Abstract
ASYMMETRIC LEAVES2 (AS2) is one of the key genes required for specifying leaf adaxial identity during leaf adaxial–abaxial polarity establishment. Previous data have shown that, in leaf development, AS2 is directly repressed by an abaxially located transcription factor KANADI1 (KAN1), so that the AS2 transcripts are restricted only in the adaxial leaf domain. It is shown here that, different from the spatial repression by KAN1, the quantitative repression of AS2 in the adaxial domain is also critical for ensuring normal leaf pattern formation. By analysing two gain-of-function as2 mutants, as2-5D and isoginchaku-2D (iso-2D), it is shown that the similar AS2-over-expressed phenotypes of these mutants reflect two different kinds of AS2 misexpression patterns. While as2-5D causes disruption of a KAN1-binding site at the AS2 promoter leading to derepression of AS2 in the abaxial side but without changing its expression level of a leaf, iso-2D results in over-expression of AS2 but without altering its adaxial expression pattern. In addition, it was found that, in iso-2D, levels of histone H3 lysine 27 trimethylation (H3K27me3) and H3K4me3 at the AS2 locus are significantly reduced and increased, respectively, compared with those in the wild type and as2-5D. These results suggest that during leaf patterning, quantitative control of the AS2 expression level might involve epigenetic regulations.
Key words: Arabidopsis, AS2, epigenetic regulation, histone modification, leaf development, polarity formation.
Introduction
Leaf primordia emerge from the peripheral zone of the shoot apical meristem (SAM), and start to establish polarity along the adaxial–abaxial, proximodistal, and mediolateral axes immediately after their initiation (Waites and Hudson, 1995; McConnell and Barton, 1998; Bowman et al., 2002). Among them, the establishment of the adaxial–abaxial axis, which is required for subsequent lamina growth and asymmetric development, is of primary importance (Waites and Hudson, 1995; McConnell and Barton, 1998; Bowman et al., 2002), and differentiation of cells along this axis leads to the formation of leaves facilitating photosynthesis (Waites and Hudson, 1995; McConnell and Barton, 1998; Bowman et al., 2002). During the past decade, a number of factors which play important roles in leaf adaxial–abaxial polarity establishment in Arabidopsis have been identified (reviewed by Byrne, 2006; Xu et al., 2007; Husbands et al., 2009; Moon and Hake, 2011).
Genes that specify leaf identity in the adaxial domain include the HD-ZIP III family members PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV) (Talbert et al., 1995; McConnell and Barton, 1998; Zhong and Ye, 1999; McConnell et al., 2001; Otsuga et al., 2001). In addition, two putative transcription factor genes ASYMMETRIC LEAVES1 (AS1) and AS2 are also critical in promoting cell differentiation in the adaxial leaf domain (Byrne et al., 2000; Iwakawa et al., 2002; Sun et al., 2002; Xu et al., 2002, 2003; Lin et al., 2003). On the other hand, the YABBY (YAB) family genes FILAMENTOUS FLOWER (FIL) and YAB3 (Siegfried et al., 1999), the KANADI (KAN) family genes KAN1 and KAN2 (Eshed et al., 1999, 2001; Kerstetter et al., 2001), and the AUXIN RESPONSE FACTOR (ARF) family genes ARF3 (also called ETT) and ARF4 (Pekker et al., 2005) specify the abaxial leaf domain. Small RNAs are also involved in leaf adaxial–abaxial patterning. MicroRNA165 and 166 (miR165 and miR166) (Rhoades et al., 2002; Emery et al., 2003; Juarez et al., 2004; Kidner and Martienssen, 2004; Mallory et al., 2004; Williams et al., 2005) and trans-acting small interfering RNA tasiR-ARF from the TAS3 gene (Yoshikawa et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Xu et al., 2006) post-transcriptionally target the HD-ZIP III and ARF genes transcripts, respectively, during leaf polarity formation. Recent studies also demonstrated that genes that promote cell proliferation in the leaf are also required for adaxial–abaxial polarity formation (Yuan et al., 2010; Horiguchi et al., 2011; Wang et al., 2011; Xu et al., 2012).
The putative transcription factor gene AS2 encodes a AS2/LOB-domain protein which forms a protein complex with the MYB-domain transcription factor AS1 to specify the adaxial leaf domain (Byrne et al., 2000; Iwakawa et al., 2002; Sun et al., 2002; Xu et al., 2002, 2003; Lin et al., 2003). AS2 expression is restricted only to the adaxial leaf domain (Iwakawa et al., 2002, 2007) and this AS2 pattern is caused by an abaxially located transcription factor, KAN1, which binds to the AS2 promoter in the abaxial leaf domain to repress AS2 directly (Eshed et al., 1999, 2001; Kerstetter et al., 2001; Wu et al., 2008). as2-5D is a gain-of-function as2 mutant that displayed phenotypes resembling transgenic plants that over-express AS2 (Wu et al., 2008). It was reported that, in the as2-5D mutant, a KAN1-binding site at the AS2 promoter is disrupted, and thus the abaxial expression of AS2 fails to be normally repressed (Wu et al., 2008).
To understand better the regulation of AS2 during leaf polarity formation, another gain-of-function AS2 mutant, isoginchaku-2D (iso-2D), which is caused by the insertion of a T-DNA vector carrying cauliflower mosaic virus (CaMV) 35S enhancers at the AS2 locus (Nakazawa et al., 2003), was investigated. It was found that, different from the defective KAN1 repression in as2-5D, iso-2D causes AS2 over-expression and the drastically increased AS2 transcripts are only accumulated in the leaf adaxial domain. Our data indicate that, similar to the spatial control by KAN1, the quantitative control of AS2 expression is also critical for leaf axial patterning.
Materials and methods
Plant materials and growth conditions
Arabidopsis mutants iso-2D and as2-5D are in the Columbia-0 (Col-0) background (Nakazawa et al., 2003; Wu et al., 2008). Plant growth conditions are according to our previous methods (Xu et al., 2003).
Scanning electron microscopy (SEM), sectioning, in situ hybridization, and GUS staining
SEM and thin-section analyses were carried out according to the methods described previously by Xu et al. (2003). In situ hybridization was performed according to the protocol described previously (Drews et al., 1991; Long and Barton, 1998; Li et al., 2005), and 14-d-old seedlings were used in in situ hybridization. The AS2 probe was made from a full-length cDNA clone in the pBluescript plasmid. The GUS and FIL probes were made as described previously (Li et al., 2005; Yao et al., 2009). The colour reaction for the detection of the digoxigenin (DIG)-labelled AS2 probes was carried out for 3 weeks at room temperature because of the low levels of AS2 transcripts, while that for detection of the DIG-labelled GUS and FIL probes was carried out for 2 d and 16h, respectively. Primers used in plasmid constructions are listed in Supplementary Table S1 at JXB online. GUS staining and plant tissue sectioning were performed as previously described (Xu and Shen, 2008; He et al., 2012).
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and chromatin immunoprecipitation (ChIP)
Total RNA was extracted from the first pair of rosette leaves or shoot apexes of 14-d-old wild-type and mutant plants and cDNA preparation was according to the method described previously by Xu et al. (2003). The ChIP experiment was performed as previously described (Xu et al., 2008), using leaves from the 20-d-old wild-type and mutant plants for chromatin extraction. Immunoprecipitation was performed by using the anti-trimethyl-Histone H3 (lys27) antibody (Cat. 07-449, Millipore, USA) or the rabbit polyclonal to Histone H3 (tri methyl K4) antibody (Cat. ab8580, Abcam, UK). Primers used in the PCR reaction are listed in Supplementary Table S1 at JXB online.
Construction of transgenic plants
A DNA fragment of about 4kb containing the AS2 promoter (–3990 to –1 prior to ATG) was PCR amplified from wild-type Col-0 or as2-5D and were subcloned into the SalI and BamHI restriction sites of the pBI101 vector to result in the AS2 pro :GUS and mAS2 pro :GUS plasmids, respectively. The 35S pro :AS2 pro :GUS and 35S pro :mAS2 pro :GUS plasmids were constructed by fusing a DNA fragment containing the 35S promoter to the 5′ end at the SalI site of AS2 pro :GUS and mAS2 pro :GUS, respectively. These plasmids were introduced into wild-type Col-0 by Agrobacterium-mediated transformation using the GV3101 strain. Primers used in the molecular cloning are listed in Supplementary Table S1 at JXB online.
Results
as2-5D and iso-2D displayed similar leaf developmental defects
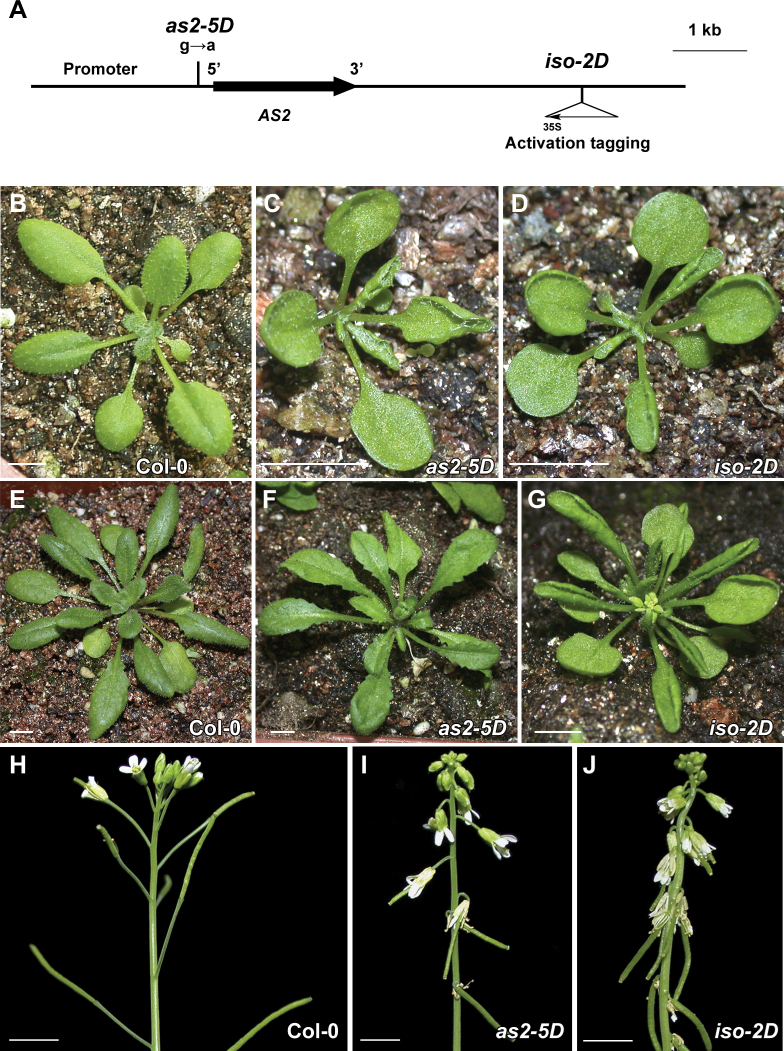
The iso-2D mutant carries an activation-tagging T-DNA insertion containing 4×35S enhancers at a position more than 3kb away from the 3΄ end of the AS2 coding region (Fig. 1A) (Nakazawa et al., 2003). Compared with the wild-type Col-0 (Fig. 1B), the previously characterized as2-5D (Wu et al., 2008) and the iso-2D mutants showed similar developmental defects at the seedling stage before the 9th leaf was formed (Fig. 1C, D). The phenotypic severity became weaker in as2-5D at subsequent plant developmental stages than that in iso-2D. For example, both mutant seedlings showed up-curled rosette leaves (Fig. 1C, D) and down-pointing flowers and siliques (Fig. 1I, J). These are the typical AS2 over-expression phenotypes first observed in the 35S pro :AS2 transgenic plants (Lin et al., 2003; Xu et al., 2003). However, at the late developmental stages, leaves of as2-5D became flat gradually (Fig. 1F), whereas those of iso-2D kept severely up-curled (Fig. 1G). In addition, the angles between siliques and stems were larger in as2-5D than in iso-2D, indicating that this inflorescence phenotype in as2-5D is also weaker (Fig. 1I, J).
Fig. 1.
as2-5D and iso-2D mutants both show AS2-over-expression phenotypes. (A) Structure of the AS2 gene. In as2-5D, a nucleotide substitution results in a disrupted KAN1 binding site in the AS2 promoter. The iso-2D mutation is caused by the activation tagging of 35S enhancers. (B–D) Phenotypes of 21-d-old Col-0 (B), as2-5D (C), and iso-2D (D) seedlings. (E–G) Phenotypes of 30-d-old Col-0 (E), as2-5D (F), and iso-2D (G) seedlings. (H–J) Inflorescence phenotypes of 45-d-old Col-0 (H), as2-5D (I), and iso-2D (J) plants. Note that although both as2-5D and iso-2D mutants show the AS2-over-expression phenotypes, the iso-2D phenotypes are usually more severe in the later plant developmental stages. Bars=5mm in (B)–(J).
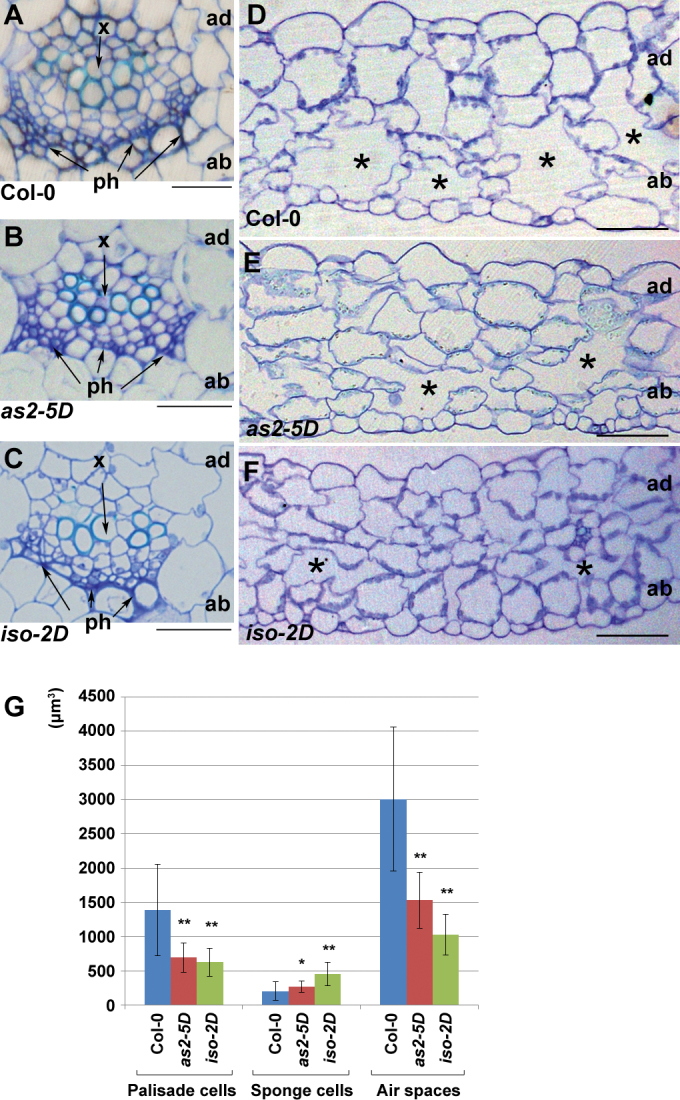
Transverse sectioning was then performed to analyse vascular and mesophyll patterns of these two mutants. The vascular patterns of both as2-5D and iso-2D petioles were indistinguishable from that of the wild type, showing that the xylems were on the adaxial pole and phloems on the abaxial pole (Fig. 2A–C). However, the mesophyll patterns in as2-5D and iso-2D leaves were altered. In the mature wild-type leaves, the adaxially located palisade mesophyll cells appear large and densely packed; whereas the abaxial spongy mesophyll cells are relatively small and are separated by large air spaces (Fig. 2D). In the as2-5D leaves, the average size of palisade cells became slightly smaller but that of the spongy mesophyll cells was enlarged with the apparently reduced size of air spaces (Fig. 2E, G). In the iso-2D leaves, the reduced size of the adaxial palisade cells and the enlarged size of the abaxial sponge cells became even more pronounced with very small air spaces in the abaxial domain, so that the abaxial leaf domain looked like the adaxial domain (Fig. 2F, G). These mesophyll phenotypes are similar to those described for the adaxialized leaves (Kerstetter et al., 2001; Lin et al., 2003; Grigg et al., 2005).
Fig. 2.
Transverse section analyses of leaf petioles and blades. (A–C) Transverse sections of Col-0 (A), as2-5D (B), and iso-2D (C) petioles. There were no obvious defects observed in the mutant petioles. (D–F) Transverse sections of Col-0 (D), as2-5D (E), and iso-2D (F) rosette leaves. Asterisks indicate air spaces. ad and ab, leaf adaxial and abaxial sides, respectively. x, xylem; ph, phloem. Bars = 50 µm in (A)–(F). (G) Quantitative analyses of the cell size and the air space size. The third or fourth rosette leaves from 21-d-old plants were used in sectioning analysis and sections at a position about a quarter of the leaf length from the proximal end of five leaves each were analysed. Cells and air spaces between the first and the second grade branches were scored using the software Image J (http://rsb.info.nih.gov/ij/). Bars show s.d. *, P <0.05; **, P <0.01.
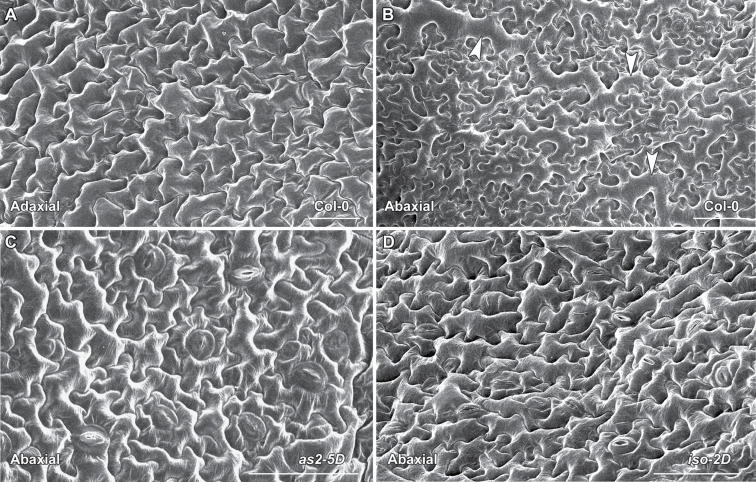
Leaf epidermal cells of the mutants were then analysed using SEM. The adaxial epidermis of the wild type was composed of uniformly sized cells (Fig. 3A), and the abaxial epidermis was characterized with small pavement cells mixed with long and large cells (Fig. 3B) (McConnell and Barton, 1998). Although the adaxial epidermis appeared normal, patches of the adaxially featured epidermal cells with a uniform cellular size were observed on the abaxial surfaces of both as2-5D and iso-2D leaves (Fig. 3C, D). All these results from morphological characterization of as2-5D and iso-2D indicate that both mutants have similar developmental defects while phenotypic abnormalities in iso-2D are usually stronger.
Fig. 3.
SEM analysis of leaf epidermal cells. (A, B) The wild-type Col-0 leaf epidermal cells on the adaxial (A) or the abaxial (B) side. Arrowheads in (B) indicate the long and large cells which appear only on the leaf abaxial surface. (C, D) The abaxial leaf surface of both as2-5D (C) and iso-2D (D) contains patches of cells that are similar to the wild-type adaxial epidermal cells, and the long and large abaxially featured cells were not observed in these patches analysed. Bars = 50 µm in (A)–(D).
The as2-5D and iso-2D leaves differ in expression levels of leaf-polarity controlling genes
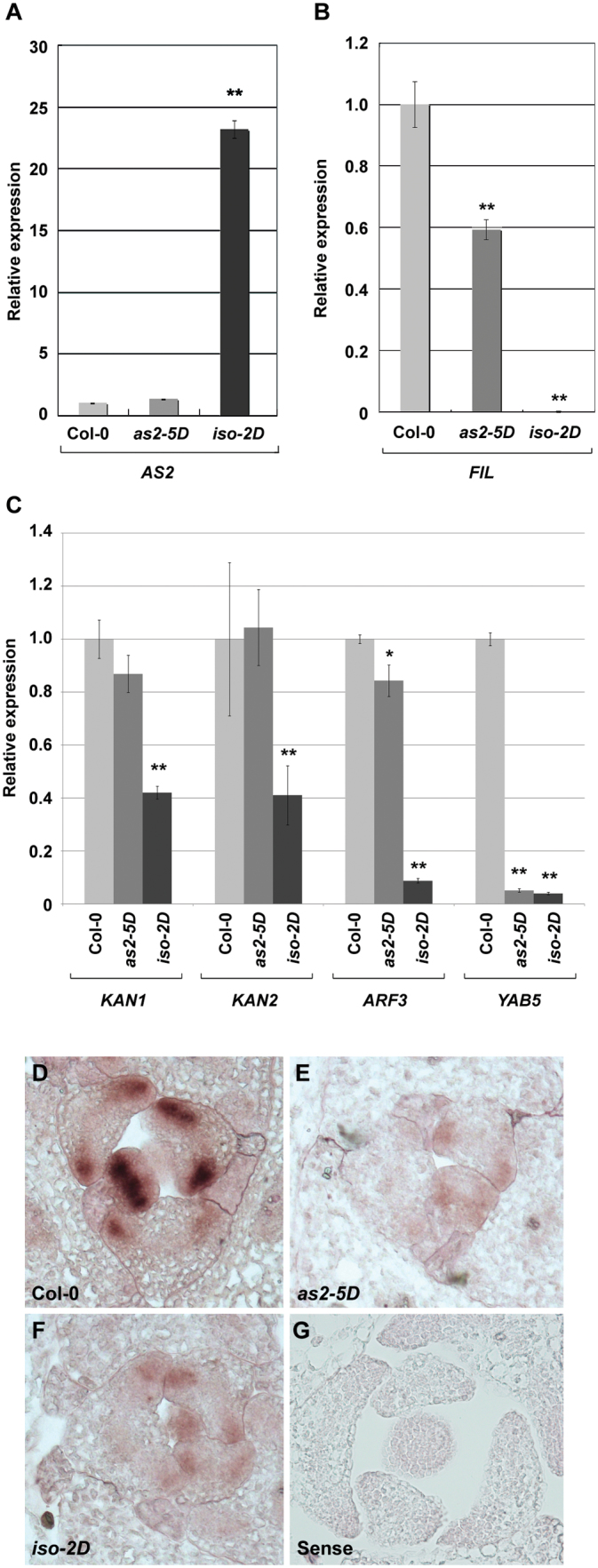
To investigate further how these two mutations affect AS2 expression levels, AS2 transcript levels in as2-5D and iso-2D were analysed by qRT-PCR using mature leaves. To our surprise, the total AS2 transcript level in as2-5D leaves was barely changed compared with that in the wild type, whereas that in iso-2D leaves was dramatically elevated (Fig. 4A). qRT-PCR was also performed in order to examine the expression levels of a leaf polarity marker gene FIL which is normally expressed in the abaxial domain of wild-type leaves (Siegfried et al., 1999). Compared with that in the wild type, the FIL expression level was reduced in both iso-2D and as2-5D at different levels in the mature leaves (Fig. 4B). While the as2-5D leaves showed a reduction of about 40%, FIL expression in the iso-2D leaves was not detected (Fig. 4B). These results are consistent with phenotypic observations that iso-2D has more severe defects in mature leaves than as2-5D in later seedling stages. In addition to the FIL gene, our analysis was extended to several other genes that are known to promote leaf abaxial identity, including KAN1, KAN2, ARF3, and YAB5. Our results showed that expression levels of ARF3 and YAB5 were reduced in the as2-5D and iso-2D leaves to different extents and expression levels of KAN1 and KAN2 were reduced only in the iso-2D leaves (Fig. 4C).
Fig. 4.
as2-5D and iso-2D leaves contain different expression levels of polarity genes. (A, B) qRT-PCR to analyse mature leaves for AS2 (A) and FIL (B) expression levels in Col-0, as2-5D, and iso-2D. (C) qRT-PCR analyses of leaf polarity controlling genes KAN1,KAN2, ARF3, and YAB5 in Col-0, as2-5D, and iso-2D mature leaves. The qRT-PCR results were normalized to that produced by the primers at ACTIN, and the value of the wild type was arbitrarily fixed at 1.0. Bars show s.e. * and **, significant difference by t-test (*, P <0.05; **, P <0.01). (D–F) In situ hybridization to analyse developing leaf primordia for FIL expression patterns using an antisense FIL probe on transverse sections of Col-0 (D), as2-5D (E), and iso-2D (F) shoot apices. (G) The FIL sense probe control.
An in situ hybridization experiment was also performed to analyse FIL transcripts in the leaf primordia of the two mutants. Our data showed that the FIL transcript level appeared markedly decreased in leaf primordia of both as2-5D and iso-2D (Fig. 4E, F) compared with that in the wild type (Fig. 4D), and no hybridization signals were detected in the sense control (Fig. 4G). These results provide a molecular basis for these two mutants to produce the adaxialized leaves.
AS2 is up-regulated dramatically only in the adaxial leaf domain in iso-2D
To investigate the mechanism by which iso-2D affects leaf polarity formation, the distribution patterns of the AS2 transcripts were examined at the early leaf developmental stages by in situ hybridization. In wild-type leaf primordia, AS2 transcripts were mainly detected in the L1 layer cells of the adaxial side, showing relatively low hybridization signals that were discontinuously distributed (Fig. 5A). This AS2 expression pattern is similar to that reported previously (Iwakawa et al., 2007). However, although the AS2 transcripts in the as2-5D leaves were also present mainly in the L1 layer cells, both adaxial and abaxial L1 layer cells contained the AS2 signals with an intensity similar to that in the wild-type leaves (Fig. 5B). The AS2 expression pattern in the as2-5D mutant is consistent with that using GUS staining of the AS2-5D pro :GUS transgenic plants (Wu et al., 2008). Different from the AS2 distribution in as2-5D leaves, AS2 was strongly expressed only in the adaxial leaf domain of iso-2D, with the strongest hybridization signals in the outermost layers (Fig. 5C). An obvious difference of the hybridization signals between the two mutants is in the L1 layer cells of the abaxial leaf side. Compared with as2-5D (Fig. 5B), the abaxial L1 layer cells of iso-2D leaf primordia lacked a hybridization signals (Fig. 5C). As a control, the sense AS2 probe detected no hybridization signals (Fig. 5D).
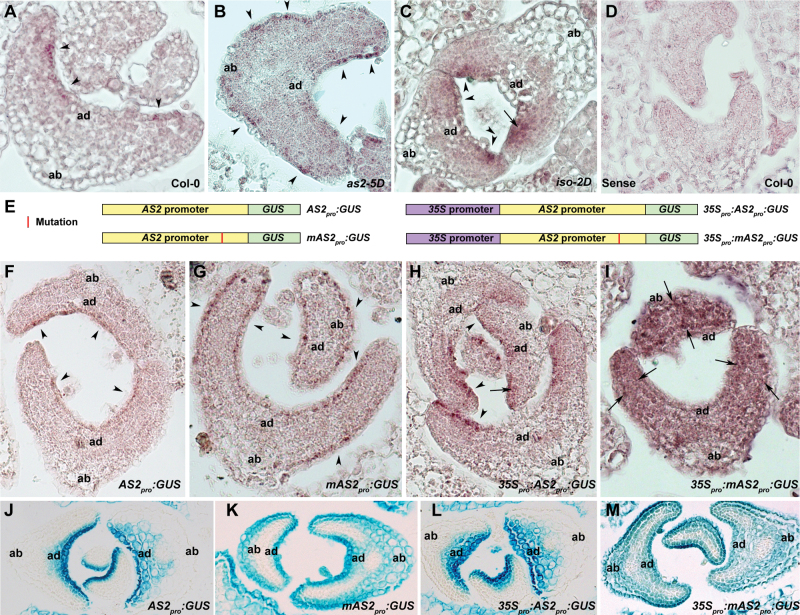
Fig. 5.
Analyses of the AS2 expression patterns. (A–C) In situ hybridization using an antisense AS2 probe on transverse sections of leaf primordia to show AS2 expression patterns in wild-type Col-0 (A), as2-5D (B), and iso-2D (C). (D) The sense probe control. (E) Diagrams of structures of AS2 pro :GUS, mAS2 pro :GUS, 35S pro :AS2 pro :GUS, and 35S pro :mAS2 pro :GUS constructs. (F–I) In situ hybridization using an antisense GUS probe on transverse sections of leaf primordia to show GUS transcript distributions in AS2 pro :GUS/Col-0 (F), mAS2 pro :GUS/Col-0 (G), 35S pro :AS2 pro :GUS/Col-0 (H), and 35S pro :mAS2 pro :GUS/Col-0 (I) transgenic plants. Note that five independent transgenic lines for each construct were analysed and the results were consistent, and shown are the results from one of the five lines analysed. (J–M) GUS staining to analyse AS2 expression. Transverse sections of leaf primordia after GUS staining show GUS distributions in AS2 pro :GUS/Col-0 (J), mAS2 pro :GUS/Col-0 (K), 35S pro :AS2 pro :GUS/Col-0 (L), and 35S pro :mAS2 pro :GUS/Col-0 (M) transgenic plants. ad and ab, leaf adaxial and abaxial sides, respectively. Arrowheads and arrows indicate the AS2 transcripts in the L1 and L2 layers, respectively.
The AS2 expression signals detected by in situ hybridization were relatively weak. To confirm that these are the true hybridization signals of AS2 transcripts, three different GUS fusions were also constructed as it was expected that the GUS transcripts are more stable than those of AS2 and thus may be easy to detect. These three fusions included: (i) the GUS coding region is driven by the AS2 promoter (AS2 pro :GUS); (ii) the GUS coding region is driven by the mutated AS2 promoter as that in the as2-5D mutant (mAS2 pro :GUS); and (iii) a 35S promoter is fused to the 5΄ end of AS2 pro :GUS (35S pro : AS2 pro :GUS) (Fig. 5E). Hence, the in situ hybridization signals detected by the GUS probe in transgenic lines carrying these three fusions represent AS2 expression in the wild-type, as2-5D, and iso-2D leaves, respectively. Our data showed that the distribution patterns of GUS signals were fully consistent with that of the endogenous AS2 transcripts detected by in situ hybridization (Fig. 5F–H). For instance, GUS signals were present in the adaxial L1 layer of the AS2 pro :GUS/Col-0 (Fig. 5F), the entire L1 layer of the mAS2 pro :GUS/Col-0 (Fig. 5G), and more strongly in the adaxial L1 layer of the 35S pro :AS2 pro :GUS/Col-0 (Fig. 5H) leaves. GUS distributions were also analysed by GUS staining (Fig. 5J–L) and it was found that the GUS distribution pattern between in situ hybridization and staining analyses is consistent. The only difference between the two methods is that the GUS signals by GUS staining are not as concentrated as those by in situ hybridization.
To test the respective effects of as2-5D and iso-2D mutations on the AS2 regulation, the 35S pro :mAS2 pro :GUS/Col-0 transgenic plants were constructed with a genetic background equivalent to that of the as2-5D iso-2D double mutant (Fig. 5E). In situ hybridization and GUS staining analyses both showed that the GUS signals were present in the entire leaf primordium (Fig. 5I, M), indicating that the mechanisms controlling AS2 expression in as2-5D and iso-2D are different.
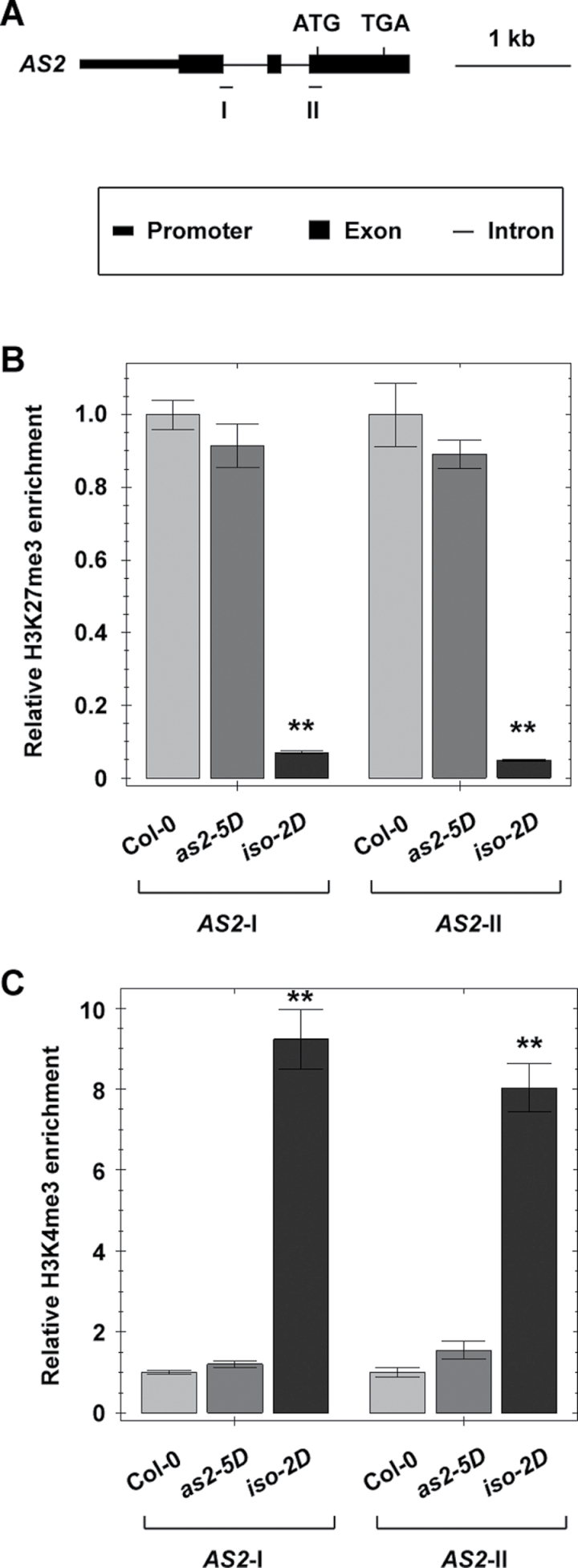
Histone methylation patterns are changed at the AS2 locus in the iso-2D mutant
Histone methylations, especially the histone H3 lysine 27 trimethylation (H3K27me3) and H3K4me3, are usually thought to be important in regulating gene expression in the euchromatin region (Liu et al., 2010). In epigenetic gene regulations, H3K27me3 and H3K4me3 are also considered as markers to define the repressive and active states of chromatin regions, respectively (Zhang et al., 2007, 2009; Roudier et al., 2011). To test whether AS2 over-expression in the iso-2D mutant is also related to epigenetic regulations, a ChIP assay was first performed to analyse H3K27me3 and H3K4me3 levels, with two pairs of PCR primers corresponding to two separate regions in the AS2 gene (Fig. 6A). Our result showed that the H3K27me3 level in the AS2 gene was significantly reduced in the iso-2D but not in the as2-5D leaves (Fig. 6B). By contrast, the H3K4me3 level was significantly increased in iso-2D, but again not in the as2-5D leaves (Fig. 6C). These results indicate that molecular mechanisms in regulating AS2 in the two mutants are different, and also suggest that the increased AS2 expression level in iso-2D may involve epigenetic regulations.
Fig. 6.
iso-2D leaves carry altered levels of histone modification markers H3K27me3 and H3K4me3 at the AS2 locus. (A) Diagram of the AS2 gene with primer positions (I and II) in ChIP analysis. (B, C) Compared with those in Col-0 and as2-5D, the H3K27me3 level in the iso-2D mutant was dramatically reduced (B) whereas the H3K4me3 level was elevated (C) at the AS2 locus. The ChIP results were normalized to those produced by the primers at PI (B) and ACTIN (C). Values of the wild type were arbitrarily fixed at 1.0. Bars show s.e. **, significant difference by t-test (P <0.01).
Discussion
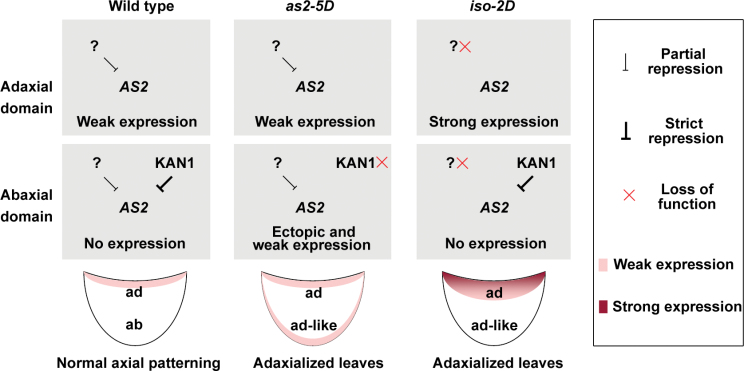
Because AS2 plays a critical role in specifying leaf adaxial identity, regulation of the AS2 gene must be important for leaf axial patterning. Based on previous knowledge and the results obtained in this study, models are proposed to explain the regulation of the AS2 gene in as2-5D and iso-2D mutants during leaf polarity formation (Fig. 7). In wild-type leaves, AS2 is expressed in the adaxial domain because of the abaxially located KAN1 proteins. In addition, the AS2 expression level in wild-type leaves is very low, possibly due to the presence of some ubiquitously located not-yet-known factor(s) that represses AS2 (Fig. 7, left column). Disruption of the KAN1-binding site in as2-5D leads to ectopic expression of AS2 to the L1 layer on the abaxial leaf side, causing the AS2-over-expression phenotypes (Wu et al., 2008). However, because the ubiquitously located factors function well in repressing AS2, expression of the AS2 gene is still kept at a low level in L1 cells of both the adaxial and abaxial sides (Fig. 7, middle column). In iso-2D, however, the insertion of 35S enhancers blocks the function of the ubiquitously located factors, resulting in over-expression of AS2. Nevertheless, since the abaxially located KAN1 protein is still functional, the dramatically increased AS2 expression is only limited in the adaxial leaf domain (Fig. 7, right column). In conclusion, it is proposed that, during leaf patterning, AS2 is regulated both spatially and quantitatively in the entire leaf and both types of regulations are critical for the establishment of the leaf adaxial–abaxial polarity.
Fig. 7.
Model for the AS2 expression patterns in the early stages of leaf development in Col-0, as2-5D, and iso-2D. The question mark indicates some proposed not-yet-known factors that are ubiquitously located in entire leaves to repress AS2 expression. Action of the factors ensures AS2 expression at a low level.
It was noticed that FIL repression occurs in both mutants, but to a greater extent in iso-2D. AS2 expression driven by its native promoter is weakened after hte establishment of the leaf adaxial–abaxial polarity (see Supplementary Fig. S1 at JXB online), so that repression of FIL by AS2 may also be weaker. Because AS2 expression in the as2-5D mutant is driven by its native promoter, its expression level should be reduced along with leaf maturation. By contrast, the presence of 35S enhancers in the iso-2D allele ensures that AS2 expression is maintained at high levels during all stages of leaf development which presumably explains the persistent repression of FIL. Our data showed that the leaf abaxially promoting genes ARF3 and YAB5 were also repressed in mature leaves of both as2-5D and iso-2D mutants, and KAN1 and KAN were repressed in iso-2D. Compared with the as2-5D phenotypes, the iso-2D phenotypes are more severe. This is consistent with the analysed abaxially promoting genes that showed greater repression in iso-2D than in as2-5D. These results are also consistent with the previous suggestions: ARF3 is a direct repressive target of the AS1–AS2 complex (Iwasaki et al., 2013), and AS2 and KAN genes may mutually repress each other’s transcription (Wu et al., 2008).
Our in situ hybridization by analysing AS2 expression showed that, in wild-type leaves, AS2 transcripts are mainly concentrated in the L1 layer of the adaxial side. However, in iso-2D leaves, the AS2 transcripts clearly form a gradient with the most concentrated part in the outmost cells. It is possible that the wild-type leaves may also possess the AS2 transcript gradient, but the current techniques fail to detect it because of the very low level of AS2 expression. The spatial and quantitative regulations of AS2 together may facilitate the formation of a transcript gradient from the outmost adaxial epidermis to the inner cell layers. During organ patterning, formation of such a gradient of certain key regulatory factors could be a common mechanism. For example, the Drosophila Decapentaplegic morphogen gradient is essential for wing disc formation (Schwank and Basler, 2010). An additional example is the abaxially located miR165 and miR166 in Arabidopsis. Different from the gradient of AS2 transcripts, the miR165 and miR166 gradients are present in the abaxial leaf domain with the highest level in the outmost epidermis (Yao et al., 2009).
How AS2 is quantitatively regulated is not yet known. Among many possible genetic pathways, epigenetic regulation could be one that plays roles in the quantitative regulation of AS2. Histone methylations have long been known to control gene expression and, in plants, several reports have provided evidence that histone methylations are involved in the quantitative regulation of gene expression (Jiang et al., 2008; Schatlowski et al., 2008; Liu et al., 2010). More importantly, a recent study has demonstrated that a number of loci corresponding to the leaf polarity-controlling genes, including AS2, are modified by H3K27me3 (Lafos et al., 2011). All these data suggest the possibility that epigenetic regulation may be involved in the control of the expression of leaf polarity genes. In this study, it is shown that the levels of the epigenetic markers H3K27me3 and H3K4me3 closely correlate with changes of AS2 expression in the iso-2D leaves. Although this could be an explanation for the low level AS2 expression in wild-type leaves, the possibility cannot be ruled out that the altered histone modification may simply be an indirect consequence of altered transcriptional regulation at the AS2 locus. Thus, more detailed analysis is needed in the future to elucidate the molecular mechanism of the quantitative AS2 regulations.
Supplementary data
Supplementary data can be found at JXB online.
SupplementaryTable S1. List of primers used in this study.
Supplementary Fig. S1. AS2 expression level is reduced in mature leaves. The qRT-PCR results were normalized to that produced by the primers at ACTIN, and the value of AS2 in the mature leaves was arbitrarily fixed at 1.0. Bars show SE **, P <0.01.
Acknowledgements
We thank M Matsui, and RS Poethig for the Arabidopsis mutant seeds used in this study and W Liang for helpful discussion of the manuscript. This work was supported by grants from National Basic Research Program of China (973 Program, 2012CB910503) and the National Natural Science Foundation of China (31071064).
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. 2006. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Current Biology 16, 927–932 [DOI] [PubMed] [Google Scholar]
- Bowman JL, Eshed Y, Baum SF. 2002. Establishment of polarity in angiosperm lateral organs. Trends in Genetics 18, 134–141 [DOI] [PubMed] [Google Scholar]
- Byrne ME. 2006. Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genetics 2, e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis . Nature 408, 967–971 [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. 1991. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991–1002 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology 13, 1768–1774 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Bowman JL. 1999. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis . Cell 99, 199–209 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. 2001. Establishment of polarity in lateral organs of plants. Current Biology 11, 1251–1260 [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. 2006. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis . Current Biology 16, 939–944 [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. 2006. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Current Biology 16, 933–938 [DOI] [PubMed] [Google Scholar]
- Grigg SP, Canales C, Hay A, Tsiantis M. 2005. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis . Nature 437, 1022–1026 [DOI] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L. 2012. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genetics 8, e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H. 2011. ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant and Cell Physiology 52, 112–124 [DOI] [PubMed] [Google Scholar]
- Husbands AY, Chitwood DH, Plavskin Y, Timmermans MC. 2009. Signals and prepatterns: new insights into organ polarity in plants. Genes and Development 23, 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. 2007. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. The Plant Journal 51, 173–184 [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, et al. 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology 43, 467–478 [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Takahashi H, Iwakawa H, et al. 2013. Dual regulation of ETTIN (ARF3) gene expression by AS1–AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial–abaxial partitioning in Arabidopsis . Development 140, 1958–1969 [DOI] [PubMed] [Google Scholar]
- Jiang D, Wang Y, He Y. 2008. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3, e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. 2004. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. 2001. KANADI regulates organ polarity in Arabidopsis . Nature 411, 706–709 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. 2004. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84 [DOI] [PubMed] [Google Scholar]
- Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, Schubert D. 2011. Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genetics 7, e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. 2005. The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. The Plant Cell 17, 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. 2003. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial–abaxial patterning. The Plant Cell 15, 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Lu F, Cui X, Cao X. 2010. Histone methylation in higher plants. Annual Review of Plant Biology 61, 395–420 [DOI] [PubMed] [Google Scholar]
- Long JA, Barton MK. 1998. The development of apical embryonic pattern in Arabidopsis . Development 125, 3027–3035 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. 2004. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO Journal 23, 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. 1998. Leaf polarity and meristem formation in Arabidopsis . Development 125, 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 [DOI] [PubMed] [Google Scholar]
- Moon J, Hake S. 2011. How a leaf gets its shape. Current Opinion in Plant Biology 14, 24–30 [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, Suzuki K, Muto S, Matsui M. 2003. Activation tagging, a novel tool to dissect the functions of a gene family. The Plant Journal 34, 741–750 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. 2001. REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal 25, 223–236 [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. 2005. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. The Plant Cell 17, 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. 2002. Prediction of plant microRNA targets. Cell 110, 513–520 [DOI] [PubMed] [Google Scholar]
- Roudier F, Ahmed I, Berard C, et al. 2011. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO Journal 30, 1928–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatlowski N, Creasey K, Goodrich J, Schubert D. 2008. Keeping plants in shape: polycomb-group genes and histone methylation. Seminars in Cell and Development Biology 19, 547–553 [DOI] [PubMed] [Google Scholar]
- Schwank G, Basler K. 2010. Regulation of organ growth by morphogen gradients. Cold Spring Harbor Perspectives in Biology 2, a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. 1999. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development 126, 4117–4128 [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhou Q, Zhang W, Fu Y, Huang H. 2002. ASYMMETRIC LEAVES1, an Arabidopsis gene that is involved in the control of cell differentiation in leaves. Planta 214, 694–702 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. 1995. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana . Development 121, 2723–2735 [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A. 1995. phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus . Development 121, 2143–2154 [Google Scholar]
- Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X. 2011. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis . Journal of Experimental Botany 62, 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. 2005. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132, 3657–3668 [DOI] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. 2008. KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2 . Proceedings of the National Academy of Sciences, USA 105, 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Huang W, Li Y, Wang H, Huang H, Cui X. 2012. Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis . The Plant Journal 69, 792–808 [DOI] [PubMed] [Google Scholar]
- Xu L, Shen W-H. 2008. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis . Current Biology 18, 1966–1971 [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. 2003. Novel as1 and as2 defects in leaf adaxial–abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107 [DOI] [PubMed] [Google Scholar]
- Xu L, Yang L, Huang H. 2007. Transcriptional, post-transcriptional and post-translational regulations of gene expression during leaf polarity formation. Cell Research 17, 512–519 [DOI] [PubMed] [Google Scholar]
- Xu L, Yang L, Pi L, Liu Q, Ling Q, Wang H, Poethig RS, Huang H. 2006. Genetic interaction between the AS1–AS2 and RDR6–SGS3–AGO7 pathways for leaf morphogenesis. Plant and Cell Physiology 47, 853–863 [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen W-H. 2008. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana . Molecular and Cellular Biology 28, 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Liang W, Huang H. 2002. The Arabidopsis AS2 gene encoding a predicted leucine-zipper protein is required for the leaf polarity formation. Acta Botanica Sinica 44, 1194–1202 [Google Scholar]
- Yao X, Wang H, Li H, Yuan Z, Li F, Yang L, Huang H. 2009. Two types of cis-acting elements control the abaxial epidermis-specific transcription of the MIR165a and MIR166a genes. FEBS Letters 583, 3711–3717 [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. 2005. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis . Genes and Development 19, 2164–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Luo D, Li G, Yao X, Wang H, Zeng M, Huang H, Cui X. 2010. Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. The Plant Journal 64, 331–342 [DOI] [PubMed] [Google Scholar]
- Zhang X, Bernatavichute YV, Cokus S, Pellegrini M, Jacobsen SE. 2009. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana . Genome Biology 10, R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. 2007. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis . PLoS Biology 5, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 1999. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. The Plant Cell 11, 2139–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.