR for C. jejuni from fecal aliquot B (scale of 1 to 4 based on amplicon intensity).
Nested RTQ-PCR for C. lanienae from fecal aliquot A using primer set 2 (1, ≤1; 2, >1 and ≤2; 3, >2 and ≤3; and 4, >3 log CFU/g).
Conventional nested PCR for C. lanienae from fecal aliquot A (scale of 1 to 4 based on amplicon intensity).
Conventional nested PCR for C. lanienae from fecal aliquot B (scale of 1 to 4 based on amplicon intensity).
Conventional nonnested PCR for C. jejuni, C. coli, and C. hyoilei from fecal aliquot A. This primer set also detects C. lanienae.
Nonnested RTQ-PCR for C. jejuni from fecal aliquot A (1, ≤1; 2, >1 and ≤2; 3, >2 and ≤3; and 4, >3 log CFU/g).
Nonnested RTQ-PCR for C. lanienae from fecal aliquot A using RTQ-PCR primer set 2 (1, ≤1; 2, >1 and ≤2; 3, >2 and ≤3; and 4, >3 log CFU/g).
Discrepancy between RTQ-PCR and conventional PCR (aliquots A and B) for C. jejuni.
Discrepancy between conventional PCR for aliquots A and B for C. jejuni.
Discrepancy between nested and nonnested RTQ-PCRs for C. jejuni.
C. lanienae was isolated on Karmali agar from the feces of only four animals (20%). With nested RTQ-PCR, 10 (50%) and 7 (35%) samples were positive for C. lanienae using primer sets 1 and 2, respectively (Fig. 5B and C). Estimates of C. lanienae populations obtained with nested RTQ-PCR were substantially larger (3.7 to 6.5 log cells/g) relative to plating. In no instance did nested RTQ-PCR indicate the presence of C. lanienae in cattle feces, contrary to conventional nPCR (Table 3). Furthermore, there were no conspicuous differences between the two fecal aliquots in detection frequency using conventional nPCR. In contrast to nested RTQ-PCR, nonnested RTQ-PCR resulted in a substantially higher number of samples in which an amplicon was detected; 20 and 19 fecal samples were positive using primer sets 1 and 2, respectively (Fig. 5B and C). This also contrasted with the results of conventional PCR and plating on Karmali agar (Table 3).
DISCUSSION
Campylobacter species are a very fastidious group of bacteria, and culture-based methods (i.e., enrichment and/or direct plating) often do not reflect species diversity and abundance. This is illustrated in the present study by our inability to detect C. lanienae on CCDA. To date, RTQ-PCR for Campylobacter species has been restricted to the quantification of C. jejuni in pure cultures (24, 35) or indirectly from foods by using enrichment culture (28). Detecting bacteria directly in herbivore feces is problematic, since there are large quantities of PCR inhibitors, such as polyphenolic compounds (15); there are large numbers of nontarget bacteria, causing specificity problems; and there are often small numbers of target taxa, resulting in sensitivity concerns. Inglis and Kalischuk (8) found that the QIAamp DNA stool minikit effectively removed inhibitors present in cattle feces, as indicated by amplification of an internal control designed to amplify under the same conditions as a 16S ribosomal DNA primer set for campylobacters.
In the present study, RTQ-PCR primers were designed for both C. jejuni and C. lanienae. Primers for C. jejuni were designed to target the single-copy mapA gene, whereas primers for C. lanienae targeted the three-copy 16S rRNA gene. A decision was made to use an intercalating dye system (SYBR Green) because it is very economical in relation to probes and molecular beacons. While SYBR Green is as sensitive as other fluorescent reporters, nonspecific binding of SYBR Green to PCR products and to primer dimers will result in false positives, particularly for products late in the amplification process (Ambion, Inc., Austin, Tex., website [http://www.ambion.com/techlib/tn/81/813.html]). For example, Hein et al. (7) observed that quantification of Staphylococcus aureus in artificially inoculated cheeses with SYBR Green was 10 times less sensitive than with a fluorogenic TaqMan probe. The reduced sensitivity in this instance could be attributed to nonspecific binding of the SYBR Green. The primers that we designed for the C. jejuni mapA gene produced minimal dimers, and they did not produce appreciable nonspecific binding. Furthermore, the primers were found to be very specific for C. jejuni in cattle feces, and they provided a limit of quantification of 19 genome copies. Although we applied RTQ-PCR to C. jejuni in a very complex substrate, the limit of quantification that we obtained was comparable to the 12-genome-copy sensitivity limit observed by Sails et al. (28) for C. jejuni cells in enrichment culture.
To allow direct comparisons with plate counts, we assessed the relationship between log genome copies and numbers of CFU per gram in cattle feces artificially inoculated with C. jejuni. We observed a strong relationship between these two variables, and coefficients of correlation typically approached 1. Furthermore, the relationship was relatively consistent across three independent replicates. Hein et al. (7) also observed high coefficients of correlation (r2 ≥ 0.98) between log genome copies and log CFU of S. aureus in cheeses. For C. jejuni, we observed that the slopes approached 1 (0.91 ± 0.05 and 0.85 ± 0.02). However, the y intercepts were <0, indicating a location shift (i.e., the numbers of CFU per gram in feces were smaller than predicted by genome copies, but the difference was constant across cell densities in feces). Approximately one genome copy of C. jejuni represented ∼100 CFU/g of feces, and the 100-fold differential between numbers of genome copies and CFU per gram may have been due to poor extraction, inefficiency in amplification, and/or the presence of PCR inhibitors. The last possibility is the least likely because we used an internal control to qualitatively assess whether PCR inhibitors were sufficiently removed to allow amplification, and the slopes and y intercepts were equal between nested and nonnested PCRs (i.e., if PCR inhibitors were present, a dilution effect would be expected with nPCR). Regardless of the reason, the limit of quantification of C. jejuni in cattle feces was relatively high (3 × 103 CFU/g). While the quantification sensitivities that we observed for C. jejuni were less than can typically be obtained by microbiological methods, RTQ-PCR substantially reduced the time required for detection. To run nonnested RTQ-PCR requires ca. 2 h for 96 samples: ≈1 h for PCR setup and ≈1 h for the RTQ-PCR step for 96 samples. This time does not include the procedure for extraction of DNA from feces using the QIAamp DNA stool minikit, which is relatively slow and labor-intensive (ca. 1.5 h for 10 samples). However, once samples are extracted, DNAs can be aliquoted and stored frozen until they can be conveniently processed, a major advantage over microbiological methods. One of the limitations of the PCR-based method that we employed is related to the small amount of fecal material processed (0.2 g). This contrasts with the 2.5 g that we used for culture-based assessments. Very little is known about the spatial distribution of campylobacters in feces. In small samples, cell aggregation could provide an erroneous indication of biomass. It may be possible to increase the amount of feces sampled, but increased sample size may overload the purification step to remove PCR inhibitors. Aggregation of C. jejuni is supported by its detection in fecal aliquot B but not in aliquot A in two instances, and we are investigating the distribution of Campylobacter species in cattle feces.
C. lanienae was originally isolated from abattoir workers exposed to pigs and cattle in Switzerland (19), but its pathogenicity and virulence for humans are unknown. Recently, six isolates of the bacterium were recovered from swine but not from cattle (33). On the basis of this observation, Sasaki et al. (33) concluded that swine are the primary source of the bacterium. Inglis and Kalischuk (8) isolated C. lanienae from cattle feces, but they found that cattle strains of the bacterium are very fastidious and do not grow on many of the commercial media commonly used to isolate campylobacters; most commercially available media tend to select for C. jejuni and C. coli and inhibit the growth of other species (1, 2, 4, 8, 17). In a subsequent study using PCR-based detection, Inglis et al. (9) demonstrated that C. lanienae was commonly associated (≈50% of the 380 fecal samples tested) with cattle. In the present study, we were unable to isolate C. lanienae L52 from inoculated feces, further illustrating its fastidiousness. Initially, we relied on extrapolations of cell densities in feces based on enumeration of CFU in the inocula. Given that ≈86% of the DNA is lost during the extraction procedure and that we processed only 200 mg of fecal material, it became obvious that the enumeration of CFU from inocula grossly underestimated the cell density in feces; initially, we calculated that the minimum limit of quantification was ≈14 to 22 CFU/g of feces. This necessitated that we directly enumerate cells in inocula using a Petroff-Hausser chamber. Campylobacters are very small and motile, and extreme care was required to obtain an accurate estimate of cell density with this method. Using the Petroff-Hausser chamber, we determined that dilution plating of C. lanienae, but not C. jejuni, substantially underestimated cell densities in inocula. This information was subsequently used to obtain a true measure of C. lanienae cell density in the feces.
Limited information on the genetics of C. lanienae is available. As a result, the RTQ-PCR primers that we designed for C. lanienae targeted the 16S rRNA gene. Others also have targeted the 16S rRNA gene to quantify bacteria in feces (20). Similar to other Campylobacter species (13), we found that C. lanienae L52 possessed three copies of the 16S rRNA gene. Since it is imperative that certain requirements be met in designing RTQ-PCR primers using SYBR Green (Plant-Microbe Genomics Facility at The Ohio State University website [http://www.biosci.ohio-state.edu/∼pmgf/procedures_rt-pcr.pdf]), we encountered considerable difficulty in designing C. lanienae-specific primers based on 16S rRNA sequences. Two primer sets were designed. While these primer sets met the criteria for RTQ-PCR primers, they were not predicted to be highly specific for C. lanienae based on BLAST analyses. The nonspecificity of these primers for C. lanienae applied without nesting was confirmed in both inoculated and uninoculated cattle feces. Measurement of the ratio of specific to nonspecific PCR products by determination of the melting peak integration has been used to assess specificity (26). However, we found that nonspecific PCR products amplified by both C. lanienae nonnested RTQ-PCR primer sets possessed the appropriate melting points, and melting peak integration alone could not be used to confirm specificity.
The specific quantification of C. lanienae required nesting. Nested quantitative PCR has been described previously (5, 22), but it has not been widely adopted as an RTQ-PCR strategy. Inglis and Kalischuk (8) modified the primer set of Logan et al. (19) based on the 16S rRNA gene and used nPCR to detect C. lanienae in cattle. For nested RTQ-PCR, it is imperative that primary amplification take place entirely in the exponential phase (5), and here we utilized 22 cycles based on a maximum theoretical density of 105 genome copies of C. lanienae. We obtained very sensitive quantification of C. lanienae in inoculated feces by using nested RTQ-PCR; we were able to detect as few as 1.8 genome copies of C. lanienae DNA (≈250 CFU/g of feces) regardless of which RTQ-PCR primer set was used. This is substantially more sensitive than has been observed by others targeting the 16S rRNA gene of bacteria in feces (20), and in several instances, we detected C. lanienae in feces that were deemed to be culture negative for the species. A strong relationship was observed between log genome copies and cell density in inoculated feces (r2 = 0.98), and the dynamic range of quantification (102 to 108 cells/g) was consistent across replicates, indicating that this is a very robust method. As with C. jejuni, linear slopes approached 1 but the y intercepts were <0, indicating a location shift.
Nested RTQ-PCR generally indicated that C. lanienae populations in feces were larger than was indicated by culturing on Karmali agar, an observation similar to those made with other bacteria and media in situ (6, 7). Given the fastidiousness of cattle strains of C. lanienae, it is not surprising that plating would underestimate population sizes. Cell aggregation in cattle feces may also have contributed to the underestimation of C. lanienae density that we observed, since a CFU may have originated from more than one bacterial cell. Other Campylobacter species enter a viable but, nonculturable (VNC) state (11, 27), and this bias has been attributed to the quantification of dead and stressed unculturable bacterial cells through PCR (6, 21). While nesting was a prerequisite for the accurate quantification of C. lanienae, the inclusion of an additional PCR step did increase the processing time by ca. 2 h (i.e., ≈1 h for PCR setup and ≈1 h for the primary PCR step) and increased the cost of processing due to the need for additional reagents and labor.
In contrast to C. lanienae, nested RTQ-PCR did not substantively increase either the specificity or the sensitivity of C. jejuni quantification. However, nesting substantially decreased threshold values. Relying on large threshold values increases variation (14) and can lead to false-positive values due to nonspecific binding late in the amplification process (http://www.ambion.com/techlib/tn/81/813.html). Furthermore, nPCR provides intrinsic PCR product carryover, generally improves robustness, and lowers the limit of detection (5). This is at the expense of increased cost and the possibility of contamination (14). Using nested RTQ-PCR, a substantially lower limit of quantification was observed for C. lanienae (1.8 genome copies) than for C. jejuni (25 genome copies). Although we do not have direct evidence, the considerably lower limit of quantification observed for C. lanienae suggests that targeting a multicopy gene, at least in part, increases the sensitivity of detection. To test this possibility, we are developing RTQ-PCR primers that target multicopy genes of C. jejuni with the goal of increasing the quantification sensitivity for the bacterium by using RTQ-PCR.
In conclusion, the suitability of RTQ-PCR for rapid quantification of C. jejuni and C. lanienae cells in a microbiologically complex substrate was demonstrated using SYBR Green. It was shown that (i) RTQ-PCR primers targeting the single-copy mapA gene were highly specific for C. jejuni, and the sensitivity of quantification was 19 genome copies; (ii) nested RTQ-PCR targeting the three-copy 16S rRNA gene was necessary for specific quantification of C. lanienae, and the limit of quantification was 1.8 genome copies; and (iii) RTQ-PCR provided quantification of C. jejuni in feces comparable to that of plating, but the number of genome copies of the more fastidious C. lanienae was substantially higher than was indicated by culturing. While this study demonstrated the suitability of RTQ-PCR for the direct quantification of C. jejuni and C. lanienae in cattle feces, the technology is applicable to other substrates. For example, we are utilizing these methods to study the colonization of the GI tracts of cattle by campylobacters and to identify factors that influence their release in feces.
Acknowledgments
We thank the following people at the Agriculture and Agri-Food Canada Research Centre at Lethbridge: Hilma Busz for conducting conventional nPCR, Jenny Gusse for her assistance with the Southern blot analysis and PFGE, and Toby Entz for his statistical advice. We also thank two anonymous reviewers for their comments on the manuscript.
This study was supported by a grant from the Canada-Alberta Beef Industry Development Fund (CABIDF).
Footnotes
Contribution 03036 from the Agriculture and Agri-Food Canada Research Centre, Lethbridge, Alberta, Canada.
REFERENCES
- 1.Atabay, H. I., and J. E. L. Corry. 1998. The isolation and prevalence of campylobacters from dairy cattle using a variety of methods. J. Appl. Microbiol. 84:733-740. [DOI] [PubMed] [Google Scholar]
- 2.Baylis, C. L., S. MacPhee, K. W. Martin, T. J. Humphrey, and R. P. Betts. 2000. Comparison of three enrichment media for the isolation of Campylobacter spp. from foods. J. Appl. Microbiol. 89:884-891. [DOI] [PubMed] [Google Scholar]
- 3.Denis, M., C. Soumet, K. Rivoal, G. Ermel, D. Blivet, G. Salvat, and P. Colin. 1999. Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 29:406-410. [DOI] [PubMed] [Google Scholar]
- 4.Engberg, J., S. L. W. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haff, L. A. 1994. Improved quantitative PCR using nested primers. PCR Methods Appl. 3:332-337. [DOI] [PubMed] [Google Scholar]
- 6.Hein, I., D. Klein, A. Lehner, A. Bubert, E. Brandl, and M. Wagner. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37-46. [DOI] [PubMed] [Google Scholar]
- 7.Hein, I., A. Lehner, P. Rieck, K. Klein, E. Brandl, and M. Wagner. 2001. Comparison of different approaches to quantify Staphylococcus aureus cells by real-time quantitative PCR and application of this technique for examination of cheese. Appl. Environ. Microbiol. 67:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inglis, G. D., and L. D. Kalischuk. 2003. Use of PCR for direct detection of Campylobacter species in cattle feces. Appl. Environ. Microbiol. 69:3435-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inglis, G. D., L. D. Kalischuk, and H. Busz. 2003. A survey of Campylobacter species shed in faeces of beef cattle using polymerase chain reaction. Can. J. Microbiol. 49:655-661. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, C., C. Li, T. Ha, D. A. Ferguson, D. S. Chi, J. J. Laffan, and E. Thomas. 1998. Identification of H. pylori in saliva by a nested PCR assay derived from a newly cloned DNA probe. Dig. Dis. Sci. 43:1211-1218. [DOI] [PubMed] [Google Scholar]
- 11.Jones, D. M., E. M. Sutcliffe, and A. Curry. 1991. Recovery of viable but non-culturable Campylobacter jejuni. J. Gen. Microbiol. 137:2477-2482. [DOI] [PubMed] [Google Scholar]
- 12.Karmali, M. A., A. E. Simor, M. Roscoe, P. C. Fleming, S. S. Smith, and J. Lane. 1986. Evaluation of a blood-free, charcoal-based, selective medium for the isolation of Campylobacter organisms from feces. J. Clin. Microbiol. 23:456-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klappenbach, J. A., P. R. Saxman, J. T. Cole, and T. M. Schmidt. 2001. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein, D. 2002. Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 8:257-260. [DOI] [PubMed] [Google Scholar]
- 15.Koonjul, P. K., W. F. Brandt, J. M. Farrant, and G. G. Lindsey. 1999. Inclusion of polyvinylpyrrolidone in the polymerase chain reaction reverses the inhibitory effects of polyphenolic contamination of RNA. Nucleic Acids Res. 27:915-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J. Wiley & Sons, Chichester, United Kingdom.
- 17.Lawson, A. J., D. Linton, J. Stanley, and R. J. Owen. 1997. PCR detection and speciation of Campylobacter upsaliensis and Campylobacter helveticus in human faeces, and comparison with cultural methods. J. Appl. Microbiol. 83:375-380. [DOI] [PubMed] [Google Scholar]
- 18.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logan, J. M., A. Burnens, D. Linton, A. J. Lawson, and J. Stanley. 2000. Campylobacter lanienae sp. nov., a new species isolated from workers in an abattoir. Int. J. Syst. Evol. Microbiol. 50:865-872. [DOI] [PubMed] [Google Scholar]
- 20.Malinen, E., A. Kassinen, T. Rinttiä, and A. Palva. 2003. Comparison of real-time PCR with SYBR Green I or 5′-nuclease assays and dot-blot hybridization with rDNA-targeted oligonucleotide probes in quantification of selected faecal bacteria. Microbiology 149:269-277. [DOI] [PubMed] [Google Scholar]
- 21.Mäntynen, V., S. Neimelä, S. Kaijalainen, T. Pirhonen, and K. Linström. 1997. MPN-PCR-quantification method for staphylococcal enterotoxin c1 gene from fresh cheese. Int. J. Food Microbiol. 36:618-623. [DOI] [PubMed] [Google Scholar]
- 22.Max, N., K. Wolf, B. Spike, E. Thiel, and U. Keilholz. 2001. Nested quantitative real time PCR for detection of occult tumor cells. Recent Results Cancer Res. 158:25-31. [DOI] [PubMed] [Google Scholar]
- 23.McOrist, A. L., M. Jackson, and A. R. Bird. 2002. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J. Microbiol. Methods 50:131-139. [DOI] [PubMed] [Google Scholar]
- 24.Nogva, H. K., A. Bergh, A. Holck, and K. Rudi. 2000. Application of the 5′-nuclease PCR assay in evaluation and development of methods for quantitative detection of Campylobacter jejuni. Appl. Environ. Microbiol. 66:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during polymerase chain reaction. Anal. Biochem. 254:154-160. [DOI] [PubMed] [Google Scholar]
- 27.Rollins, D. M., and R. R. Colwell. 1986. Viable but nonculturable stage of Campylobacter jejuni and its role in survival in the natural aquatic environment. Appl. Environ. Microbiol. 52:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sails, A. D., A. J. Fox, F. J. Bolton, D. R. A. Wareing, and D. L. A. Greenway. 2003. A real-time PCR assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 69:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salma, S. M., E. Newnham, N. Chang, and D. E. Taylor. 1995. Genome map of Campylobacter fetus subsp. fetus ATCC 27374. FEMS Microbiol. Lett. 132:239-245. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Saruta, K., T. Matsunaga, M. Kono, S. Hoshina, S. Kanemoto, O. Sakai, and K. Machida. 1997. Simultaneous detection of Streptococcus pneumoniae and Haemophilus influenzae by nested PCR amplification from cerebrospinal fluid samples. FEMS Immunol. Med. Microbiol. 19:151-157. [DOI] [PubMed] [Google Scholar]
- 32.SAS Institute Inc. 1999. User's guide, version 8.0. SAS Institute, Cary, N.C.
- 33.Sasaki, Y., T. Fujisawa, K. Ogikubo, T. Ohzono, K. Ishihara, and T. Takahashi. 2003. Characterization of Campylobacter lanienae from pig feces. J. Vet. Med. Sci. 65:129-131. [DOI] [PubMed] [Google Scholar]
- 34.Verdin, E., C. Saillard, A. Labbe, J. M. Bove, and M. Kobisch. 2000. A nested PCR assay for detection of Mycoplasma hyopneumoniae in tracheobronchiolar washings from pigs. Vet. Microbiol. 76:31-40. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, D. L., S. R. Abner, T. C. Newman, L. S. Mansfield, and J. E. Linz. 2000. Identification of ciprofloxacin-resistant Campylobacter jejuni by use of fluorogenic PCR assay. J. Clin. Microbiol. 38:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]