Abstract
Tomato, melon, grape, peach, and strawberry primarily accumulate soluble sugars during fruit development. In contrast, kiwifruit (Actinidia Lindl. spp.) and banana store a large amount of starch that is released as soluble sugars only after the fruit has reached maturity. By integrating metabolites measured by gas chromatography–mass spectrometry, enzyme activities measured by a robot-based platform, and transcript data sets during fruit development of Actinidia deliciosa genotypes contrasting in starch concentration and size, this study identified the metabolic changes occurring during kiwifruit development, including the metabolic hallmarks of starch accumulation and turnover. At cell division, a rise in glucose (Glc) concentration was associated with neutral invertase (NI) activity, and the decline of both Glc and NI activity defined the transition to the cell expansion and starch accumulation phase. The high transcript levels of β-amylase 9 (BAM9) during cell division, prior to net starch accumulation, and the correlation between sucrose phosphate synthase (SPS) activity and sucrose suggest the occurrence of sucrose cycling and starch turnover. ADP-Glc pyrophosphorylase (AGPase) is identified as a key enzyme for starch accumulation in kiwifruit berries, as high-starch genotypes had 2- to 5-fold higher AGPase activity, which was maintained over a longer period of time and was also associated with enhanced and extended transcription of the AGPase large subunit 4 (APL4). The data also revealed that SPS and galactinol might affect kiwifruit starch accumulation, and suggest that phloem unloading into kiwifruit is symplastic. These results are relevant to the genetic improvement of quality traits such as sweetness and sugar/acid balance in a range of fruit species.
Key words: Actinidia deliciosa, AGPase, berry development, enzyme profiling, fruit quality, glucose, metabolite profiling, neutral invertase, planteose, primary metabolism, starch, sucrose phosphate synthase, transcript profiling.
Introduction
Kiwifruit (Actinidia Lindl. spp.) is one of the few new fruit crops successfully adopted by international markets in recent years. It has become a high value export crop for a number of countries including New Zealand, Italy, and Chile. The success of kiwifruit has been largely based on a single cultivar, Actinidia deliciosa ‘Hayward’, which has a distinctive appearance and flavour, compared with other fruit types, as well as health benefits from high concentrations of vitamin C and fibre (Ferguson and Ferguson, 2003). In the past decade, several new kiwifruit cultivars have been developed and released to complement sales of ‘Hayward’ fruit in world markets. However, to accelerate the development of further new cultivars, a greater understanding of the metabolic control of major quality traits in kiwifruit is required.
Sweetness is the single most important quality trait for kiwifruit as it influences overall fruit flavour (sugar/acid balance, perception of volatiles), consumer acceptability, and grower returns (Harker et al., 2009; Nardozza et al., 2011a ). While sucrose (Suc) and planteose are the major sugars transported to fruit during development (Klages et al., 2004), adequate storage of soluble sugars as starch/dry matter (DM) reserves is essential to produce an acceptably flavoured kiwifruit berry (Richardson et al., 1997). Starch metabolism in kiwifruit is a complex, dynamic process, characterized by simultaneous starch accumulation and degradation (Wegrzyn and MacRae, 1995). Net starch accumulation does not occur until the fruit has completed cell division (Richardson et al., 2004), while net starch degradation mainly occurs after fruit harvest and, by the time the fruit is eating ripe, almost all starch has been converted into soluble sugars (MacRae et al., 1992).
Metabolic profiling has been applied to study developmental changes in several fruits, such as tomato (Solanum lycopersicum L.) (Carrari et al., 2006; Steinhauser et al., 2010), grape (Vitis vinifera L.) (Deluc et al., 2007; Dai et al., 2013), strawberry [Fragaria×ananassa (Weston) Duchesne ex Rozier (pro sp.)] (Fait et al., 2008), melon (Cucumis L.) (Dai et al., 2011), and peach [Prunus persica (L.) Batsch] (Lombardo et al., 2011). All these fruits have been demonstrated to differ from one another not only from physiological and botanical points of view, but also at the metabolic level. What was clearly identified in these studies was the need to extend metabolite investigations to a wide range of fruit species, especially those where the metabolism differs significantly from the tomato model. Kiwifruit vines bear fleshy fruit that are botanically classified as berries and therefore share characteristics with the model berry, tomato. However, kiwifruit differs from tomato in several important ways. Kiwifruit accumulate large amounts of starch over a long period of time, store only limited amounts of soluble sugars, and the development of osmotic pressure is mainly driven by organic acid accumulation (Nardozza et al., 2010). Kiwifruit is also different from grape, melon, and apple (Malus Mill.) which accumulate all or most carbon as soluble sugars (Deluc et al., 2007; Dai et al., 2011; Li et al., 2012), and from peach and strawberry which do not produce berries (being a stone fruit and an aggregate accessory fruit, respectively), and also do not accumulate significant amounts of starch (Fait et al., 2008; Lombardo et al., 2011). Current knowledge of kiwifruit primary metabolism is limited to the commercial cultivars ‘Hayward’ and ‘Hort16A’ (Richardson et al., 2011; Moscatello et al., 2011).
This comprehensive analysis of the Suc to starch metabolic pathway during kiwifruit development combines data on primary metabolites, transcripts, and enzyme activities to identify changes between developmental stages and regulatory steps that affect carbohydrate accumulation. This work examines the hallmarks of starch accumulation in the kiwifruit berry by investigating why certain kiwifruit genotypes are able to accumulate twice the amount of starch during fruit development compared with others (Nardozza et al., 2010). The majority of fruit metabolic studies published to date, for any crop, focus on one cultivar. The novel approach adopted in this work was to compare genotypes contrasting in fruit size and starch concentration. The results show that ADP-glucose pyrophosphorylase (AGPase) is a key enzyme involved in kiwifruit starch accumulation. Sucrose phosphate synthase (SPS) and galactinol (Gol) also appear to have roles in maximizing starch accumulation. Novel aspects of carbon metabolism in kiwifruit were also revealed, including that changes in glucose (Glc) levels and neutral invertase (NI) activity mark the transition to net starch accumulation, the presence of planteose and Gol in fruit, and the potential involvement of SPS/sucrose synthase (SUSY) and β-amylase 9/α-amylase 2 (BAM9/AMY2) in Suc cycling and starch turnover, respectively.
Materials and methods
Plant material, sampling, and physical measurements
Actinidia deliciosa (A. Chev.) C.F. Liang et A.R. Ferguson var. deliciosa genotypes characterized in this study were selected from a larger group of 24 genotypes (Nardozza et al., 2010). In the 2007 harvest year (HY), fruit samples representative of all fruit tissue types were collected from 10 selected and replicated genotypes (Nardozza et al., 2010) at four fruit developmental stages (Supplementary Tables S1, S2 available at JXB online), snap-frozen in liquid nitrogen, and stored at –80 °C for transcript analysis.
In the 2009 HY, a subset of four A. deliciosa genotypes was selected to give one individual each representing the extremes in starch concentration (high and low) and fruit size (large and small) (Fig. 1E; Supplementary Fig. S1B at JXB online). This experimental design minimizes the dilution effect of different rates of fresh weight (FW) growth that could hamper the identification of factors affecting starch accumulation. Fruit outer pericarp samples were collected in six biological replicates, at five different fruit developmental stages [14, 28, 56, 98, and 154 days after anthesis (DAA); Supplementary Tables S1, S3 at JXB online], snap-frozen in liquid nitrogen, and stored at –80 °C for metabolite, enzyme activity, and transcript analysis. FW, dry weight (DW), and DM were measured on sampled fruit as described in Nardozza et al. (2010).
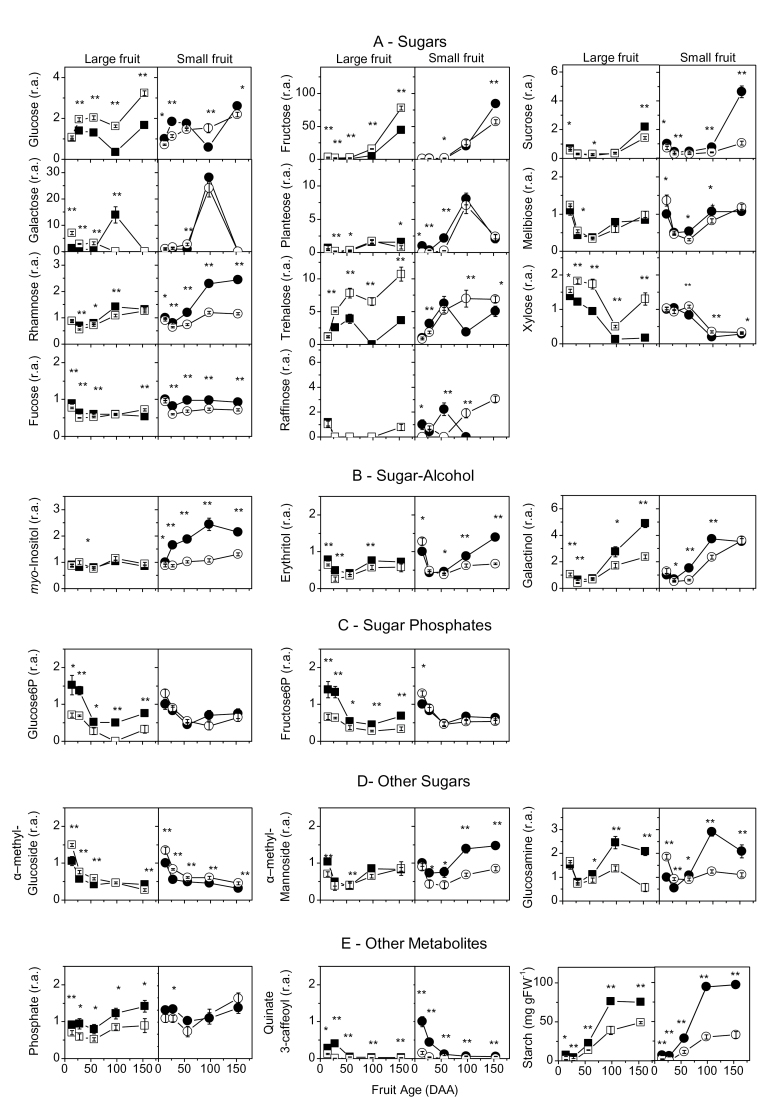
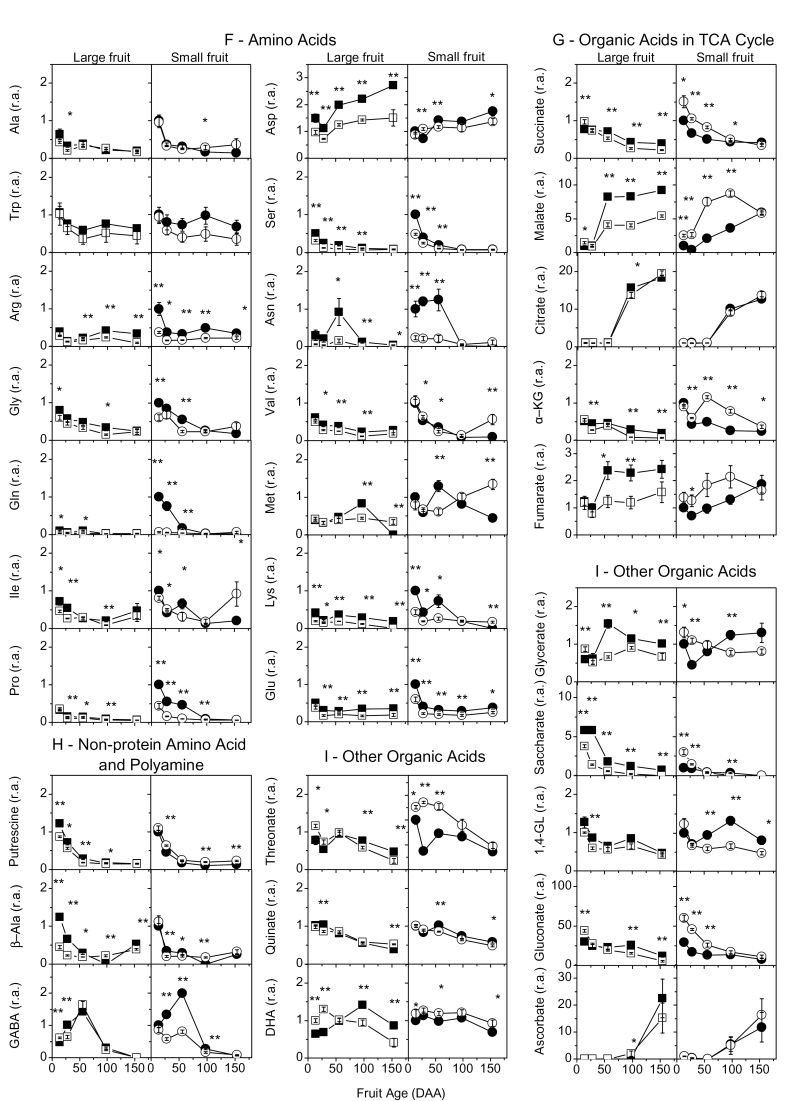
Fig. 1.
Changes in metabolite concentrations during fruit development of kiwifruit berries. Four genotypes were examined: G3, large fruit and high starch (filled squares); G25, large fruit and low starch (open squares); G30, small fruit and high starch (filled circles), and G17, small fruit and low starch (open circles). The 51 metabolites identified by GC-TOF-MS plus starch were subdivided into the following classes: A, sugars (Glc, Fru, sucrose, Gal, planteose, melibiose, Rha, Tre, Xyl, Fuc, raffinose); B, sugar-alcohols (myo-inositol, erythritol, galactinol); C, phosphate sugars (Glc6P, Fru6P); D, other sugars (α-methyl glucoside, α-methyl mannoside, glucosamine); E, other metabolites (phosphate, quinate 3-caffeoyl, starch); F, amino acids (alanine, tryptophan, arginine, glycine, glutamine, isoleucine, proline, aspartate, serine, asparagine, valine, methionine, lysine, glutamine); G, organic acids in the TCA cycle (succinate, malate, citrate, α-KG, fumarate); H, non-protein amino acids and polyamine (putrescine, β-alanine, GABA); I, other organic acids (threonate, quinate, DHA, gluconate, ascorbate, glycerate, saccharate, 1,4-GL). For each developmental stage, t-test significance levels are reported: *P < 0.05; **P < 0.01. Where the symbol is missing, the difference is not statistically significant. Fruit age is in days after anthesis (DAA); 1,4-GL, glucaric acid 1,4-lactone; DHA, dehydroascorbate; GABA, γ-aminobutyric acid; α-KG, α-ketoglutarate. Values are means of six biological replicates ±SEM. All data are expressed as relative amount (r.a.), with the exception of starch which is expressed as milligrams per gram fresh weight (mg g FW–1), and normalized to sample G30 at 14 DAA. Data refer to 2009 outer pericarp samples.
The developmental stages were chosen to include key times when FW and starch curves were changing (Nardozza et al., 2010). Supplementary Table S3 at JXB online shows their correspondence to the BBCH scale (Richardson et al., 2011). Growth slowing (56 DAA and BBCH stage 75) represents the transition from cell division to cell expansion (Hopping, 1976).
Profiling of primary metabolites
Polar metabolites were extracted following the method of Roessner et al. (2001) optimized for tomato fruit (Roessner-Tunali et al., 2003). Samples were analysed by gas chromatography-time of flight-mass spectrometry (GC-TOF-MS) using a high-throughput platform and the protocol of Lisec et al. (2006). A 5 µl aliquot of extract was analysed to identify compounds present at high concentration [i.e. Glc, fructose (Fru), Suc, myo-inositol, citrate, quinate, and malate]. For metabolites at lower concentration, a second 50 µl aliquot was analysed. The retention time index standard mixture was a combination of fatty acid methyl esters (FAMEs). Mass spectra were cross-referenced with those of authentic standards in the Golm Metabolome Database (Kopka et al., 2005; Schauer et al., 2005). Metabolite quantification was based on the relative peak response area of each chromatogram (Fait et al., 2008). The relative metabolite concentrations were determined on the basis of an internal standard (ribitol), normalized to sample G30 at 14 DAA and expressed as relative amounts (r.a.).
For soluble sugar (Glc, Fru, Suc, myo-inositol, planteose, and galactose) quantification, a third aliquot of extract was analysed by ion chromatography (IC). The methanol was blown off using a stream of nitrogen gas, and samples were redissolved in ultrapure water and analysed on a DIONEX ICS-3000 Reagent-Free™ IC (RFIC™) system with a CarboPac PA20 column (Trought et al., 2011). Starch was analysed from the pellet obtained after the extraction of polar metabolites as described in Smith et al. (1992). Starch and sugar concentrations were expressed as milligrams per gram FW (mg g FW –1).
Carbohydrate and amino acid enzyme extractions and assays
Enzyme activities were measured by a semi-automated robot-based platform for multiple enzyme assays (Gibon et al., 2004; Steinhauser et al., 2010). With this sensitive method, a large number of enzyme activities can be measured from the same extract and no crude extract desalting is required.
Samples were ground in liquid nitrogen and aliquots (~40mg FW) of frozen powder were extracted after addition of 10mg of polyvinylpolypyrrolidone and vigorous vortexing in 500 µl of ice-cold extraction buffer (Supplementary Table S4 at JXB online). The crude extract was centrifuged for 10min at 3500rpm at 4 °C. Extract aliquots were further diluted with extraction buffer to obtain the optimal dilution for each enzyme (Supplementary Table S5 at JXB online). Enzyme activities were assayed immediately with established protocols (Gibon et al., 2004; Steinhauser et al., 2010) (Supplementary Table S6 at JXB online), giving higher priority to more unstable enzymes (i.e. SPS and SUSY).
To estimate the contribution of each measured enzyme in starch synthesis, the correlation coefficients of individual enzymes were calculated. Starch and enzyme values for all genotypes during net starch accumulation (56–154 DAA) were normalized on a zero to one scale. Then a regression analysis slope for each enzyme was calculated.
Transcript analysis
RNA was isolated from 2g of fruit tissues as described in Chang et al. (1993) and DNase treated (DNA-free™; Ambion-Invitrogen). cDNA was synthesized using SuperScript™ III Reverse Transcriptase (Invitrogen). Gene-specific primers (Supplementary Table S7 at JXB online) were designed using Primer3 software (Rozen and Skaletsky, 2000) to a stringent set of criteria (Bulley et al., 2009). Relative transcripts were quantified on a LightCycler 480 (Roche) real-time PCR detection system using a LightCycler 480 SYBR Green master mix (Roche) (Nieuwenhuizen et al., 2009). Elongation factor 1α (EF1α) and protein phosphatase 2A (PP2A) were the housekeeping genes. Target gene transcripts were normalized to PP2A (Bulley et al., 2009) using the 2–ΔCt method (Ren et al., 2007).
Statistical analysis
Statistical analysis of the metabolite, IC sugar, starch, and the enzyme activity data was performed using Microsoft® Excel two-samples t-tests assuming unequal variances. All the correlation analyses were performed in Microsoft® Excel, by calculating Pearson’s product–moment (r) and its probability (P). The correlation analysis was run in pairs of genotypes from the same starch group including the genotypes’ mean values of all developmental stages. Significant correlations (P < 0.05) were visualized in matrices using Multi Experiment Viewer software version 4.8.1 (Saeed et al., 2003). The large number of correlations tested may increase false-positive rates, but correction for an inflated Type-1 error may cause a reduction in power due to the small sample size (n=10). Therefore, heat maps show all correlations whose unadjusted P-value is <0.05, also allowing a fair comparison with literature data (Carrari et al., 2006; Fait et al., 2008; Lombardo et al., 2011) where no adjustment for multiple testing was carried out. Non-corrected and false discovery rate-corrected r critical values are shown in Supplementary Table S8 at JXB online.
Transcript data (Supplementary Fig. S3 at JXB online) were analysed with physical measurements (FW, DW, DM; 2007 HY), growth indexes [FW relative growth rate (RGR) and DW RGR], and metabolites [starch, Fru, Glc, Suc, myo-inositol, galactose (Gal); 2005 HY] by a redundancy analysis (RDA), a principal component analysis (PCA) of multivariate regression predicted values that integrates independent data sets (Stewart and Love, 1968). RDA plots are interpreted in the same way as a PCA (Zuur et al., 2007). The RDA was chosen to account for samples collected in different years. RGRs were calculated as described in Moscatello et al. (2011).
Results
Metabolite profiling in A. deliciosa genotypes during fruit development
Fifty-one metabolites were identified by GC-TOF-MS (Fig. 1) and subdivided into nine groups: sugars (A), sugar-alcohols (B), sugar phosphates (C), other sugars (D), other metabolites (E), amino acids (F), non-protein amino acids (G), organic acids in the tricarboxylic acid (TCA) cycle (H), and other organic acids (I).
Suc, Fru, Glc, and myo-inositol were the major soluble sugars identified by GC-TOF-MS (Fig. 1A, B), and quantified by IC (Supplementary Fig. S1E–H at JXB online), whereas all the other sugars detected had lower concentrations. Planteose, a major sugar in kiwifruit leaves (Klages et al., 2004), was also detected in fruit tissues (Fig. 1A). Starch and sugar concentrations declined during cell division and then increased again during cell expansion (Fig. 1A, B, E), while Glc peaked during cell division prior to net starch accumulation. Trehalose (Tre) had a Glc-like pattern. Rhamnose (Rha), melibiose, erythritol, and Gol had starch-like patterns (Fig. 1A, B, E). High-starch genotypes (G3 and G30) had significantly more Suc, Rha, erythritol, Gol, glucosamine, and quinate-3-caffeoyl than low-starch genotypes (G25 and G17) (Fig. 1A, B, D, E). In contrast, low-starch genotypes had significantly more Tre, xylose (Xyl), and α-methyl-glucoside than high-starch genotypes (Fig. 1A, D).
Glutamate, asparagine, glutamine, and γ-aminobutyric acid (GABA) decreased during cell expansion, whereas aspartate increased (Fig. 1F, H). Malate and citrate increased in concentration during cell expansion (Fig. 1G). GABA followed a Glc-like pattern (Fig. 1H) whereas aspartate, arginine, tryptophan, and citrate showed starch-like patterns (Fig. 1F, G). Quinate decreased throughout fruit development (Fig. 1I). Tryptophan, arginine, aspartate, glycine, and glutamine were significantly higher in high-starch genotypes than in low-starch genotypes, as were serine and asparagine, but generally only at cell division (Fig. 1F).
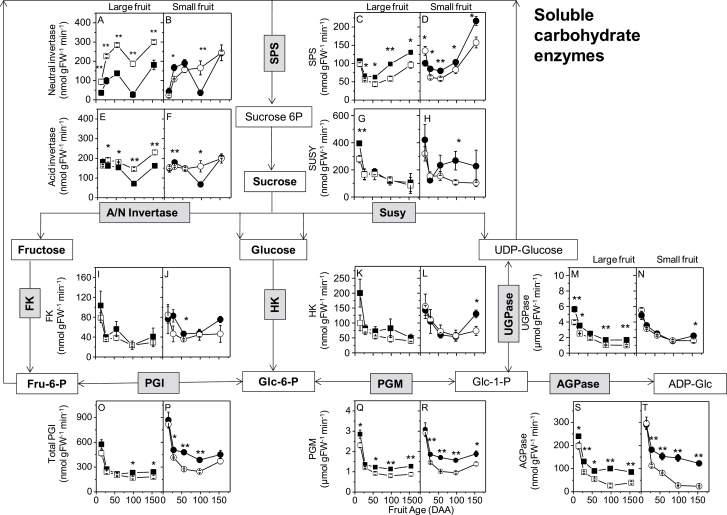
Carbohydrate and amino acid enzymes during fruit development
The majority of the selected soluble enzymes of Suc, starch, and amino acid metabolism were cytosolic. NI activity (Fig. 2A, B) had a Glc-like pattern, whilst SPS activity (Fig. 2C, D) followed a similar trend to Suc, starch, and DM. Most of the enzyme activities measured in the Suc and starch biosynthetic pathways were higher at cell division than in later stages: e.g. SUSY (Fig. 2G, H), hexokinase (HK; Fig. 2K, L), UDP-Glc pyrophosphorylase (UGPase; Fig. 2M, N), phosphoglucoisomerase (PGI; Fig. 2O, P), phosphoglucomutase (PGM; Fig. 2Q, R), and AGPase (Fig. 2S, T). SPS was the only sugar biosynthetic enzyme with activity levels consistently higher in high-starch genotypes (G3 and G30) than in low-starch genotypes (G25 and G17) (Fig. 2C, D). Total PGM and the first enzyme of starch synthesis in the plastid, AGPase, were significantly higher in high-starch genotypes than in low-starch genotypes (Fig. 2Q–T).
Fig. 2.
Kiwifruit soluble enzyme activities associated with Suc to starch metabolism. The graph for each enzyme activity is placed in proximity to the enzyme in the simplified pathway chart: A–B, neutral invertase; C–D, sucrose phosphate synthase; E–F, acid invertase; G–H, sucrose synthase; I–J, fructokinse; K–L, hexokinase; M–N, UDP-Glc pyrophosphorylase; O–P, total phosphoglucose isomerase; Q–R, phosphoglucomutase; S–T, ADP-Glc pyrophosphorylase. t-test significance levels are reported: *P < 0.05; ** P < 0.01. Where the symbol is missing, the difference is not statistically significant. Four genotypes were examined: G3, large fruit and high starch (filled squares); G25, large fruit and low starch (open squares); G30, small fruit and high starch (filled circles), and G17, small fruit and low starch (open circles). Enzyme codes: AGPase, ADP-Glc pyrophosphorylase; A/N invertase, acid and neutral invertase; FK, fructokinase; HK, hexokinase; PGM, phosphoglucomutase; PGI, phosphoglucose isomerase (total); SPS, sucrose phosphate synthase; SuSy, sucrose synthase; UGPase, UDP-Glc pyrophosphorylase. Metabolite abbreviations: ADP-Glc, ADP-glucose; Fru-6-P, fructose-6-phosphate; Glc-1-P, glucose-1-phosphate; Glc-6-P, glucose-6-phosphate. Fruit age is in days after anthesis (DAA). Values are means of four biological replicates ±SEM. Data refer to 2009 outer pericarp samples. White boxes, metabolites; grey boxes, enzymes; bold text, measured metabolites.
To test which enzymes may have a key role in fruit starch accumulation, the correlation coefficients were estimated for the enzymes of the Suc to starch pathway: AGPase, 0.23; SUSY, 0.10; PGM, 0.19; PGI, 0.13; UGPase, –0.06; HK, 0.14; and fructokinase (FK), 0.12. Enzymes outside the Suc to starch pathway, but important for carbohydrate dynamics, gave the following coefficients: SPS, 0.50; acid invertase (AI), –0.20; NI, –0.33.
All three amino acid metabolism enzymes measured fell 3-fold during cell division (Supplementary Fig. S2 at JXB online). Only shikimate dehydrogenase (SDH) increased again during cell expansion. Both glutamate dehydrogenase (GLDH) and aspartate aminotransferase (AST) activities were higher in high-starch genotypes than in low-starch genotypes.
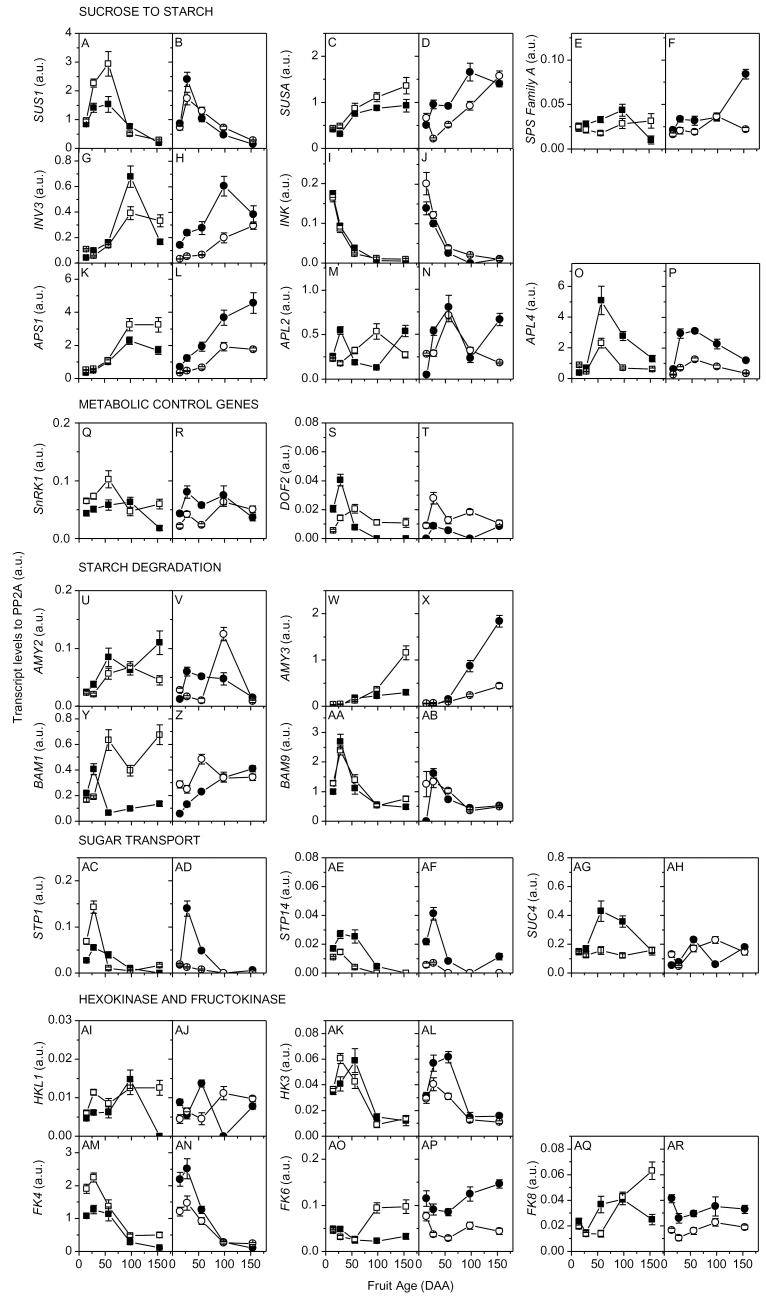
Transcript analysis of gene families impacting cytosolic sugar concentrations
The PFR Actinidia EST (expressed sequence tag) database (Crowhurst et al., 2008) was mined for genes potentially impacting cytosolic sugar concentrations. Thirty-three genes were identified, many of which belonged to multigene families. Actinidia genes were named after the nearest Arabidopsis homologue, as identified by phylogenetic analysis (data available on request): α-amylase (AMY1, AMY2, AMY3), AGPase (APL1, APL2, APL4, APS1), AI (INV3, CWINV1), β-amylase (BAM1, BAM2, BAM3, BAM8, BAM9), Dof zinc finger transcription factor (DOF2), FK (FK4, FK6, FK8), hexose transporter (STP1, STP3, STP14), HK (HK1, HK3, HKL1), NI (INVD, INVE, INVK), sucrose non-fermenting kinase (SnRK1), sucrose transporter (SUC3, SUC4), and SUSY (SUSA, SUS1, SUS6).
Based on the TargetP predicted location of the enzymes from transcripts (Emanuelsson et al., 2007) and published literature, AMY2, BAM9, SUSA, SUS1, INK, APL1, APL2, APL4, SUC3, SUC4, STP1, STP3, STP7, INV3, FK4, FK6, and SPS Family A (SPSA) were cytosolic, AMY3, BAM3, APS1, DOF2, and HKL1 were plastidial, CWINV1 and AMY1 were apoplastic, and HK1 was associated with the mitochondria. A homologue of the tomato LIN5 cell wall invertase, reported to be critical in phloem Suc unloading (Fridman et al., 2004), is absent from the PFR Actinidia EST database. In addition, it was not possible to isolate this isoform from fruit tissue by qPCR and sequencing (data not shown). Kiwifruit cell wall invertase CWINV1 was only expressed in vegetative tissue (data not shown).
Gene transcription patterns in whole fruit tissue from five high- and five low-starch genotypes (2007 HY; Supplementary Table S2 at JXB online) were surveyed during fruit development (Supplementary Fig. S3 at JXB online). Genes present in fruit tissues, showing differential transcript abundance between high- and low-starch genotypes, or overall high expression levels, were analysed further. The transcript abundance of 21 genes meeting these criteria (plus SPSA) was measured in the same samples used for metabolic and enzyme activity profiling (2009 HY; Fig. 3). SUS1 (Fig. 3A, B), APL2 (Fig. 3M, N), BAM9 (Fig. 3AA, AB), HK3 (Fig. 3AK, AL), and FK4 (Fig. 3AM, AN) peaked early in fruit development in a pattern similar to Glc. SUSA (Fig. 3C, D), SPSA (Fig. 3E, F), INV3 (Fig. 3G, H), APS1 (Fig. 3G, H), AMY3 (Fig. 3U, V), and FK6 (Fig. 3AO, AP) increased during cell expansion. APL4 (Fig. 3O, P) and STP14 (Fig. 3AE, AF) transcripts were higher in high-starch genotypes (G3 and G30) than in low-starch genotypes (G25 and G17) during cell expansion and during cell division, respectively.
Fig. 3.
Transcript levels of kiwifruit genes involved in carbohydrate metabolism. Transcript levels measured during fruit development of four Actinidia deliciosa genotypes: G3, high starch and large fruit (filled squares); G25, low starch and large fruit (open squares); G30, high starch and small fruit (filled circles); and G17, low starch and small fruit (open circles). Gene codes: sucrose synthase, (A–B) SUS1, (C–D) SUSA; sucrose phosphate synthase, (E–F) SPS Family A; vacuolar invertase, (G–H) INV3; neutral invertase, (I–J) INK; ADP-glucose pyrophosphorylase, (K–L) APS1, (M–N) APL2, (O–P) APL4; sucrose non-fermenting related kinase, (Q–R) SnRK1; DOF2 zinc finger transcription factor, (S–T) DOF2; α-amylase, (U–V) AMY2, (W–X) AMY3; β-amylase, (Y–Z) BAM1, (AA–AB) BAM9; monosaccharide transporters, (AC–AD) STP1, (AE–AF) STP14; sucrose transporter, (AG–AH) SUC4; hexokinase, (AI–AJ) HKL1, (AK–AL) HK3; fructokinase, (AM–AN) FK4, (AO–AP) FK6, (AQ–AR) FK8. Values are averages of four technical replicates of the reverse-transcribed RNA sample of six pooled biological replicates ±SEM. Housekeeping gene: protein phosphatase 2A (PP2A). Fruit age is given in days after anthesis (DAA). Data refer to 2009 outer pericarp samples.
Metabolite, enzyme, and transcript correlations in high- and low-starch genotypes
Correlations were calculated to assess the associations between metabolite, enzyme, and transcript during A. deliciosa fruit development. High- (G3 and G30) and low-starch (G25 and G17) genotypes were compared.
Significant metabolite (51 metabolites plus starch, FW, and DM) correlations (P < 0.05) are shown in Supplementary Fig. S4 at JXB online. For both genotype groups, Suc was positively correlated to DM but not to starch concentration; Tre was positively correlated to Glc; Rha was positively correlated to starch, Gol, Fru, planteose, and DM; planteose was positively correlated to Gal; and starch was positively correlated to Gol. In low-starch genotypes only, starch was positively correlated to Glc and Tre.
Enzyme activities were highly correlated to one another during development in both genotype groups (P < 0.05; Supplementary Fig. S5 at JXB online). The majority of the correlations were positive, but NI was negatively correlated to 10 other enzymes and positively correlated to AI.
Transcripts only had a limited number of significant correlations (P < 0.05; Supplementary Fig. S6 at JXB online). For both genotype groups, SUSA transcripts were positively correlated to AMY3, INV3, and APS1, as were FK4 transcripts to INK, SUS1, STP1, STP14, and HK3. SUS1 transcripts were positively correlated to BAM9 and HK3 in both high- and low-starch genotypes and to APL4 in high-starch genotypes only. SUSA transcripts were negatively correlated to BAM9 in both genotype groups.
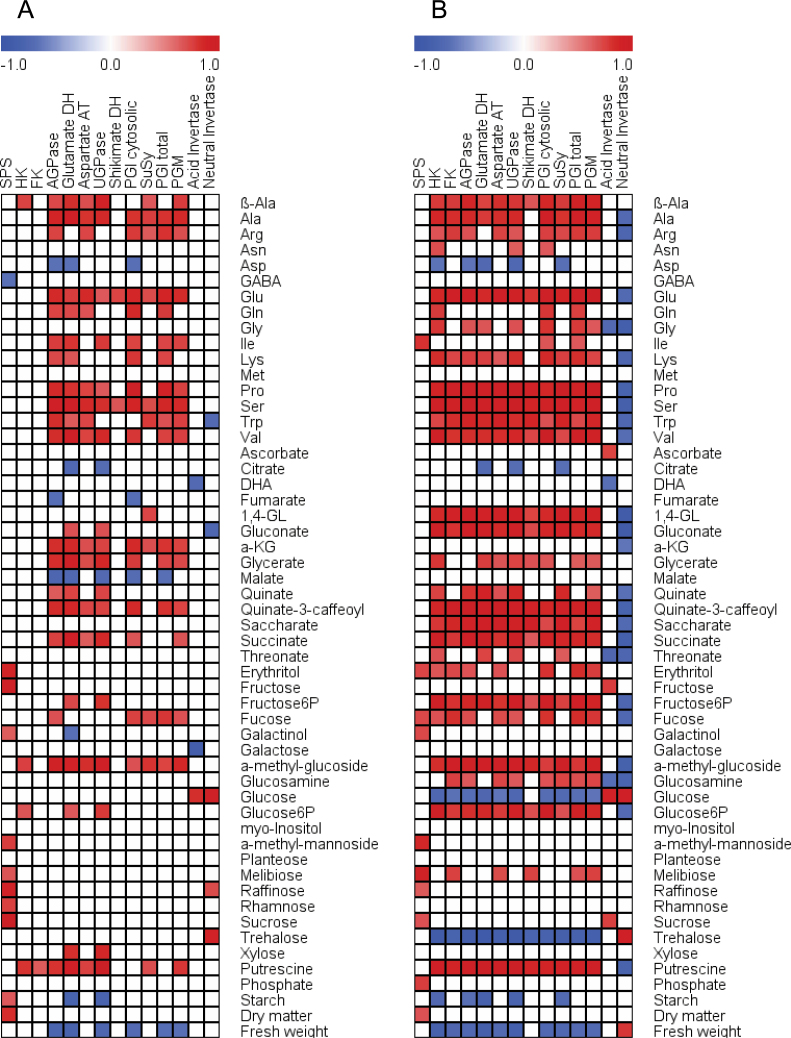
Significant metabolite to enzyme correlations (P < 0.05) are shown in Fig. 4. SPS was positively correlated to DM in both genotype groups and with Rha in high-starch genotypes. Starch was positively correlated to SPS in high-starch genotypes, and negatively correlated to GLDH and UGPase. In low-starch genotypes, there were only negative correlations between starch and the activities of AGPase, SUSY, UGPase, GLDH, and HK. For correlations between enzymes and their substrate/product, Suc was positively correlated to SPS enzyme activity, Glc6P was positively correlated to HK, and Glc was positively correlated to AI and NI activities for both genotype groups. In low-starch genotypes only, fructose 6-phosphate (Fru6P) was positively correlated to FK, Glc was negatively correlated to HK, AI was positively correlated to Suc and Fru, PGI was positively correlated to Fru6P and glucose 6-phosphate (Glc6P), and PGM was positively correlated to Glc6P. In the amino acid pathway, GLDH was positively correlated to Glc for both genotype classes, whereas it was positively correlated to α-ketoglutarate only in high-starch genotypes. Most of the enzymes correlated negatively to FW.
Fig. 4.
Metabolite to enzyme activity correlation matrix during fruit development of kiwifruit berries. (A) High-starch genotypes (G3 and G30); (B) low-starch genotypes (G25 and G17). The analysis included 51 metabolites analysed by GC-TOF-MS plus starch, fresh weight, and dry matter, and 14 enzymes. The significance threshold of P < 0.05 was used (open squares were not statistically significant). 1,4-GL, glucaric acid 1,4-lactone; DHA, dehydroascorbate; α-KG, α-ketoglutarate; AGPase, ADP-glucose pyrophosphorylase; Aspartate AT, aspartate aminotransferase; FK, fructokinase; HK, hexokinase; Glutamate DH, glutamate dehydrogenase; PGM, phosphoglucomutase; PGI, phosphoglucose isomerase; shikimate DH, shikimate dehydrogenase; SPS, sucrose phosphate synthase; SuSy, sucrose synthase; UGPase, UDP-glucose pyrophosphorylase. Data refer to 2009 outer pericarp samples.
Significant (P < 0.05) metabolite to transcript (Supplementary Fig. S7 at JXB online) and transcript to enzyme (Supplementary Fig. S8 at JXB online) correlations were scarce, with little consistency between high- and low-starch genotypes and no correlations between genes and their respective enzyme activity.
Redundancy analysis of transcript levels, and metabolite,and physical measurements
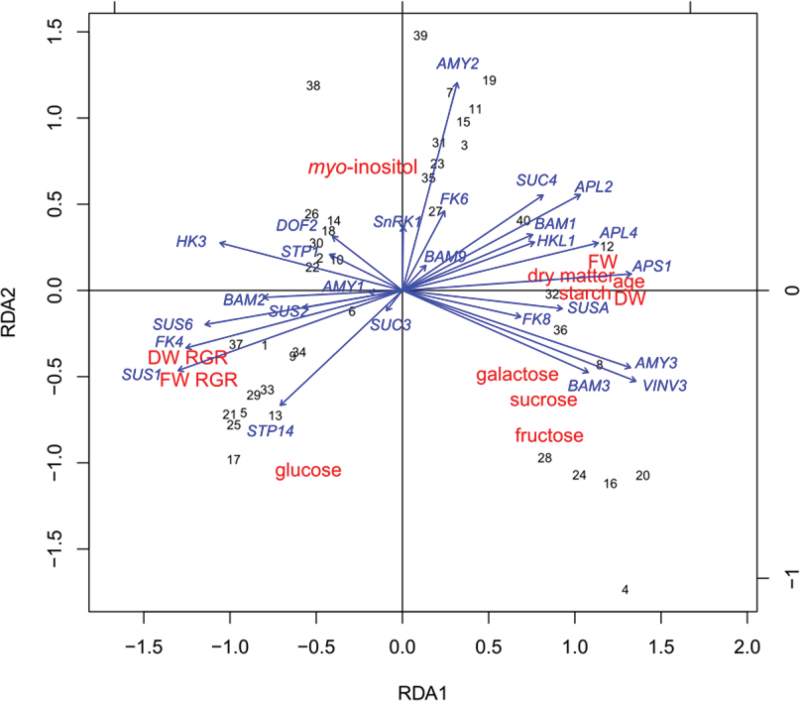
To test if the results of this study were applicable to a larger number of genotypes, the existing data sets for fruit development and carbohydrate composition (Nardozza et al., 2010) were re-analysed by RDA together with transcript abundance (Supplementary Fig. S3 at JXB online) measured for 10 genotypes with contrasting starch concentrations (Fig. 5). Starch, DM, and DW were positively correlated with dimension1 (RDA1), whereas FW RGR and DW RGR were negatively correlated with RDA1. APL4 and APS1 transcripts were connected to starch and DM, as they were largely explained by RDA1. SUS1 was connected to both FW RGR and DW RGR, whereas SUSA was connected to starch. FK4 was connected to SUS1 and again to both FW RGR and DW RGR.
Fig. 5.
Redundancy analysis of physical measurements, growth rates, metabolites, and transcript levels obtained from kiwifruit genotypes. Fruit samples used in this analysis were collected in 2005 and in 2007 (Supplementary Table S1 at JXB online) and were representative of all fruit tissue types. Gene codes: α-amylase (AMY1, AMY2, AMY3), ADP-glucose pyrophosphorylase (APL2, APL4, APS1), β-amylase (BAM1, BAM2, BAM3, BAM9), DOF zinc finger (DOF2), fructokinase (FK4, FK6, FK8), hexokinase (HK3, HKL1), sucrose non-fermenting kinase (SnRK1), monosaccharide transporter (STP1, STP14), sucrose transporter (SUC3, SUC4), sucrose synthase (SUS1, SUS6, SUSA), vacuolar invertase (INV3). Other abbreviations: DW, dry weight; FW, fresh weight; RGR, relative growth rate. Numbers represent each genotype/sampling date combination (Supplementary Table S2 at JXB online). Colour coding: red, physical measurements and metabolites; blue, gene transcript abundance; black, sample number.
Discussion
Kiwifruit metabolism is different from that of the other fruiting species examined to date as carbon is mainly stored as starch. Here it is shown that NI activity is likely to contribute to the rise in Glc concentration associated with cell division, while BAM9/AMY2 transcripts and SPS/SUSY activities are likely to be involved in starch turnover and Suc cycling, respectively, throughout fruit development. Novel compounds in kiwifruit berries, including planteose and Gol, were also identified. Gol, a minor metabolite associated with high-starch genotypes, is possibly involved in planteose metabolism. The innovative approach of comparing kiwifruit genotypes contrasting in fruit size and starch concentration allowed the identification of AGPase as a critical metabolic step that contributed to enhanced starch accumulation in high-starch genotypes. Therefore, the integration of metabolite, enzyme, and transcript data highlights novel aspects of carbon metabolism in kiwifruit that may be transferable to other starch-accumulating fruit.
Cell division and Glc accumulation
Kiwifruit show important developmental differences from other well studied fruits. At fruit set, the fruit had significant levels of starch and Suc which declined during the first 20–30 DAA. Incoming Suc, together with Suc and starch already present in the fruit at anthesis, are likely to support both cell division and accumulation of Glc. Kiwifruit cell division is characterized by a peak in Glc (Klages et al., 1998; Nardozza et al., 2010) not observed in other fruit as tomato maximize their starch accumulation (Baxter et al., 2005; Nardozza et al., 2010) and peach and melon slowly accumulate soluble carbohydrates during this phase (Dai et al., 2011; Lombardo et al., 2011). In this study, NI activity increased in parallel to the increase in Glc concentration during cell division, presumably to metabolize Suc unloaded during this period. In contrast, hydrolysis of Suc by invertase in other fruit usually occurs during rapid cell expansion, when hexoses double their osmotic contribution (Ruan and Patrick, 1995). The high FK/HK enzyme activities and FK4/HK3 transcripts early in fruit development were probably involved with the phosphorylation of Fru and Glc released by NI.
BAM9 (cytosolic) also peaked at cell division and was highly transcribed (Fig. 3AA, AB). In addition AMY2 (cytosolic) transcripts was present throughout fruit development (Fig. 4K, L). It is suggested that these amylases are involved in cytosolic starch or carbon turnover during fruit development. In Arabidopsis leaves, an involvement of BAM9 and AMY2 in the degradation of unknown cytosolic glucans was proposed (Smith et al., 2004). BAM9 may also interact with heteroglycans recently demonstrated to participate in starch turnover in potato (Solanum tuberosum L.) and Arabidopsis (Fettke et al., 2009).
Cell expansion and starch accumulation
From ~50 DAA as the fruit cells expanded, kiwifruit accumulated DM predominantly as starch (Boldingh et al., 2000; Nardozza et al., 2010), unlike tomato, where soluble sugars accumulated (Baxter et al., 2005; Nardozza et al., 2010). During this period, characterized by a decrease in Glc concentration and NI activity, starch, Suc, and SPS activity increased. The drop in Glc, a feature not apparent in other fruits, together with the decrease in GABA and Tre, may be signals for cell expansion and starch accumulation. Although most of the soluble metabolites and enzymes are generally diluted by growth (Figs 1, 2), malate and citrate increased significantly, confirming their involvement in the osmotic regulation of fruit expansion (Nardozza et al., 2010). APL4 transcripts coding for an AGPase large subunit did not increase as starch accumulation increased, suggesting that sufficient enzyme, synthesized perhaps in response to the Glc peak, was made available earlier to support full starch accumulation.
At harvest (154 DAA), kiwifruit are considered ‘mature’ but unripe (MacRae et al., 1989, 1992). A number of metabolic changes were associated with fruit approaching maturity: starch concentrations reached their maximum or plateaued (Supplementary Fig. S1E at JXB online), Suc and hexoses started to accumulate (Fig. 1A), and SPS activity (Fig. 2C, D) and AMY3 transcripts (Fig. 3W, X; Supplementary Fig. S3 at JXB online) increased significantly. As AMY3 is the only plastidial α-amylase (Streb and Zeeman, 2012), its increased transcription could indicate that fruit are mature with increased cycling between sugars in the cytosol and starch in the plastid, or, alternatively, the readiness of the fruit to use its starch to progress ripening. Unexpectedly, BAM1 transcripts, a plastidial β-amylase critical in leaf starch degradation (Streb and Zeeman, 2012), did not increase along with AMY3. However, BAM3 (also plastidial) could be a candidate for starch degradation in heterotrophic tissues as its transcripts significantly increased by 154 DAA (Supplementary Fig. S3 at JXB online) and were strongly up-regulated in ripening kiwifruit (Atkinson et al., 2011; Richardson et al., 2011).
While NI and AI activity and Glc increased at maturity, SUSY activity and SUS1 transcripts declined during starch accumulation and remained low in mature fruit, suggesting a more critical role for invertase in kiwifruit metabolism. In contrast, SUSA increased with the rise in starch, Suc, and SPS activity, becoming the dominant SUSY in mature fruit, as previously reported in kiwifruit (Richardson et al., 2004), melon (Dai et al., 2011), and citrus (Komatsu et al., 2002). These data, together with the strong positive correlation between SUSA, starch, and DM in the RDA (Fig. 5), suggest the involvement of SUSA in Suc cycling in the fruit cell rather than fruit Suc unloading.
Correlations between metabolites, enzyme activity, and transcripts
Metabolism profiling of A. deliciosa fruit showing extremes of starch accumulation revealed correlations between metabolites, enzyme activity, and transcripts. The positive and significant correlation between SPS activity and Suc (Fig. 4) suggests a role for SPS in Suc cycling. SPSA transcripts were also correlated to Suc, but only in high-starch genotypes (Supplementary Fig. S7A at JXB online). There have been various reports of correlations between SPS enzyme activity and Suc (Dai et al., 2011), or transcript levels and Suc (Xue et al., 2013). Interestingly, the Arabidopsis SPSA1 null mutant also demonstrated strong correlations between starch, Suc, and enzyme/transcript levels (Sun et al., 2011).
Metabolite to transcript or enzyme to transcript correlations (Supplementary Figs S7, S8 at JXB online) were generally infrequent, and no correlations between transcript levels and their respective enzyme activities were observed. Previous studies of metabolism profiling in other fruit or systems (Gibon et al., 2004; Steinhauser et al., 2010; Dai et al., 2011; Lombardo et al., 2011) have shown that metabolites, enzyme activities, and transcripts are often interconnected but not in a simple way. For example, a transcriptional response does not necessarily lead to a functional response so metabolites often appear more coordinated than transcripts. In particular, transcripts do not usually correlate to functionally related enzymes or metabolites (Gibon et al., 2004; Piques et al., 2009; Steinhauser et al., 2010; Dai et al., 2011); as enzyme assays do not distinguish between isoforms of multigene families, enzyme turnover is slower than transcript turnover or an enzyme might be modified post-translationally (Gibon et al., 2004; Carrari et al., 2006; Piques et al., 2009; Steinhauser et al., 2010; Dai et al., 2011).
Sugar unloading in the fruit
The present data suggest that kiwifruit phloem unloading may be symplastic throughout fruit development as cytosolic enzymes, NI and SUSY (Fig. 2A, B, G, H), were the dominant Suc-cleaving enzymes. The presence of symplasmic connectivity between phloem cells and vascular parenchyma cells (Gould et al., 2013) and the lack of both cell wall invertase activity (Moscatello et al., 2011) and the gene for a fruit-specific cell wall invertase (this study) in kiwifruit berries support this hypothesis. Tomato phloem unloading is still debated, and evidence supporting two contrasting theories has been produced using different systems. The introgression of a cell wall invertase exotic allele (LIN5) into cultivated tomato showed its critical role for Suc apoplastic unloading (Fridman et al., 2004; Zanor et al., 2009) and fruit starch accumulation (Baxter et al., 2005) early in fruit development. Conversely, elevated SUSY activity at the time of high Suc unloading was associated with Suc symplastic unloading early in fruit development (Nguyen-Quoc et al., 1999; Nguyen-Quoc and Foyer, 2001). Similarly to kiwifruit, transcript and biochemical studies also failed to detect the LIN5 homologue fruit cell wall invertase in melon and peach fruit (Dai et al., 2011; Lombardo et al., 2011).
The trisaccharide planteose, a raffinose oligosaccharide (RFO) family sugar, was found by GC-TOF-MS (Fig. 1A) and quantified in trace amounts by IC in kiwifruit flesh. Planteose was first identified in A. arguta as a short-term storage form of Suc in leaves and as a transport sugar in phloem exudates (Klages et al., 2004), but it has not been previously reported in fruit. myo-Inositol, a common Actinidia fruit sugar-alcohol, is a precursor for the synthesis of raffinose: first UDP-Gal and myo-inositol form Gol, then Suc and Gol form raffinose (Sprenger and Keller, 2000). In this study, both myo-inositol and Gol were present in A. deliciosa fruit, as well as raffinose and planteose (Fig 1A, B). Following a possible symplastic transfer of planteose to the fruit (Klages et al., 2004), planteose is broken down to the trace levels measured herein, thereby releasing Gal which may return to storage as Gol or enter the Glc pool, possibly in a similar way to raffinose (Dai et al., 2011). So far kiwifruit is the only fleshy fruit species where planteose has been identified, and planteose metabolism represents a novel research area as neither its synthetic nor its degradation pathway within kiwifruit is known, and its phloem loading and unloading mechanisms have not been characterized. Finally, a link between RFO sugar translocation and the lack of cell wall invertase is suggested, as these two conditions also characterize melon fruit (Schaffer et al., 1996; Dai et al., 2011).
What are the causes of variation in starch concentration?
Comparison of A. deliciosa high-starch genotypes (G3 and G30) with low-starch genotypes (G25 and G17) during fruit development shows that higher rates of starch accumulation are consistently associated with higher final starch concentrations, despite differences in fruit size. The 2- to 5-fold higher AGPase activity found in high-starch genotypes, which was also maintained for a longer period of time, was a determinant of high starch concentration (Fig. 2S, T). High-starch kiwifruit genotypes also had at least 2-fold higher levels of APL4 transcripts (Fig. 3O, P). AGPase is the first committed step in the starch synthesis pathway (Preiss et al., 1987), synthesizing ADPGlc from the precursor Glc1P—originating in the cytosol from cytoplasmic sugar metabolism. The AGPase involvement in starch metabolism was also confirmed in a wider selection of genotypes by the correlation between the AGPase isoforms (APL4 and APS1) and starch/DM (Fig. 5). In near-isogenic lines of tomato, the high-starch phenotype was linked to a temporal extension of transcription of an AGPase large subunit gene (AgpL) that also conferred higher AGPase activity to the high-starch tomato line (Petreikov et al., 2006). AGPase may therefore be the enzyme contributing to the starch concentration variations observed in outer pericarp small cells of these A. deliciosa genotypes (Nardozza et al., 2011b ).
SPS activity was also significantly higher in high-starch genotypes throughout fruit development (Fig. 2C, D), and the correlation coefficient suggested that the SPS contribution in starch synthesis was substantial (0.50). SPS has been associated with increased starch in other plants. Kerr et al. (1984) observed positive correlations between SPS activity and DM accumulation in soybean [Glycine max (L.) Merr.], whereas the overexpression of a maize SPS gene in tomato fruit increased starch accumulation (Nguyen-Quoc et al., 1999).
Gol curves followed a starch-like pattern and its concentration was higher in high-starch genotypes compared with low-starch genotypes (Fig. 1B). Lombardo et al. (2011) suggested that peach mesocarp Gol levels were involved in seed formation. In kiwifruit, the high outer pericarp Gol levels of high-starch genotypes could be associated with the higher numbers of seeds present in the inner pericarp of these genotypes (Nardozza et al., 2010).
Conclusion
Metabolic profiling in kiwifruit, a starch-storing fruit, has shown that A. deliciosa is a metabolic variant compared with other well studied fruit. This study identifies some features leading to high-starch kiwifruit and already evident at fruit set, for example AGPase and SPS activities, and Suc and starch concentrations. Kiwifruit accumulates Glc at cell division and its decline coincided with a loss of NI and the accumulation of starch, citrate, and malate. APL4 transcription and AGPase activity, together with SPS activity, have a role in the level of starch accumulation, as evidenced by the contrast between high- and low-starch-accumulating genotypes. This study is relevant for the genetic improvement of quality and flavour in kiwifruit as achieving high starch concentration, hence high DM, is important for consumer acceptance (Harker et al., 2009; Nardozza et al., 2011a ), while planteose and Gol metabolism in kiwifruit represent novel research areas.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Changes in starch, physical measurements, and ion chromatography quantified soluble sugars and the sugar-alcohol myo-inositol in outer pericarp during fruit development of Actinidia deliciosa genotypes (2009 samples).
Figure S2. Kiwifruit soluble enzyme activities associated with amino acid metabolism (2009 samples).
Figure S3. Heat map of fruit developmental transcript variations in 10 Actinidia deliciosa genotypes for a selection of carbohydrate metabolism genes (2007 samples).
Figure S4. Metabolite correlations in fruit outer pericarp of Actinidia deliciosa genotypes (2009 samples).
Figure S5. Enzyme activity correlations in fruit outer pericarp of Actinidia deliciosa genotypes (2009 samples).
Figure S6. Transcript correlations in fruit outer pericarp of Actinidia deliciosa genotypes (2009 samples).
Figure S7. Hierarchical cluster analysis of metabolite to transcript correlations in fruit outer pericarp of Actinidia deliciosa genotypes (2009 samples).
Figure S8. Transcript to enzyme correlations in fruit outer pericarp of Actinidia deliciosa genotypes (2009 samples).
Table S1. List of Actinidia deliciosa fruit sample types involved in the present study.
Table S2. List of Actinidia deliciosa fruit samples included in the redundancy analysis.
Table S3. Actinidia deliciosa fruit developmental stages sampled in 2009 harvest year, with relative BBCH stage and description.
Table S4. Enzyme extraction buffer composition.
Table S5. Dilution of extraction buffer to tissue fresh weight used for each kiwifruit enzymatic assay.
Table S6. Methods adopted for enzyme activity assays.
Table S7. List of primers used for transcript abundance analysis, with respective GenBank identification.
Table S8. Effects on correlation analyses of adjustment of critical significance thresholds for Pearson’s correlation coefficients to control the false discovery rate.
Acknowledgements
The authors are grateful to John McCallum and William Laing for their critical review of the manuscript, to Gail Timmerman-Vaughan for statistical advice, and to Minna Pesonen for digital art formatting. This work was supported by the New Zealand Foundation for Research, Science and Technology [Contract No. C06X0706], the New Zealand Ministry of Science and Innovation, and by the Max Planck Society. SN was supported by a post-doctoral fellowship from the Agricultural and Marketing Research and Development Trust [Contract No. 22817].
Glossary
Abbreviations:
- AGPase
ADP-glucose pyrophosphorylase
- AI
acid invertase
- AST
aspartate aminotransfarase
- DAA
days after anthesis
- DM
dry matter
- DW
dry weight
- FK
fructokinase
- Fru6P
fructose 6-phosphate
- FW
fresh weight
- G3
genotype 3
- G17
genotype 17
- G25
genotype 25
- G30
genotype 30
- GABA
γ-aminobutyric acid
- GC-TOF-MS
gas chromatography-time of flight-mass spectrometry
- Glc6P
glucose 6-phosphate
- GLDH
glutamate dehydrogenase
- Gol
galactinol
- HK
hexokinase
- NI
neutral invertase
- PFR
Plant & Food Research
- PGI
phosphoglucoisomerase
- PGM
phosphoglucomutase
- RDA
redundancy analysis
- RGR
relative growth rate
- SDH
shikimate dehydrogenase
- SPS
sucrose phosphate synthase
- SUSY
sucrose synthase
- TCA
tricarboxylic acid
- Tre
trehalose
- UGPase
UDP-glucose pyrophosphorylase.
References
- Atkinson RG, Gunaseelan K, Wang MY, et al. 2011. Dissecting the role of climacteric ethylene in kiwifruit (Actinidia chinensis) ripening using a 1-aminocyclopropane-1-carboxylic acid oxidase knockdown line. Journal of Experimental Botany 62, 3821–3835 [DOI] [PubMed] [Google Scholar]
- Baxter CJ, Carrari F, Bauke A, Overy S, Hill SA, Quick PW, Fernie AR, Sweetlove LJ. 2005. Fruit carbohydrate metabolism in an introgression line of tomato with increased fruit soluble solids. Plant and Cell Physiology 46, 425–437 [DOI] [PubMed] [Google Scholar]
- Boldingh H, Smith GS, Klages K. 2000. Seasonal concentrations of non-structural carbohydrates of five Actinidia species in fruit, leaf and fine root tissue. Annals of Botany 85, 469–476 [Google Scholar]
- Bulley SM, Rassam M, Hoser D, Otto W, Schünemann N, Wright M, MacRae E, Gleave A, Laing W. 2009. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-l-galactose guanyltransferase is a major control point of vitamin C biosynthesis. Journal of Experimental Botany 60, 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, et al. 2006. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiology 142, 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116 [Google Scholar]
- Crowhurst RN, Gleave AP, MacRae EA, et al. 2008. Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC Genomics 9, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Cohen S, Portnoy V, et al. 2011. Metabolism of soluble sugars in developing melon fruit: a global transcriptional view of the metabolic transition to sucrose accumulation. Plant Molecular Biology 76, 1–18 [DOI] [PubMed] [Google Scholar]
- Dai ZW, Leon C, Feil R, Lunn JE, Delrot S, Gomes E. 2013. Metabolic profiling reveals coordinated switches in primary carbohydrate metabolism in grape berry (Vitis vinifera L.), a non-climacteric fleshy fruit. Journal of Experimental Botany 64, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc L, Grimplet J, Wheatley M, et al. 2007. Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971 [DOI] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. 2008. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiology 148, 730–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AR, Ferguson LR. 2003. Are kiwifruit really good for you? Acta Horticulturae 610, 131–138 [Google Scholar]
- Fettke J, Malinova I, Eckermann N, Steup M. 2009. Cytosolic heteroglycans in photoautotrophic and in heterotrophic plant cells. Phytochemistry 70, 696–702 [DOI] [PubMed] [Google Scholar]
- Fridman E, Carrari F, Liu YS, Fernie AR, Zamir D. 2004. Zooming in on a quantitative trait for tomato yield using interspecific introgressions. Science 305, 1786–1789 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Blaesing OE, Hannemann J, et al. 2004. A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. The Plant Cell 16, 3304–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould N, Morrison DR, Clearwater MJ, Ong S, Boldingh HL, Minchin PEH. 2013. Elucidating the sugar import pathway into developing kiwifruit berries (Actinidia deliciosa). New Zealand Journal of Crop and Horticultural Science (in press). [Google Scholar]
- Harker FR, Carr BT, Lenjo M, et al. 2009. Consumer liking for kiwifruit flavour: a meta-analysis of five studies on fruit quality. Food Quality and Preference 20, 30–41 [Google Scholar]
- Hopping ME. 1976. Effect of exogenous auxins, gibberellins, and cytokinins on fruit development in Chinese gooseberry (Actinidia chinensis Planch.). New Zealand Journal of Botany 14, 69–75 [Google Scholar]
- Kerr PS, Huber SC, Israel DW. 1984. Effect of N-source on soybean leaf sucrose phosphate synthase, starch formation, and whole plant-growth. Plant Physiology 75, 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages KU, Boldingh HL, Cooney JM, MacRae EA. 2004. Planteose is a short-term storage carbohydrate in Actinidia leaves. Functional Plant Biology 31, 1205–1214 [DOI] [PubMed] [Google Scholar]
- Klages K, Donnison H, Boldingh H, MacRae E. 1998. myo-Inositol is the major sugar in Actinidia arguta during early fruit development. Australian Journal of Plant Physiology 25, 61–67 [Google Scholar]
- Komatsu A, Moriguchi T, Koyama K, Omura M, Akihama T. 2002. Analysis of sucrose synthase genes in citrus suggests different roles and phylogenetic relationships. Journal of Experimental Botany 53, 61–71 [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, et al. 2005. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21, 1635–1638 [DOI] [PubMed] [Google Scholar]
- Li MJ, Feng FJ, Cheng LL. 2012. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One 7, e33055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. 2006. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nature Protocols 1, 387–396 [DOI] [PubMed] [Google Scholar]
- Lombardo VA, Osorio S, Borsani J, et al. 2011. Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiology 157, 1696–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae EA, Bowen JH, Stec MGH. 1989. Maturation of kiwifruit (Actinidia deliciosa cv Hayward) from 2 orchards: differences in composition of the tissue zones. Journal of the Science of Food and Agriculture 47, 401–416 [Google Scholar]
- MacRae E, Quick WP, Benker C, Stitt M. 1992. Carbohydrate metabolism during postharvest ripening in kiwifruit. Planta 188, 314–323 [DOI] [PubMed] [Google Scholar]
- Moscatello S, Famiani F, Proietti S, Farinelli D, Battistelli A. 2011. Sucrose synthase dominates carbohydrate metabolism and relative growth rate in growing kiwifruit (Actinidia deliciosa, cv Hayward). Scientia Horticulturae 128, 197–205 [Google Scholar]
- Nardozza S, Boldingh HL, Richardson AC, Costa G, Marsh H, MacRae EA, Clearwater MJ. 2010. Variation in carbon content and size in developing fruit of Actinidia deliciosa genotypes. Functional Plant Biology 37, 545–554 [DOI] [PubMed] [Google Scholar]
- Nardozza S, Gamble J, Axten LG, Wohlers MW, Clearwater MJ, Feng J, Harker FR. 2011a. Dry matter content and fruit size affect flavour and texture of novel Actinidia deliciosa genotypes. Journal of the Science of Food and Agriculture 91, 742–748 [DOI] [PubMed] [Google Scholar]
- Nardozza S, Hallett IC, McCartney R, Richardson AC, MacRae EA, Costa G, Clearwater MJ. 2011b. Is fruit anatomy involved in variation in fruit starch concentration between Actinidia deliciosa genotypes? Functional Plant Biology 38, 63–74 [DOI] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Foyer CH. 2001. A role for ‘futile cycles’ involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. Journal of Experimental Botany 52, 881–889 [DOI] [PubMed] [Google Scholar]
- Nguyen-Quoc B, N’Tchobo H, Foyer CH, Yelle S. 1999. Overexpression of sucrose phosphate synthase increases sucrose unloading in transformed tomato fruit. Journal of Experimental Botany 50, 785–791 [Google Scholar]
- Nieuwenhuizen NJ, Wang MY, Matich AJ, et al. 2009. Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). Journal of Experimental Botany 60, 3203–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreikov M, Shen S, Yeselson Y, Levin I, Bar M, Schaffer AA. 2006. Temporally extended gene expression of the ADP-Glc pyrophosphorylase large subunit (AgpL1) leads to increased enzyme activity in developing tomato fruit. Planta 224, 1465–1479 [DOI] [PubMed] [Google Scholar]
- Piques M, Schulze WX, Höhne M, Usadel B, Gibon Y, Rohwer J, Stitt M. 2009. Ribosome and transcript copy numbers, polysome occupancy and enzyme dynamics in Arabidopsis . Molecular Systems Biology 5, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J, Bloom M, Morell MK, Knowles VI, Plaxton WC, Okita TW, Larsen R, Harmon AC, Putnam-Evans C. 1987. Regulation of starch synthesis: enzymological and genetic studies. In: Bruening G, Harada J, Kosuge T, Hollaender A, eds. Tailoring genes for crop improvement . New York: Plenum Publishing Corp., 133–152 [DOI] [PubMed] [Google Scholar]
- Ren G, Healy RA, Horner HT, Martha GJ, Thornburg RW. 2007. Expression of starch metabolic genes in the developing nectaries of ornamental tobacco plants. Plant Science 173, 621–637 [Google Scholar]
- Richardson A, Boldingh H, McAtee P, Gunaseelan K, Luo Z, Atkinson R, David K, Burdon J, Schaffer R. 2011. Fruit development of the diploid kiwifruit, Actinidia chinensis ‘Hort16A’. BMC Plant Biology 11, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AC, Marsh KB, Boldingh HL, et al. 2004. High growing temperatures reduce fruit carbohydrate and vitamin C in kiwifruit. Plant, Cell and Environment 27, 423–435 [Google Scholar]
- Richardson AC, McAneney KJ, Dawson TE. 1997. Carbohydrate dynamics in kiwifruit. Journal of Horticultural Science 72, 907–917 [Google Scholar]
- Roessner U, Willmitzer L, Fernie AR. 2001. High-resolution metabolic phenotyping of genetically and environmentally diverse potato tuber systems. Identification of phenocopies. Plant Physiology 127, 749–764 [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. 2003. Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiology 133, 84–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology 132, 365–386 [DOI] [PubMed] [Google Scholar]
- Ruan Y-L, Patrick JW. 1995. The cellular pathway of postphloem sugar transport in developing tomato fruit. Planta 196, 434–444 [Google Scholar]
- Saeed A, Sharov V, White J, et al. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Pharr DM, Madore MA. 1996. Cucurbits. In: Zamski E, Schaffer AA, eds. Photoassimilate distribution in plants and crops: source–sink relationships . New York: Marcel Dekker, 729–757 [Google Scholar]
- Schauer N, Steinhauser D, Strelkov S, et al. 2005. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Letters 579, 1332–1337 [DOI] [PubMed] [Google Scholar]
- Smith GS, Clark CJ, Boldingh HL. 1992. Seasonal accumulation of starch by components of the kiwifruit vine. Annals of Botany 70, 19–25 [Google Scholar]
- Smith SM, Fulton DC, Chia T, Thorneycroft D, Chapple A, Dunstan H, Hylton C, Zeeman SC, Smith AM. 2004. Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiology 136, 2687–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N, Keller F. 2000. Allocation of raffinose family oligosaccharides to transport and storage pools in Ajuga reptans: the roles of two distinct galactinol synthases. The Plant Journal 21, 249–258 [DOI] [PubMed] [Google Scholar]
- Steinhauser M-C, Steinhauser D, Koehl K, Carrari F, Gibon Y, Fernie AR, Stitt M. 2010. Enzyme activity profiles during fruit development in tomato cultivars and Solanum pennellii . Plant Physiology 153, 80–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D, Love W. 1968. A general canonical correlation index. Psychological Bulletin 70, 160–163 [DOI] [PubMed] [Google Scholar]
- Streb S, Zeeman SC. 2012. Starch metabolism in Arabidopsis . The Arabidopsis Book , e0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JD, Zhang JS, Larue CT, Huber SC. 2011. Decrease in leaf sucrose synthesis leads to increased leaf starch turnover and decreased RuBP regeneration-limited photosynthesis but not Rubisco-limited photosynthesis in Arabidopsis null mutants of SPSA1 . Plant, Cell and Environment 34, 592–604 [DOI] [PubMed] [Google Scholar]
- Trought MCT, Bennett JS, Boldingh HL. 2011. Influence of retained cane number and pruning time on grapevine yield components, fruit composition and vine phenology of Sauvignon Blanc vines. Australian Journal of Grape and Wine Research 17, 258–262 [Google Scholar]
- Wegrzyn T, MacRae E. 1995. Alpha-amylase and starch degradation in kiwifruit. Journal of Plant Physiology 147, 19–28 [Google Scholar]
- Xue G-P, Drenth J, Glassop D, Kooiker M, McIntyre CL. 2013. Dissecting the molecular basis of the contribution of source strength to high fructan accumulation in wheat. Plant Molecular Biology 81, 71–92 [DOI] [PubMed] [Google Scholar]
- Zanor MI, Osorio S, Nunes-Nesi A, et al. 2009. RNA interference of LIN5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiology 150, 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuur AK, Ieno EN, Smith GM. 2007. Principal component analysis and redundancy analysis. In: Zuur AK, Ieno EN, Smith GM, eds. Analysing ecological data . New York: Springer, 193–224 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.