Abstract
OsWRKY76 encodes a group IIa WRKY transcription factor of rice. The expression of OsWRKY76 was induced within 48h after inoculation with rice blast fungus (Magnaporthe oryzae), and by wounding, low temperature, benzothiadiazole, and abscisic acid. Green fluorescent protein-fused OsWRKY76 localized to the nuclei in rice epidermal cells. OsWRKY76 showed sequence-specific DNA binding to the W-box element in vitro and exhibited W-box-mediated transcriptional repressor activity in cultured rice cells. Overexpression of OsWRKY76 in rice plants resulted in drastically increased susceptibility to M. oryzae, but improved tolerance to cold stress. Microarray analysis revealed that overexpression of OsWRKY76 suppresses the induction of a specific set of PR genes and of genes involved in phytoalexin synthesis after inoculation with blast fungus, consistent with the observation that the levels of phytoalexins in the transgenic rice plants remained significantly lower than those in non-transformed control plants. Furthermore, overexpression of OsWRKY76 led to the increased expression of abiotic stress-associated genes such as peroxidase and lipid metabolism genes. These results strongly suggest that OsWRKY76 plays dual and opposing roles in blast disease resistance and cold tolerance.
Key words: Blast disease resistance, cold stress, phytoalexin, rice, transcriptional repressor, WRKY.
Introduction
Plant growth is greatly affected by various types of stresses such as pathogen attacks, insect herbivory, and abiotic environmental stresses including high salinity, drought, and excessive temperature. In order to maintain growth and productivity, plants have developed adaptive responses to these stresses. These responses are triggered by environmental cues, and the transduction of these stress signals results in the regulation of a large number of stress-associated genes (Singh et al., 2002). Biotic and abiotic stress signalling pathways have been considered to form a complicated network of synergistic and antagonistic interactions (Fujita et al., 2006; Sharma et al., 2013). A plant hormone, abscisic acid (ABA), is involved in responses to abiotic stresses such as drought, low temperature, and high salinity stress (Yamaguchi-Shinozaki and Shinozaki, 2006). Salicylic acid (SA) plays a positive role in resistance against biotrophic and hemibiotrophic pathogens (Robert-Seilaniantz et al., 2007; Thaler et al., 2012). Other plant hormones, such as jasmonic acid (JA), ethylene, and brassinosteroid, also participate in the processes of biotic and abiotic stress responses (Mauch-Mani and Mauch, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006).
Rice blast fungus (Magnaporthe oryzae, Oryzae isolate) is a hemibiotrophic pathogen that causes blast disease in rice, which accounts for major losses in the global yield of rice (Talbot, 2003). In rice, SA signalling positively regulates blast disease resistance; application of benzothiadiazole (BTH), a chemical activator of SA signalling, induces resistance in rice against M. oryzae (Schweizer et al., 1999). On the other hand, ABA interacts antagonistically with the SA signalling pathway in the interaction between rice and M. oryzae (Jiang et al., 2010), which might cause low temperature-induced blast disease susceptibility (Koga et al., 2004). A wide range of transcriptional alterations of gene expression occurs when the rice interacts with the fungus. Previous studies have identified several families of transcription factors (TFs) that are involved in responses to infections by blast fungus. They are ERF/AP2 (Liu et al., 2012), NAC (Nakashima et al., 2006), bHLH (Kim et al., 2012), and WRKY (Pandey and Somssich, 2009).
WRKY TFs are plant-specific families of zinc finger transcription factors that are characterized by a conserved DNA-binding WRKY domain. Most WRKY TFs bind to a consensus cis-element termed the W-box (TTGACT/C), which is found in the promoters of many defence-associated genes, and regulates their transcription (Eulgem et al., 2000; Rushton et al., 2010). In rice, >100 WRKY genes have been identified in the genome (Xie et al., 2005; Ross et al., 2007), and at least 20 WRKY genes are transcriptionally regulated in response to M. oryzae inoculation, implying that they are important for the defence response (Ryu et al., 2006; Bagnaresi et al., 2012). In particular, OsWRKY45 has been demonstrated to play a crucial role in mediating BTH-induced resistance to M. oryzae, and Xanthomonas oryzae pv. oryzae, the bacterial causal agent of rice leaf blight (Shimono et al., 2007, 2012). OsWRKY45 has transcriptional activator activity and is involved in the up-regulation of various defence-associated genes. Similarly, other WRKY domain-containing transcriptional activators, including OsWRKY53 (Chujo et al., 2007), OsWRKY31 (Zhang et al., 2008), and OsWRKY30 (Peng et al., 2012), are postulated to regulate positively the defence against M. oryzae. In various plant species, some WRKY transcriptional activators play important roles in disease resistance, whereas some others that have transcriptional repressor activity are also transcriptionally up-regulated by pathogen infection and are considered to play a role as negative regulators of disease resistance. For example, Arabidopsis AtWRKY7 has repressor activity and is negatively involved in resistance to the bacterial pathogen Pseudomonas syringae (Kim et al., 2006). In barley, HvWRKY1 and HvWRKY2, which encode transcriptional repressors, are involved in the down-regulation of basal defence responses triggered by pathogen-associated molecular patterns (Shen et al., 2007).
Recent studies have suggested important roles for WRKYs in abiotic stress responses (Pandey and Somssich, 2009; Rushton et al., 2010). For example, HvWRKY38 is a positive regulator of the drought stress response (Marè et al., 2004; Xiong et al., 2010). In Arabidopsis, three closely related WRKY genes, AtWRKY18, 40, and 60, are involved in both biotic and abiotic stress responses by regulating the signalling of the stress-associated plant hormones SA, JA, and ABA (Xu et al., 2006; Chen et al., 2010; Shang et al., 2010). Another Arabidopsis WRKY gene, AtWRKY25, negatively regulates the SA-mediated defence response to P. syringae (Zheng et al., 2007), but positively regulates responses to high salinity (Jiang and Deyholos, 2009) and high temperature (Li et al., 2011) stresses. Thus, WRKY proteins are probably involved in a variety of stress responses, and their roles apparently vary depending on the stress.
OsWRKY76 encodes a rice WRKY TF of group IIa possessing a single WRKY domain (Eulgem et al., 2000; Xie et al., 2005; Ross et al., 2007). The expression of OsWRKY76 is increased by M. oryzae inoculation and treatment with BTH, suggesting that it is also involved in the response to blast disease (Ryu et al., 2006; Shimono et al., 2007; Bagnaresi et al., 2012). Previous studies demonstrated that the overexpression of OsWRKY76 in rice causes reduced resistance to rice leaf blight disease (Seo et al., 2011). On the basis of an interactome analysis performed using yeast two-hybrid assays, Seo et al. (2011) reported that OsWRKY76 probably regulates cellular responses to both biotic and abiotic stresses. However, little is known about the detailed molecular and physiological functions of OsWRKY76. Here, it is shown that OsWRKY76 is a transcriptional repressor with DNA binding activity to the W-box element, and is localized in the nucleus. The in vivo biological functions of OsWRKY76 in biotic and abiotic stresses were also investigated using transgenic rice plants overexpressing OsWRKY76 (W76-OX).
Materials and methods
Plants and pathogens
Rice plants (Oryza sativa L. cv. Nipponbare) carrying the blast resistance gene Pia [Nipponbare (Pia)] were used in this study. Rice plants were grown in a chamber under a 14h light (28 °C) and 10h dark (24 °C) cycle in hydroponic culture, as previously described in Tanabe et al. (2006). Magnaporthe oryzae isolates Ina86-137 (MAFF 101511, race 007.0) and P91-15B (001.0) were used as virulent and avirulent strains, respectively, for Nipponbare (Pia).
Stress and chemical treatments
Fifteen-day-old rice plants were used to examine the effects of stress and chemical treatment on the expression of OsWRKY76. To examine the effect of rice blast inoculation, conidia were washed and suspended at 1×105 cells ml–1 in sterile water, and sprayed on the plants, which were then incubated at 24 °C in the dark for 24h, followed by 14h light and 10h dark cycles. For wound treatment, the fourth leaf blade was chopped into 1mm long pieces with a knife and incubated on moist filter paper at 24 °C in the dark. Cold treatment was performed by incubation at 4 °C in the dark. Chemical treatment was performed by spraying the plants with 500 μM BTH containing 0.02% (v/v) Tween-20 and 0.5% (v/v) dimethylsulphoxide (DMSO), or with 50 μM ABA containing 0.02% (v/v) Silwet L-77 and 0.1% (v/v) ethanol. The solvents of BTH or ABA were used for the mock control in each case. The fourth leaf blade was used for quantitative reverse transcription–PCT (qRT–PCR) analysis.
Measurement of ion leakage
For measurement of ion leakage, 3cm long leaf segments were excised from the centre of each leaf blade, chopped into 5mm lengths, and incubated in 1ml of distilled water for 1h with moderate shaking. The conductivity was measured using a B-173 conductivity meter (Horiba, Kyoto, Japan) before and after the leaves were autoclaved. Ion leakage was presented as the ratio of conductivity values before and after autoclaving.
Transient expression of green fluorescent protein (GFP)-fused OsWRKY76 in rice cells
The coding region of OsWRKY76 was fused with the synthetic GFP gene (Niwa et al., 1999), and inserted between the XbaI and SacI sites of pBI221 (Clontech, Palo Alto, CA, USA). As a control, the 35S-GFP was used. For the transient expression assay, 1 μg of plasmid DNA was introduced into rice leaf sheath cells using the PDS-1000/He particle delivery system (BioRad, Hercules, CA, USA). As a gene expression marker and subcellular cytosol and nuclear localization marker, the plasmid containing the Discosoma red fluorescent protein (DsRed) gene was co-expressed. After overnight incubation in the dark, leaf sheath cells were observed under a fluorescence microscope.
Gel mobility shift assay
The coding sequence of OsWRKY76 was inserted between the BamHI and HindIII sites of pMALc2x (New England Biolabs, Beverly, MA, USA) and introduced into Escherichia coli JM109. OsWRKY76 fused to maltose-binding protein (MBP) was purified using amylose resin (New England Biolabs), according to the manufacturer’s instructions. Nucleotide sequences of the probes used for DNA-binding assays are as follows: WB (5′-AACTTTGACCAATCTTTCAAGTA-3′) and mWB (5′-AACTTTGAACAATCTTTCAAGTA-3′). The W-box or mutated W-box core sequence are underlined. A gel mobility shift assay was performed using the DIG Gel Shift Kit 2nd generation (Roche Diagnostics GmbH, Mannheim, Germany).
Luciferase gene (LUC) reporter assay
The GUS gene in pBI221 was replaced with the coding sequence of OsWRKY76. The plasmid expressing the GAL4 DNA-binding (DB) domain was used as a control effector. The 23bp W-box-containing oligonucleotide sequence used in the gel mobility shift assay was multimerized five times (5W) to construct 35S-5W-TATA-LUC-NOS (35S-5W-LUC). In co-transfection assays, 2 μg of the reporter plasmid, 2 μg of the effector plasmid, and 0.04 μg of the internal control plasmid (pPTRL) were mixed and introduced into suspension-cultured rice cells (the Oc cell line) by particle bombardment. Transformed rice cells were incubated for 24h at 24 °C in the dark. The luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Luminescence was measured using a TD20/20 luminometer (Turner Designs, Sunnyvale, CA, USA).
RNA isolation and qRT–PCR
Total RNA was extracted using Sepasol RNA I Super (Nacalai Tesque, Kyoto, Japan). First-strand cDNA was synthesized using the PrimeScript™ RT reagent kit (TaKaRa Co., Ltd, Ohtsu, Japan). RT–PCR was performed using 1× SYBR Premix Ex Taq II (TaKaRa). The primer sequences are listed in Supplementary Table S1 available at JXB online. The relative levels of gene expression were quantified using MX3000P (Stratagene, La Jolla, CA, USA). The data were normalized to those of the elongation factor gene eEF-1α (Jain et al., 2006).
Rice transformation
For the overexpression of OsWRKY76 in rice, the maize polyubiquitin promoter (Ubi-1), the full-length cDNA fragment of OsWRKY76 (AK068337), and the NOS terminator were inserted between the HindIII and PacI sites of the binary vector pZH1 (Shimono et al., 2007), and the resultant vector was introduced into rice via Rhizobium radiobacter strain EHA105. Rice transformation was performed as described (Toki et al., 2006).
Fungal inoculation and microscopic observation
To evaluate disease symptoms, the fifth leaf blades were detached from rice plants at the 5.6-leaf stage and placed on moistened filter paper in Petri dishes. Washed conidia suspended at 1×105 ml–1 in sterile water were sprayed on the leaf blades, followed by incubation at 25 °C in the dark for 24h, then under 14h light and 10h dark cycles. Blast disease development was quantified by measurement of M. oryzae genomic DNA (encoding 28S rRNA) relative to rice genomic DNA (encoding the eEF-1α gene) using quantitative genomic PCR analysis (Zellerhoff et al., 2006). The primer sequences are listed in Supplementary Table S1 at JXB online. Data are presented relative to the value in leaves receiving immediate inoculation with conidia, which was taken as 1.
Plant response to the blast fungus at an early stage was observed in leaf sheaths under a microscope. Sheaths of the fifth leaves of rice plants at the 5.6-leaf stage were detached and inoculated with a suspension of conidia (1×105 ml–1), and then incubated at 25 °C in the dark. After fixation in formalin-aceto-alcohol (formaldehyde–acetic acid–ethanol–water, 5:5:45:45, v/v/v/v), the level of infection was evaluated for intact appressoria under a light microscope. The samples were scored as ‘no invasion’, ‘invasion of one cell’, or ‘invasion of two or more cells’, corresponding to appressoria that penetrated into no rice cells beneath the appressorium, one cell, or more than one cell, respectively (Tanabe et al., 2006).
Determination of phytoalexins
The sheaths of the fifth leaves at the 5.6-leaf stage were detached and inoculated with Ina86-137 followed by extraction with 79% (v/v) ethanol containing 14% (v/v) water, 7% (v/v) acetonitrile, and 0.1% (v/v) acetic acid at 96 °C for 20min. Samples were analysed for the simultaneous determination of momilactones, phytocassanes, and sakuranetin using HPLC–mass spectrometry (Shimizu et al., 2008). Phytoalexin levels were determined using combinations of the precursor and product ions (m/z 317/299 for phytocassanes A, D, and E, m/z 335/317 for phytocassane B, m/z 319/301 for phytocassane C, m/z 315/271 for momilactone A, m/z 331/269 for momilactone B, and m/z 287/167 for sakuranetin) in the multiple-reaction monitoring mode.
Microarray analysis
Sheaths of the fifth leaf at the 5.6-leaf stage were used for the microarray experiment. For rice blast fungus inoculation, excised leaf sheaths were inoculated with a compatible strain of M. oryzae (Ina86-137) as described above and incubated at 25 °C in the dark for 36h. Cold treatment was performed by incubation at 4 °C in the dark for 36h. Leaf sheaths incubated at 25 °C in the dark for 36h were used as untreated controls. Total RNA was isolated using the RNeasay Plant Mini Kit (Qiagen, Valencia, CA, USA). Cy3-labelled complementary RNA was prepared from 200ng of total RNA and hybridized to a rice 44K oligo microarray based on the Rice Annotation Project (RAP) according to the manufacturer’s instructions (Agilent Technologies, Palo Alto, CA, USA). The slide images were scanned with a DNA microarray scanner (Agilent) using the manufacture’s Feature Extraction software.
Data were analysed using R/Bioconductor (http://www.bioconductor.org/, last accessed on 23 August 2013). Three independent leaf sheaths were used for each sample as biological replicates. After log2 transformation and global normalization, the genes for which the expression was altered significantly were selected by applying a t-test [one-way analysis of variance (ANOVA) Welch t-test, P = 0.01]. Data were z-score transformed across each gene and presented as a heat map represented by two-dimensional hierarchical clustering.
Results
Up-regulation of OsWRKY76 under both biotic and abiotic stresses and by stress-related chemicals
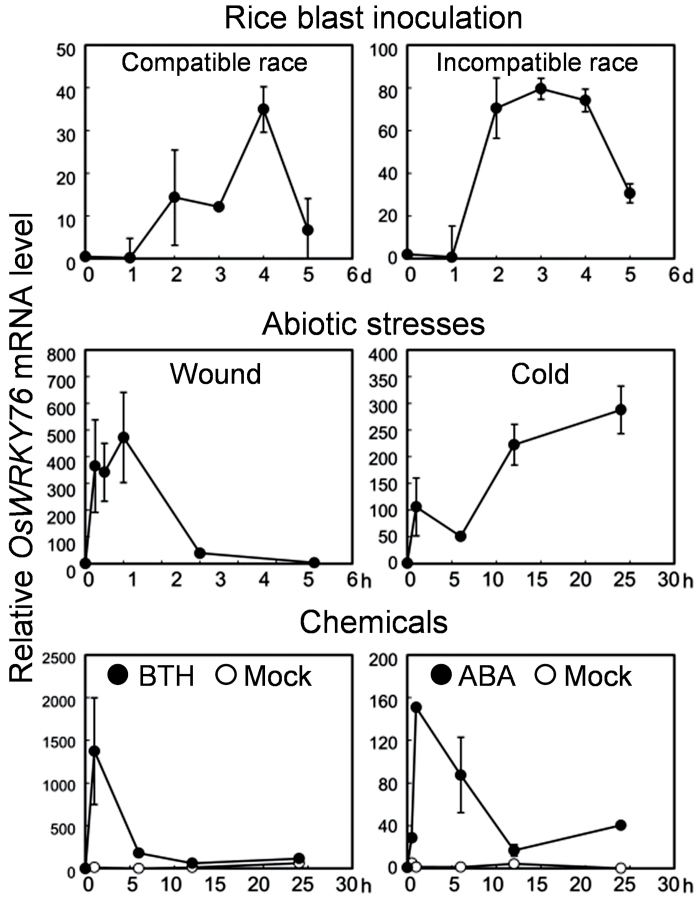
Previous analyses demonstrated that OsWRKY76 is up-regulated by M. oryzae inoculation and treatment with BTH (Ryu et al., 2006; Shimono et al., 2007; Bagnaresi et al., 2012). To characterize the expression pattern of OsWRKY76 further in detail, the effects of a variety of stresses and stress-related chemicals on the expression of OsWRKY76 over time were examined by qRT–PCR analysis (Fig. 1). In untreated or mock-treated rice plants, the expression of OsWRKY76 was nearly undetectable. The expression of OsWRKY76 was induced by both compatible and incompatible strains of M. oryzae at 2 days after inoculation (dai). The levels of transcripts from OsWRKY76 remained high until 4 dai, then decreased at 5 dai. The expression of OsWRKY76 was rapidly induced by wounding and cold treatment. The expression of OsWRKY76 was drastically induced by the application of BTH. ABA treatment also induced the accumulation of OsWRKY76 transcripts within 1h. These results indicate that OsWRKY76 is responsive to both biotic and abiotic stresses.
Fig. 1.
Expression of OsWRKY76 in the response to biotic and abiotic stresses and stress-associated chemicals. OsWRKY76 transcripts in leaf blades were measured by quantitative RT–PCR analysis. Transcription levels are expressed as the ratio to the level of transcript at 0h. Data are represented as mean values ±standard error (SE) value for three replicates.
Nuclear localization of OsWRKY76
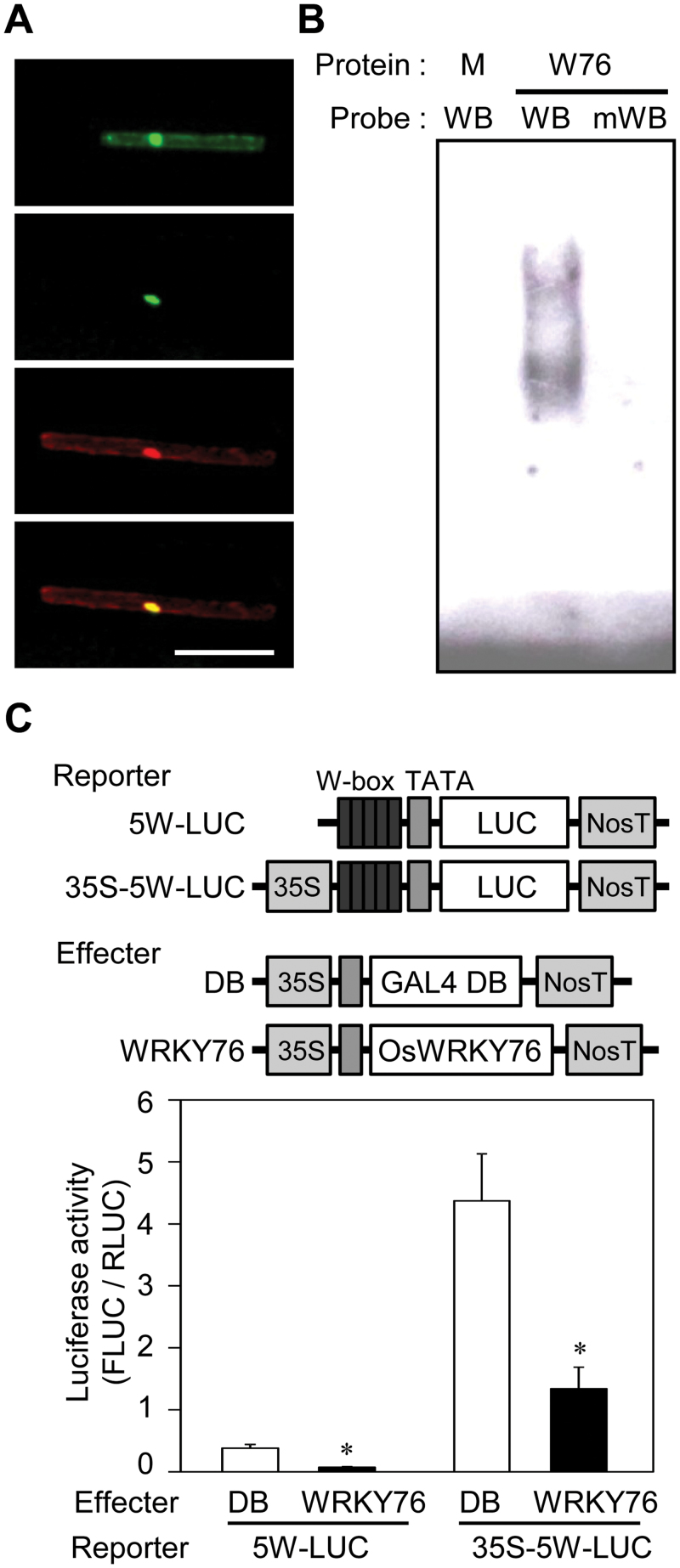
To investigate the subcellular localization of OsWRKY76, expression plasmids carrying GFP, GFP-fused full-length OsWRKY76 (OsWRKY76–GFP), or DsRed under the control of the Cauliflower mosaic virus 35S promoter were co-introduced into rice leaf sheath cells by particle bombardment. As shown in Fig. 2A, GFP and DsRed were detected in the nucleus and cytosol, whereas OsWRKY76–GFP was detected only in the nucleus. Therefore, it is concluded that the OsWRKY76 protein is targeted to the nucleus.
Fig. 2.
W-box-mediated transcriptional repressor activity of OsWRKY76. (A) Nuclear localization of OsWRKY76. Recombinant plasmids were transiently expressed in rice leaf sheath cells via particle bombardment and observed under a microscope. Bar = 30 μm. Top to bottom: fluorescence image of GFP control, OsWRKY76–GFP, co-expressed DsRed control, merged image of OsWRKY76–GFP and DsRed. Bar = 30 μm. (B) Binding of the OsWRKY76 protein to the W-box-containing sequence. Recombinant OsWRKY76 protein fused to MBP (W76) and MBP control (M) were used for the gel mobility shift assay. DIG-labelled W-box core sequence (WB) and mutated WB (mWB) were used as probe. (C) LUC reporter assay of W-box-mediated luciferase activity by OsWRKY76 in cultured rice cells. The data are represented as mean values ±SE for three independent experiments. Significantly lower LUC activity of OsWRKY76 compared with that of the DB control is denoted by asterisks (*P < 0.05 by t-test).
Sequence-specific binding of OsWRKY76 to the W-box element
To determine the sequence-specific DNA-binding activity in vitro, OsWRKY76 was prepared as a fusion protein with MBP in E. coli and tested for its binding activity by a gel mobility shift assay. As shown in Fig. 2B, the MBP-fused OsWRKY76 protein bound to the digoxigenin (DIG)-labelled W-box core sequence (WB), but MBP control did not. Furthermore, MBP–OsWRKY76 did not bind to the mutated WB with a single base substitution. The specific interaction between MBP–OsWRKY76 and WB was effectively blocked by an excess amount of unlabelled WB but not by mutated WB (Supplementary Fig. S1 at JXB online). These results demonstrate that OsWRKY76 binds specifically to the W-box element.
W-box-mediated transcriptional repressor activity of OsWRKY76
The activity of OsWRKY76 as a transcriptional regulator was tested using a chimeric effector/reporter assay (Fig. 2C). The reporter plasmids, 5W-LUC and 35S-5W-LUC, containing the firefly LUC were introduced into cultured rice cells with an effector plasmid carrying either OsWRKY76 or the GAL4 DB domain. The LUC activities of 5W-LUC and 35S-5W-LUC were considerably reduced when co-introduced with 35S-promoter-driven OsWRKY76 compared with 35S-driven GAL4-DB, indicating that OsWRKY76 acts as a transcriptional repressor via binding to the W-box.
Overexpression of OsWRKY76 increases susceptibility to the blast fungus in rice
To characterize the biological function of OsWRKY76, transgenic rice plants constitutively overexpressing OsWRKY76 under the control of the maize ubiquitin promoter were produced. Three independent lines of transgenic plants (W76-OX#01, 03, and 06) grown in a growth chamber accumulated OsWRKY76 transcripts at a >100-fold higher level without morphological changes compared with the parental line (Supplementary Fig. S2 at JXB online), and were fertile.
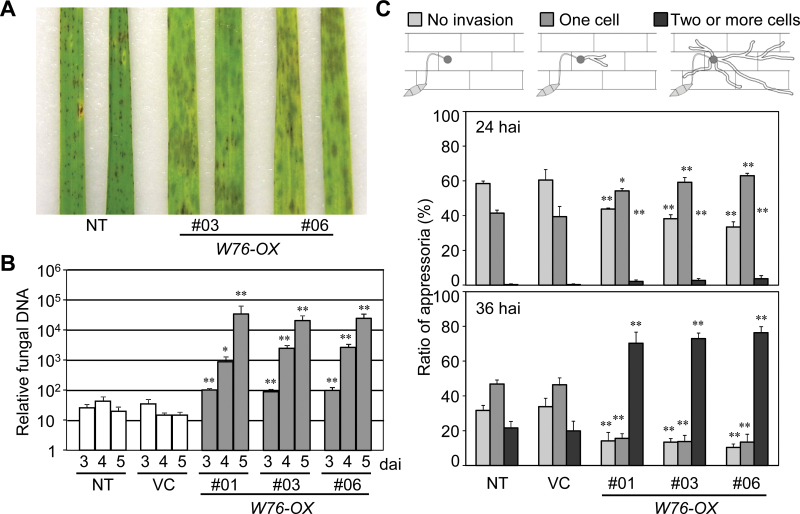
The effect of overexpression of OsWRKY76 on disease resistance to rice blast fungus was examined. Transgenic plants showed more severe symptoms, which often blasted the whole plant, compared with non-transformed control (NT) plants both in the excised leaves (Fig. 3A) and in the intact plants (Supplementary Fig. S3A at JXB online). Observations of cross-sections of the inoculated leaf blades revealed that in NT plants, the spread of mycelia was generally limited to the spaces between motor cells and vascular tissues at 4 dai. In W76-OX plants, however, the mycelia vigorously spread into parenchymatous tissues in addition to motor cells and vascular tissues, causing a collapse of the parenchyma, and hyphae already spread over the leaf surface at 4 dai (Supplementary Fig. S3B). Quantitative genomic PCR analysis demonstrated that >100-fold higher levels of fungal DNA were detected from W76-OX plants compared with NT plants (Fig. 3B). Microscopic observation of the inoculated leaf sheaths demonstrated that W76-OX plants allow penetration and elongation of infectious hyphae at significantly higher frequencies compared with NT plants (Fig. 3C; Supplementary Fig. S3C). In addition, W76-OX plants allowed greater penetration of an incompatible strain of M. oryzae, although the incompatibility was not overcome (Supplementary Fig. S4). These results indicate that the constitutive overexpression of OsWRKY76 suppresses basal resistance to M. oryzae to a large extent.
Fig. 3.
Effects of constitutive overexpression of OsWRKY76 on susceptibility to a compatible strain of M. oryzae. (A) Disease symptoms on leaf blades at 4 dai. (B) Development of blast disease in leaf blades evaluated by quantitating M. oryzae genomic DNA. The amount of M. oryzae 28S rDNA relative to rice genomic eEF1α DNA was determined by quantitative PCR analysis. Values are represented as mean values ±SE for 12 leaf blades. Significantly higher values compared with those of the non-transformed control are denoted by asterisks (*P < 0.05, **P < 0.01 by Dunnett’s test on log-transformed data). (C) Invasion rate of the infectious hyphae in rice leaf sheath. Upper illustrations show the classification of patterns of invasion. Data are expressed as the ratio of the invasion pattern of infectious hyphae per 200 appressoria. Values are represented as mean values ±SE for five leaf sheaths. Significantly different values compared with those of the non-transformed control are denoted by asterisks (*P < 0.05, **P < 0.01 by Dunnett’s test).
Overexpression of OsWRKY76 improves tolerance to cold stress
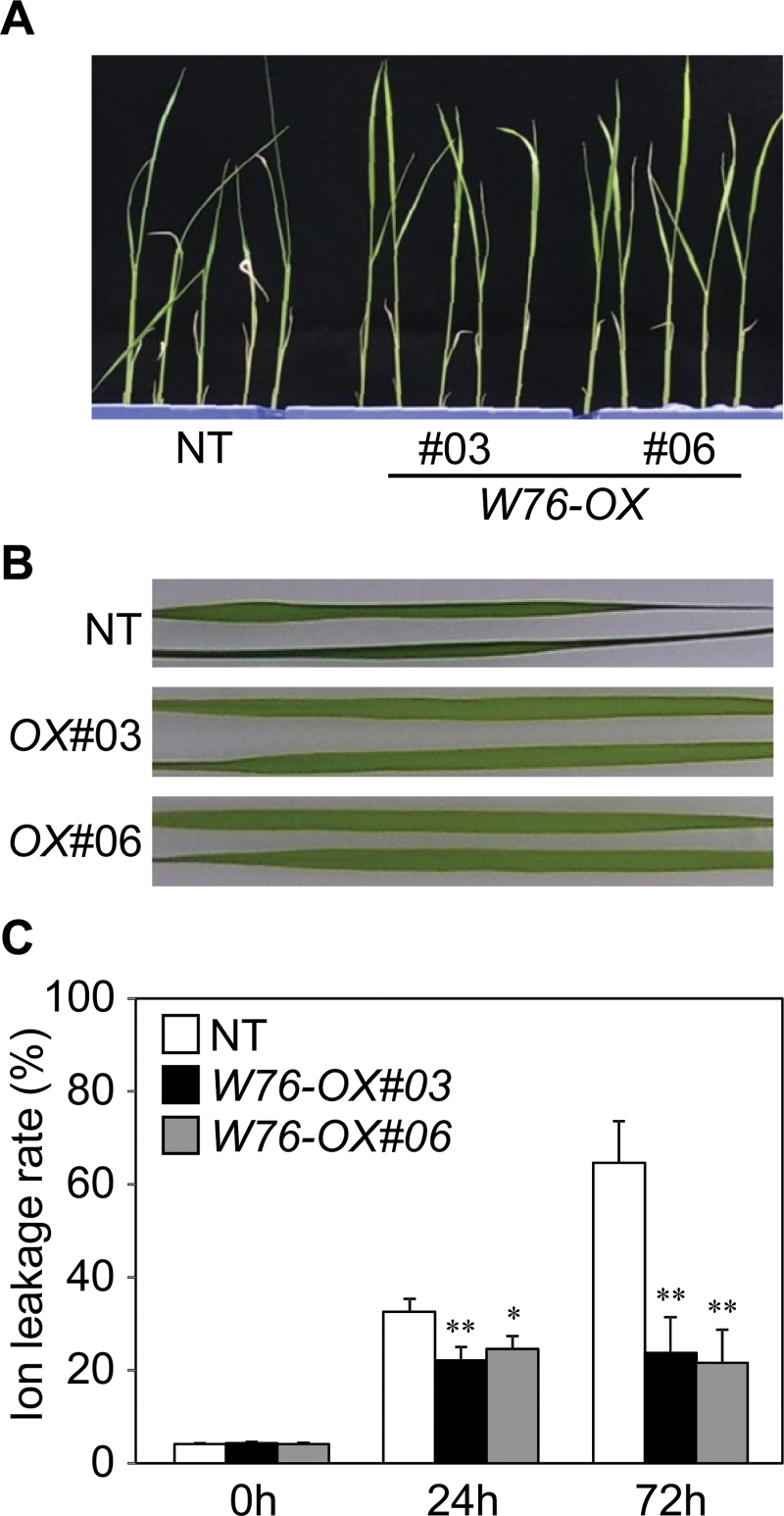
The expression of OsWRKY76 was induced by cold and ABA treatment as shown in Fig. 1. ABA mediates tolerance to low temperature in plants (Mauch-Mani and Mauch, 2005; Chinnusamy et al., 2007); thus, the cold tolerance of W76-OX was examined. Following incubation at 4 °C for 72h, NT plants presented a severely wilted appearance, while W76-OX appeared normal (Fig. 4A, B). To determine the effect of cold stress on membrane stability, an ion leakage test was performed. After a 24h incubation at 4 °C, ion leakage from leaves had drastically increased, whereas in W76-OX, the cold-induced increase in ion leakage was significantly lower than that in the NT plants, and remained at similar levels even after 72h (Fig. 4C). Therefore, it is likely that constitutive overexpression of OsWRKY76 confers cold tolerance to rice via protection of membranes from damage.
Fig. 4.
Tolerance of transgenic plants overexpressing OsWRKY76 to cold stress. (A) Plants after treatment with cold stress. Fourteen-day-old plants were incubated at 4 °C in the dark for 72h. (B) Leaf blades after the cold treatment for 72h. (C) Ion leakage rate in the leaf blades after cold treatment. Values are represented as mean values ±SE for eight plants. Significantly lower values compared with the non-transformed control are denoted by asterisks (*P < 0.1, **P < 0.05 by Dunnett’s test). Experiments were carried out three times, and a representative result is shown.
Genome-wide profiling of gene expression in W76-OX
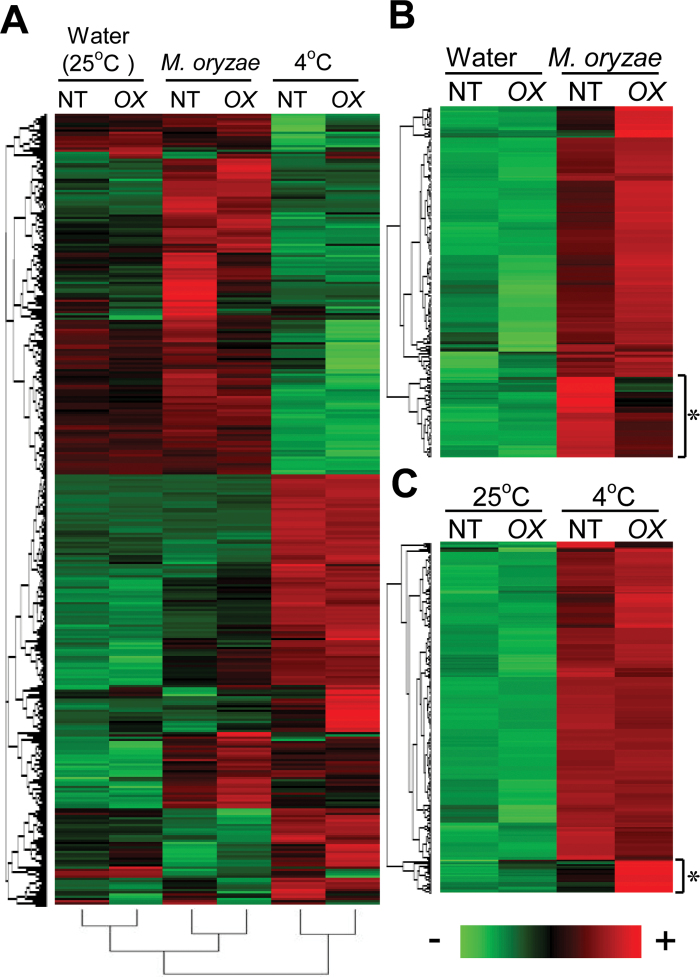
To gain an insight into the role of OsWRKY76 in blast susceptibility and cold tolerance, microarray analysis was performed. The fifth leaf sheaths of NT and W76-OX seedlings were inoculated with a compatible strain of M. oryzae or incubated at 4 °C for 36h. Leaf sheaths incubated at 25 °C for 36h were used as unstressed controls. After statistical processing, 1160 genes were selected by applying the filter of the average max/min difference being >6 (P < 0.01). An overview of a heat map with two-dimensional hierarchical clustering (Fig. 5A; Supplementary Table S2 at JXB online) revealed the following information. First, the number of genes that were up-regulated in response to inoculation of M. oryzae or cold treatment was larger than that of genes counteracted by these stresses. Secondly, a large number of genes showed opposing expression patterns between blast infection and cold treatment. Thirdly, gene expression profiles of NT and W76-OX plants in unstressed leaf sheaths were more similar to each other than those in blast-inoculated and cold-treated sheaths, indicating that the overexpression of OsWRKY76 affects the expression of other genes upon exposure to these stresses. To identify the effect of OsWRKY76 overexpression on the alteration of gene expression, 208 (Fig. 5B) and 338 (Fig. 5C) genes were next selected out of the 1160 genes that were at least 6-fold up-regulated by blast inoculation and cold treatment, respectively (P < 0.01) in either NT or W76-OX plants. A hierarchical clustering tree revealed the genes whose expression profiles were significantly different between NT and W76-OX plants. These genes might be involved in enhanced susceptibility to M. oryzae and tolerance to low temperature of W76-OX plants.
Fig. 5.
Gene expression profile of transgenic plants overexpressing OsWRKY76 (W76-OX#03) in response to blast inoculation and cold stress. Lists of these genes are shown in Supplementary Tables S2, S3, and S4 at JXB online. (A) Result of microarray analysis presented in a heat map of z-scores. (B) Effect of OsWRKY76 overexpression on the expression of blast disease-responsive genes in the leaf sheath at 36 hai. Genes with suppressed expression by OsWRKY76 overexpression are indicated by a square bracket and an asterisk. (C) Effect of OsWRKY76 overexpression on the expression of cold-responsive genes in the leaf blades incubated at 4 °C for 36h. Genes up-regulated by OsWRKY76 overexpression are indicated by a square bracket and an asterisk.
Overexpression of OsWRKY76 suppresses the activation of defence-associated genes
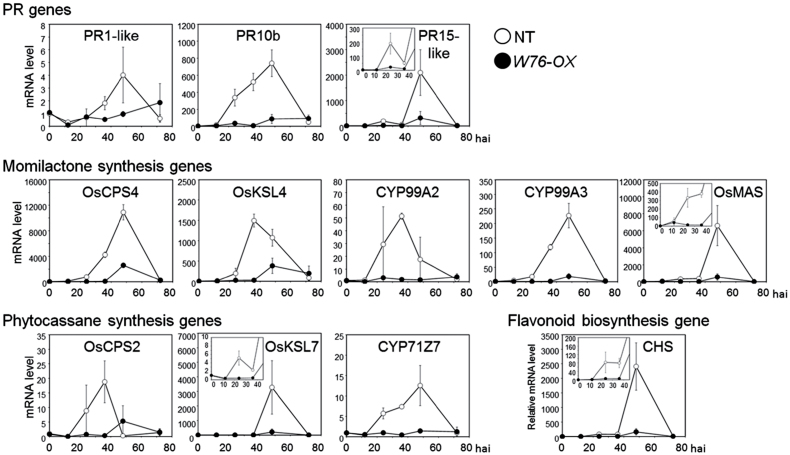
Among the 208 genes selected as up-regulated by blast inoculation either in NT or in W76-OX plants (Fig. 5B), 48 genes were counteracted in W76-OX plants, including several PR genes (Supplementary Table S3 at JXB online). To confirm the results of the microarray analysis further, qRT–PCR analysis was performed for PR1, PR10b, and PR15. As shown in Fig. 6, transcriptional induction of all these genes was drastically suppressed in W76-OX plants.
Fig. 6.
Effect of overexpression of OsWRKY76 on the expression of PR genes and phytoalexin synthesis genes. Sheaths of the fifth leaves of NT and W76-OX#03 were detached, inoculated with a suspension of conidia, and incubated at 25 °C in the dark. The time-course of gene expression was determined by qRT–PCR analysis. Transcription levels are expressed as the ratio to the level of transcript at 0h in NT. Data are represented as mean values ±SE for three replicates.
Another striking suppression was seen for genes encoding enzymes involved in phytoalexin biosynthesis. Two major classes of diterpenoid phytoalexins in rice, momilactones and phytocassanes, are synthesized from geranylgeranyl diphosphate (GGDP) through multiple steps (Supplementary Fig. S5 at JXB online), and multiple genes have been identified as those encoding the enzymes of momilactones and phytocassanes (Okada, 2011). The results of microarray analysis showed that, in W76-OX plants, the activation of five genes for momilactone synthesis (OsCPS4, OsKSL4, CYP99A2, CYP99A3, and OsMAS) and three genes for phytocassane synthesis (OsCPS2, OsKSL7, and CYP71Z7) was strongly suppressed (Supplementary Fig. S5); this was further confirmed by qRT–PCR in two independent W76-OX lines (Fig. 6; Supplementary Fig. S6). In addition, the blast-induced expression of the gene for chalcone synthase (CHS, Os04g0103900), one of the key enzymes in flavonoid synthesis, was also suppressed in W76-OX plants (Fig. 6; Supplementary Fig. S6).
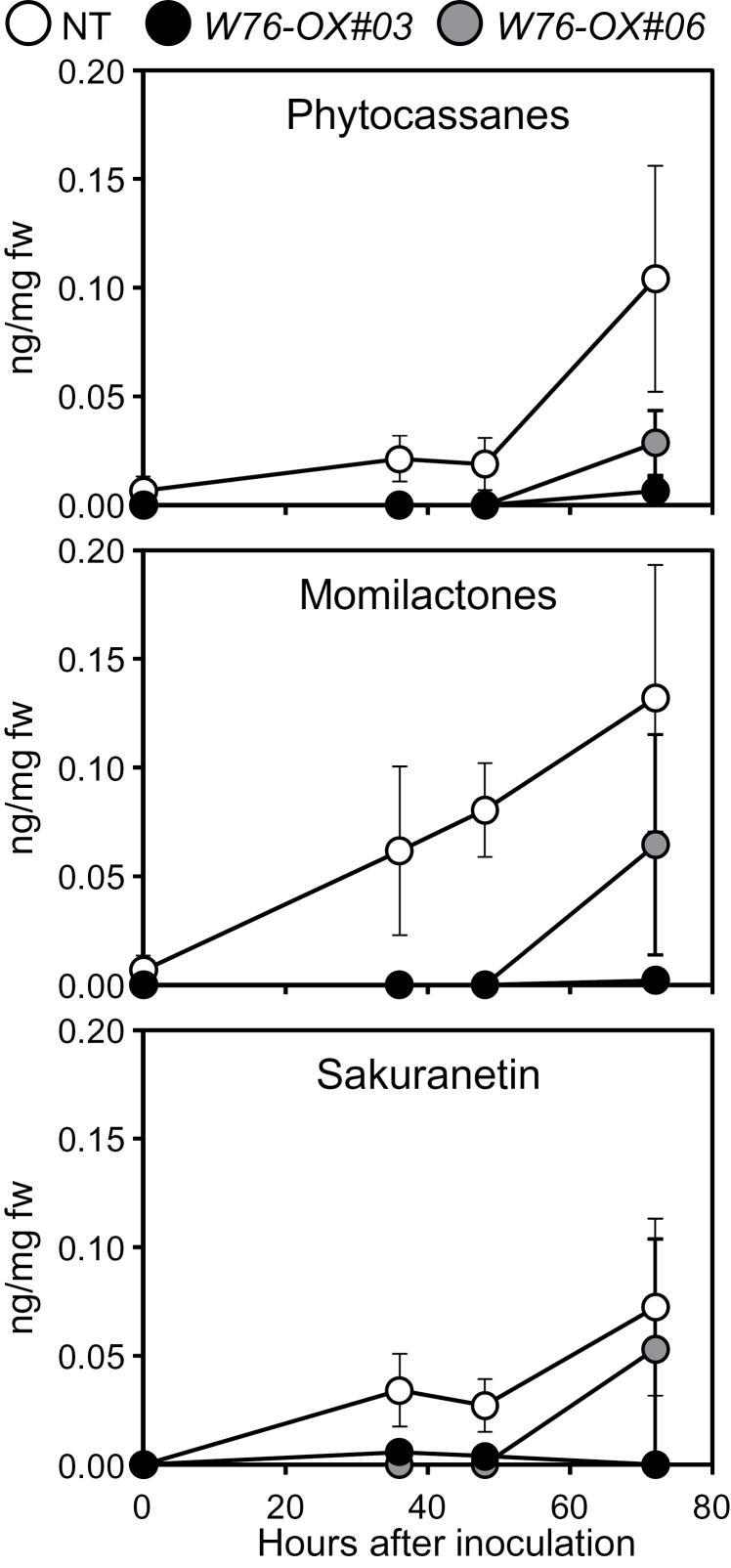
Overexpression of OsWRKY76 reduces the blast-induced accumulation of phytoalexins
Next, the levels of the two classes of diterpenoid phytoalexins, phytocassanes and momilactones, and a flavonoid phytoalexin, sakuranetin, were determined in rice leaf sheaths after inoculation with the blast fungus. Accumulation of all three phytoalexins was observed at 36h after inoculation (hai) in NT plants, whereas in W76-OX plants, it was greatly suppressed (Fig. 7).
Fig. 7.
Effects of overexpression of OsWRKY76 on the accumulation of phytoalexins in response to M. oryzae inoculation. The sheaths of the fifth leaves were detached, inoculated with a suspension of conidia, and incubated at 25 °C in the dark.
Overexpression of OsWRKY76 causes the up-regulation of abiotic stress-associated genes
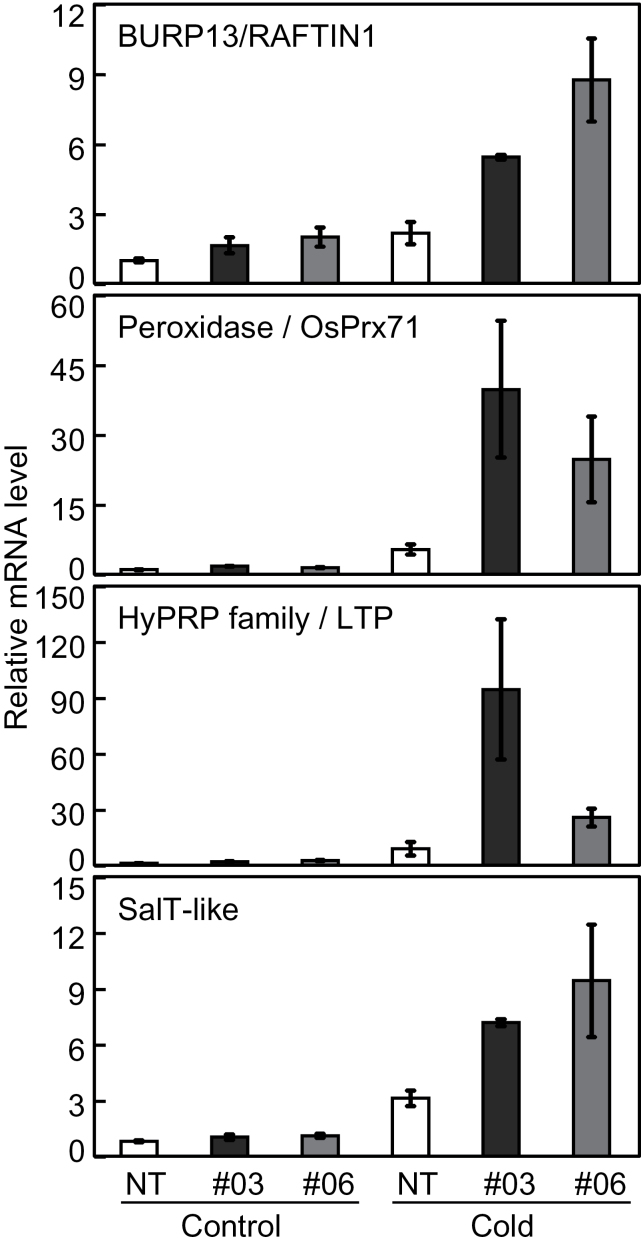
Microarray analysis identified a total of 338 low temperature-responsive genes in either NT or W76-OX plants (Fig. 5C). Among them, 30 genes exhibited up-regulation, and five genes were counteracted in W76-OX plants in response to cold treatment (Supplementary Table S4 at JXB online). It was difficult to designate functions for the five counteracted genes, while the 30 up-regulated genes included those for antioxidant enzymes such as peroxidases (OsPrx16/17, OsPrx39, OsPrx71, and OsPrx74; Passardi et al., 2007) and glutathione S-transferase (OsGSTU5; Soranzo et al., 2004). Microarray analysis also detected three genes for lipid transfer proteins as up-regulated genes in W76-OX plants. Among these, Os04g0554800 and Os04g0644400 belong to the hybrid proline-rich protein (HyPRP) family and are homologues of Arabidopsis EARLI1 (Bubier and Schläppi, 2004) and AZI1 (Xu et al., 2011), which confers cold tolerance to Arabidopsis possibly via the maintenance of membrane stability. The pollen development-related BURP domain gene OsBURP13/OsRAFTIN1 (Wang et al., 2003), which is very similar to the stress-associated gene RD22 of Arabidopsis (Abe et al., 1997), was also up-regulated in W76-OX plants.
QRT-PCR analyses were performed for OsPrx71, OsBURP13/OsRAFTIN1, a HyPRP family gene, and a SalT-like gene. As shown in Fig. 8, all the tested genes showed up-regulation in W76-OX plants following cold treatment.
Fig. 8.
Effect of overexpression of OsWRKY76 on the expression of cold stress-related genes. Transcription levels of NT and W76-OX lines were measured by qRT–PCR analysis. Data are expressed as the ratio to the level of transcript in NT at 25 °C.
Discussion
The transcription factor OsWRKY76, a rice group IIa WRKY gene, was identified as a blast- and BTH-responsive gene (Ryu et al., 2006; Shimono et al., 2007). The results presented here clearly show that OsWRKY76 is a transcriptional repressor involved in both fungal disease resistance and cold stress tolerance. OsWRKY76 has two putative nuclear localization signals and a DNA-binding domain, but does not have any known plant transcriptional repressor domains such as an EAR motif (Ohta et al., 2001). Thus, it is unknown how OsWRKY76 suppresses the transcription of its target genes. Several studies identified the transcriptional regulator activity of group IIa WRKY proteins. OsWRKY71 (Chujo et al., 2008) and OsWRKY28 (Chujo et al., 2013), closely related paralogues of OsWRKY76, encode proteins that exhibit transcriptional repressor activity in cultured rice cells. On the other hand, Chen et al. (2010) tested the transcriptional regulator activity of three WRKYs of group IIa in Arabidopsis using stable transformants, and found that AtWRKY18 and 60 are transcriptional activators while AtWRKY40 is a transcriptional repressor. Cotton GaWRKY1 activates the promoter of the sesquiterpene phytoalexin synthesis gene CAD1-A in transgenic Arabidopsis (Xu et al., 2004). Therefore, not all group IIa WRKY genes encode transcriptional repressors, although the experimental systems used to test the activity differ in each report.
Overexpression of OsWRKY76 enhanced the susceptibility of rice plants to the blast fungus (Fig. 3), indicating that it is a negative regulator of disease resistance. Seo et al. (2011) also reported that the overexpression of OsWRKY76 causes reduced resistance to bacterial leaf blight caused by X. oryzae. Similarly, overexpression of OsWRKY62, a closely related paralogue of OsWRKY76, also causes enhanced susceptibility to X. oryzae (Peng et al., 2008). Delteil et al. (2012) and Chujo et al. (2013) showed that OsWRKY28 also acts as a negative regulator of resistance to M. oryzae. On the other hand, among genes encoding WRKY transcriptional activators in rice, the overexpression of OsWRKY45 (Shimono et al., 2007), OsWRKY53 (Chujo et al., 2007), OsWRKY31 (Zhang et al., 2008), and OsWRKY30 (Peng et al., 2012) confers disease resistance. These results consistently suggest that disease resistance in rice is negatively and positively regulated by WRKY transcriptional repressors and activators, respectively.
Microarray analysis and qRT–PCR analysis revealed that, in W76-OX plants, the up-regulation of a set of genes for PR proteins and enzymes involved in phytoalexin biosynthesis was largely cancelled after inoculation with M. oryzae. Therefore, these genes might be direct or indirect targets of OsWRKY76. The promoter regions of these genes include the W-box and W-box-like sequences (Supplementary Fig. S7 at JXB online), and it was found that OsWRKY76 binds to a W-box in the PR10b promoter (data not shown). The expression of these genes is, therefore, implicated to be regulated by OsWRKY76 via the W-box sequence. In rice, the blast-responsive expression of several defence-associated genes including PR10b is mediated by the SA signalling pathway (Shimono et al., 2007), implying that OsWRKY76 participates in SA signalling. Interestingly, blast-responsive expression of genes encoding almost all of the biosynthesis enzymes of diterpenoid phytoalexins, momilactones and phytocassanes, was suppressed (Fig. 6). These genes are clustered on chromosomes 4 and 2, respectively, and exhibited synchronous expression patterns in response to N-acetylchitooligosaccharide elicitors (Okada et al., 2007). A bZIP transcription factor, OsTGAP1, is considered to regulate positively the expression of genes for diterpenoid phytoalexins (Okada et al., 2009). However, because no alteration in OsTGAP1 expression was observed in W76-OX plants, it is not clear at present how OsTGAP1 and OsWRKY76 cooperate with or counteract each other in vivo in the process of phytoalexin biosynthesis. Consistent with the expression profiles, accumulation of phytoalexins including sakuranetin was drastically reduced to low levels in W76-OX plants (Fig. 7). Because exogenously applied momilactones were shown to be toxic to M. oryzae in rice leaves at physiological concentrations (Hasegawa et al., 2010), the enhanced susceptibility to M. oryzae in W76-OX plants is possibly due to the suppressed accumulation of these phytoalexins. This idea is further supported by the observation that a knock-out line of OsCPS4, which encodes syn-copalyl diphosphate synthase responsible for the biosynthesis of momilactones and oryzalexin S, is more susceptible to the rice blast fungus (Toyomasu et al., 2013). Recently, Shimizu et al. (2012) purified an enzyme from rice leaves that catalyses the O-methylation of naringenin chalcone, forming sakuranetin (NOMT; naringenin O-methyltransferase). Microarray analysis, however, indicated that the overexpression of OsWRKY76 suppressed the expression of not OsNOMT but CHS (Os04g0103900). Because naringenin chalcone is a key precursor to a variety of flavonoids, this result implies that the overexpression of OsWRKY76 results in the reduction of several flavonoids in rice cells. OsWRKY13 has also been reported to activate genes involved in the flavonoid biosynthesis pathway and in resistance to M. oryzae (Qiu et al., 2008).
The up-regulation of OsWRKY76 upon infection with M. oryzae is paradoxical, because OsWRKY76 is a negative regulator of disease resistance. One hypothesis is that OsWRKY76, which is induced after OsWRKY45, adjusts the intensity of defence responses in cooperation with positive regulators of defence to protect the plant from the damage caused by defence responses. For example, OsWRKY76 might restrict the biosynthesis of diterpenoid phytoalexins in order to maintain the pool of the substrate, GGDP, which is also used as a substrate for other components such as carotenoids, tocopherols, and chlorophylls (DellaPenna and Pogson, 2006). To test this hypothesis, it would be necessary to compare the levels of these metabolites in OsWRKY76-overexpressing plants and OsWRKY76 knock-out or knock-down plants. Despite their efforts, the authors have not succeeded in isolating OsWRKY76 knock-out/down plants. Similar feedback-like responses in the defence response have been reported in other plant species. For example, an Arabidopsis ERF/AP2 transcriptional repressor, AtERF4, is transcriptionally up-regulated by bacterial disease but negatively regulates disease resistance (McGrath et al., 2005). A zinc-finger transcriptional repressor, ZCT, of Catharanthus roseus is induced by an elicitor and negatively regulates elicitor-induced genes involved in alkaloid biosynthesis (Pauw et al., 2004).
W76-OX plants showed improved tolerance to low temperature stress (Fig. 4), indicating that OsWRKY76 functions as a positive regulator of the cold tolerance. Similarly, transgenic grass overexpressing HvWRKY38, a predicted orthologue of OsWRKY76 in barley, shows improved tolerance to drought (Xiong et al., 2010). Overexpression of OsWRKY76 altered cold-induced electron leakage (Fig. 4C), which is a major problem causing chilling injury (Chinnusamy et al., 2007). Microarray analysis revealed that a gene encoding a lipid transfer protein of the HyPRP family, which is predicted to play a role in membrane stability (Bubier and Schläppi, 2004; Xu et al., 2011), was up-regulated by the overexpression of OsWRKY76 (Fig. 8). In W76-OX plants, genes encoding defence-related proteins such as the antioxidant enzyme peroxidase were also up-regulated (Fig. 8). It might be possible that the up-regulation of these genes resulted in increased tolerance to cold treatment in W76-OX plants. Because OsWRKY76 shows transcriptional repressor activity (Fig. 2C), it is unlikely that OsWRKY76 directly activates the expression of these cold-responsive genes; instead, it may suppress the expression of negative regulators for these genes. Alternatively, the function of OsWRKY76 might be modulated to act as an activator by interaction with other proteins. There are reports that AtWRKY18, AtWRKY40, and AtWRKY60 form both homocomplexes and heterocomplexes, and function as transcriptional activators depending on the complex (Xu et al., 2006; Chen et al., 2010).
The present results demonstrate that OsWRKY76 plays opposite roles in blast resistance and cold tolerance when overexpressed. Low temperature treatment enhances the susceptibility of rice to M. oryzae, and, in fact, blast disease causes more serious damage in cold summer in the northern part of Japan (Matsuyama and Dimond, 1974; Koga et al., 2004). Recent studies have suggested that the level of endogenous ABA is elevated by incubation at low temperature (Cuevas et al., 2008), and low temperature-induced susceptibility to M. oryzae is mediated by ABA, which antagonizes SA signalling (Mauch-Mani and Mauch, 2005; Robert-Seilaniantz et al., 2007; Ton et al., 2009; Jiang et al., 2010; Sharma et al., 2013). A similar antagonistic interaction between SA and ABA has been observed in Arabidopsis (Yasuda et al., 2008). OsWRKY76 is transcriptionally up-regulated by treatment with BTH, a chemical activator of SA signalling, and ABA (Fig. 1), implying its involvement in the SA and ABA responses. OsWRKY76 might be involved in antagonistic regulation between SA-mediated biotic and ABA-mediated abiotic stress responses. Some studies suggest that WRKY might be associated with ABA signalling in several aspects (Rushton et al., 2010). For example, AtWRKY18, 40, and 60, possible orthologues of OsWRKY76 in Arabidopsis, have been suggested to be involved in ABA-associated stress responses (Chen et al., 2010; Shang et al., 2010).
In this study, it was demonstrated that OsWRKY76 plays important roles in plant responses to biotic and abiotic stresses. However, the signalling pathways upstream and downstream of OsWRKY76 remain to be elucidated. In W76-OX plants, the expression of genes encoding stress-associated signal components, including several types of transcriptional regulators, was also affected (Fig. 5; Supplementary Table S2 at JXB online), suggesting that responses to exogenous stresses are regulated by complex transcription events. Further analysis of these genes should be valuable for the purpose of balancing disease resistance and cold tolerance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Gel mobility shift competition assay of OsWRKY76 and the W-box-containing oligonucleotide.
Figure S2. Growth of transgenic plants constitutively overexpressing OsWRKY76.
Figure S3. Effects of overexpression of OsWRKY76 on disease resistance to a compatible strain (Ina86-137) of M. oryzae.
Figure S4. Effects of overexpression of OsWRKY76 on disease resistance to an incompatible strain (P91-15B) of M. oryzae.
Figure S5. Schematic view integrating the biosynthetic pathway of diterpenoid phytoalexins with microarray data.
Figure S6. Effects of the overexpression of OsWRKY76 on the accumulation of phytoalexins in response to M. oryzae inoculation.
Figure S7. Distribution of W-box/W-box-like elements upstream (2kb) of the initiation codon of genes that are putatively regulated by OsWRKY76.
Table S1. Primers used for real-time PCR analysis.
Table S2. List of genes with significantly different expression profiles in W76-OX plants identified by microarray analysis (Fig. 5A).
Table S3. List of blast disease-responsive genes in the leaf sheath at 36 hai.
Table S4. List of cold-responsive genes in the leaf blades incubated at 4 °C for 36h.
Acknowledgements
We thank Professor H. Naitoh (Akita Prefectural University, Japan) for providing M. oryzae isolate P91-15B, and Dr M. Takagi (National Institute of Advanced Industrial Science and Technology) for providing the 35S-GAL4-TATA-LUC-NOS and pPTRL plasmids. We are deeply grateful to Ms R. Motoyama and Dr Y. Nagamura (National Institute of Agrobiological Sciences, Microarray Open Lab.) for their sophisticated assistance in microarray analysis. We thank to Ms E. Nakajima, Ms M. Kouzai, Mr K. Nakajima, and Ms H. Mochizuki for their technical assistance. The Japanese isolate of M. oryzae, Ina86-137 (MAFF 101511), was supplied by the NIAS Genebank. This research was supporeted by a grant to YN from the Japan Society for the Promotion of Science (JSPS) initiated by the Council for Science and Technology Policy (CSTP).
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. 1997. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell 9, 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnaresi P, Biselli C, Orrù L, Urso S, Crispino L, Abbruscato P, Piffanelli P, Lupotto E, Cattivelli1 L, Valè G. 2012. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS One 7, e51609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier J, Schläppi M. 2004. Cold induction of EARLI1, a putative Arabidopsis lipid transfer protein, is light and calcium dependent. Plant, Cell and Environment 27, 929–936 [Google Scholar]
- Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X. 2010. Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biology 10, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. 2007. Cold stress regulation of gene expression in plants. Trends in Plant Science 12, 444–451 [DOI] [PubMed] [Google Scholar]
- Chujo T, Kato T, Yamada K, et al. 2008. Characterization of an elicitor-induced rice WRKY gene, OsWRKY71 . Bioscience, Biotechnology, and Biochemistry 72, 240–245 [DOI] [PubMed] [Google Scholar]
- Chujo T, Miyamoto K, Shimogawa T, et al. 2013. OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Molecular Biology 82, 23–37 [DOI] [PubMed] [Google Scholar]
- Chujo T, Takai R, Akimoto-Tomiyama C, et al. 2007. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochimica et Biophysica Acta 1769, 497–505 [DOI] [PubMed] [Google Scholar]
- Cuevas JC, López-Cobollo R, Alcázar R, Zarza X, Koncz C, Altabella T, Salinas J, Tiburcio AF, Ferrando A. 2008. Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiology 148, 1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. 2006. Vitamin synthesis in plants: tocopherols and carotenoids. Annual Review of Plant Biology 57, 711–738 [DOI] [PubMed] [Google Scholar]
- Delteil A, Blein M, Faivre-Rampant O, Guellim A, Estevan J, Hirsch J, Bevitori R, Michel C, Morel JB. 2012. Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defense. Molecular Plant Pathology 13, 72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. 2000. The WRKY superfamily of plant transcription factors. Trends in Plant Science 5, 199–206 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. 2006. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Current Opinion in Plant Biology 9, 436–442 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Mitsuhara I, Seo S, Imai T, Koga J, Okada K, Yamane H, Ohashi Y. 2010. Phytoalexin accumulation in the interaction between rice and the blast fungus. Molecular Plant-Microbe Interactions 23, 1000–1011 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. 2006. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications 345, 646–651 [DOI] [PubMed] [Google Scholar]
- Jiang CJ, Shimono M, Sugano S, Kojima M, Yazawa K, Yoshida R, Inoue H, Hayashi N, Sakakibara H, Takatsuji H. 2010. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice–Magnaporthe grisea interaction. Molecular Plant-Microbe Interactions 23, 791–798 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. 2009. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology 69, 91–105 [DOI] [PubMed] [Google Scholar]
- Kim SH, Oikawa T, Kyozuka J, Wong HL, Umemura K, Kishi-Kaboshi M, Takahashi A, Kawano Y, Kawasaki T, Shimamoto K. 2012. The bHLH Rac Immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant and Cell Physiology 53, 740–754 [DOI] [PubMed] [Google Scholar]
- Kim KC, Fan B, Chen Z. 2006. Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae . Plant Physiology 142, 1180–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Dohi K, Mori M. 2004. Abscisic acid and low temperatures suppress the whole plant-specific resistance reaction of rice plants to the infection of Magnaporthe grisea . Physiological and Molecular Plant Pathology 65, 3–9 [Google Scholar]
- Li S, Fu Q, Chen L, Huang W, Yu D. 2011. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252 [DOI] [PubMed] [Google Scholar]
- Liu D, Chen X, Liu J, Ye J, Guo Z. 2012. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. Journal of Experimental Botany 63, 3899–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama N, Dimond AE. 1973. Effect of nitrogenous fertilizer on biochemical processes that could affect lesion size of rice blast. Phytopathology 63, 1202–1203 [Google Scholar]
- Marè C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. 2004. Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Molecular Biology 55, 399–416 [DOI] [PubMed] [Google Scholar]
- Mauch-Mani B, Mauch F. 2005. The role of abscisic acid in plant–pathogen interactions. Current Opinion in Plant Biology 8, 409–414 [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. 2005. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiology 139, 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, et al. 2007. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. The Plant Journal 51, 617–630 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H. 1999. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. The Plant Journal 18, 455–463 [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. The Plant Cell 13, 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Okada K, Miyamoto K, Koga J, Shibuya N, Nojiri H, Yamane H. 2009. OsTGAP1, a bZIP transcription factor, coordinately regulates the inductive production of diterpenoid phytoalexins in rice. Journal of Biological Chemistry 284, 26510–26518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H. 2007. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Molecular Biology 65, 177–187 [DOI] [PubMed] [Google Scholar]
- Okada K. 2011. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Bioscience, Biotechnology, and Biochemistry 75, 1219–1225 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. 2009. The role of WRKY transcription factors in plant immunity. Plant Physiology 150, 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauw B, Hilliou FA, Martin VS, et al. 2004. Zinc finger proteins act as transcriptional repressors of alkaloid biosynthesis genes in Catharanthus roseus . Journal of Biological Chemistry 279, 52940–52948 [DOI] [PubMed] [Google Scholar]
- Passardi F, Theiler G, Zamocky M, et al. 2007. PeroxiBase: the peroxidase database. Phytochemistry 68, 1605–1611 [DOI] [PubMed] [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. 2012. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498 [DOI] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. 2008. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Molecular Plant 1, 446–458 [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Xie W, Liu H, Li X, Xiong L, Wang S. 2008. Rice gene network inferred from expression profiling of plants overexpressing OsWRKY13, a positive regulator of disease resistance. Molecular Plant 1, 538–551 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Navarro L, Bari R, Jones JD. 2007. Pathological hormone imbalances. Current Opinion in Plant Biology 10, 372–379 [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QJ. 2007. The WRKY gene family in rice (Oryza sativa). Journal of Integrative Plant Biology 49, 827–842 [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ. 2010. WRKY transcription factors. Trends in Plant Science 15, 247–258 [DOI] [PubMed] [Google Scholar]
- Ryu HS, Han M, Lee SK, Cho JI, Ryoo N, Heu S, Lee YH, Bhoo SH, Wang GL, Hahn TR, Jeon JS. 2006. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Reports 25, 836–847 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Schlagenhauf E, Schaffrath U, Dudler R. 1999. Different patterns of host genes are induced in rice by Pseudomonas syringae, a biological inducer of resistance, and the chemical inducer benzothiadiazole (BTH). European Journal of Plant Pathology 105, 659–665 [Google Scholar]
- Seo YS, Chern M, Bartley LE, et al. 2011. Towards establishment of a rice stress response interactome. PLoS Genetics 7, e1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Vleesschauwer DD, Sharma MK, Ronald PC. 2013. Elucidating stress regulatory crosstalk in rice. Molecular Plant 6, 250–260 [DOI] [PubMed] [Google Scholar]
- Shang Y, Yan L, Liu ZQ, et al. 2010. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. The Plant Cell 22, 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. 2007. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Jikumaru Y, Okada A, et al. 2008. Effects of a bile acid elicitor, cholic acid, on the biosynthesis of diterpenoid phytoalexins in suspension-cultured rice cells. Phytochemistry 69, 973–981 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Lin F, Hasegawa M, Okada K, Nojiri H, Yamane H. 2012. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. Journal of Biological Chemistry 287, 19315–19325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Koga H, Akagi A, et al. 2012. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Molecular Plant Pathology 13, 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. 2007. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. The Plant Cell 19, 2064–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. 2002. Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology 5, 430–436 [DOI] [PubMed] [Google Scholar]
- Soranzo N, Sari Gorla M, Mizzi L, De Toma G, Frova C. 2004. Organisation and structural evolution of the rice glutathione S-transferase gene family. Molecular Genetics and Genomics 271, 511–521 [DOI] [PubMed] [Google Scholar]
- Talbot NJ. 2003. On the trail of a cereal killer: exploring the biology of Magnaporthe grisea . Annual Review of Microbiology 57, 177–202 [DOI] [PubMed] [Google Scholar]
- Tanabe S, Okada M, Jikumaru Y, Yamane H, Kaku H, Shibuya N, Minami E. 2006. Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N-acetylchitooligosaccharide. Bioscience, Biotechnology, and Biochemistry 70, 1599–1605 [DOI] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. 2012. Evolution of jasmonate and salicylate signal crosstalk. Trends in Plant Science 17, 260–270 [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. 2006. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. The Plant Journal 47, 969–976 [DOI] [PubMed] [Google Scholar]
- Ton J, Flors V, Mauch-Mani B. 2009. The multifaceted role of ABA in disease resistance. Trends in Plant Science 14, 310–317 [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Usui M, Sugawara C, et al. 2013. Reverse-genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiologia Plantarum (in press). [DOI] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Datla R, Selvaraj G. 2003. The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proceedings of the National Academy of Sciences, USA 100, 14487–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Zhang ZL, Zou X, Huang J, Ruas P, Thompson D, Shen QJ. 2005. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiology 137, 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, James VA, Zhang H, Altpeter F. 2010. Constitutive expression of the barley HvWRKY38 transcription factor enhances drought tolerance in turf and forage grass (Paspalumnotatum Flugge). Molecular Breeding 25, 419–432 [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z. 2006. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. The Plant Cell 18, 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Wang JW, Wang S, Wang JY, Chen XY. 2004. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiology 135, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Zhang X, Schläppi M, Xu ZQ. 2011. Cold-inducible expression of AZI1 and its function in improvement of freezing tolerance of Arabidopsis thaliana and Saccharomyces cerevisiae . Journal of Plant Physiology 168, 1576–1587 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57, 781–803 [DOI] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, et al. 2008. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. The Plant Cell 20, 1678–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellerhoff N, Jarosch B, Groenewald JZ, Crous PW, Schaffrath U. 2006. Nonhost resistance of barley is successfully manifested against Magnaporthe grisea and a closely related Pennisetum-infecting lineage but is overcome by Magnaporthe oryzae . Molecular Plant-Microbe Interactions 19, 1014–1022 [DOI] [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z. 2008. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Research 18, 508–521 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Mosher SL, Fan B, Klessig DF, Chen Z. 2007. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae . BMC Plant Biology 7, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.