Abstract
Thermoresponsive poly(N-isopropyl acrylamide) (PNIPAM) microgels were patterned on polystyrene substrates via dip coating, creating cytocompatible substrates that provided spatial control over cell adhesion. This simple dip coating method, which exploits variable substrate withdrawal speeds form particle suspension formed stripes of densely-packed PNIPAM microgels, while spacings between the stripes contained sparsely-distributed PNIPAM microgels. The assembly of three different PNIPAM microgel patterns, namely patterns composed of 50 μm stripes/50 μm spacings, 50 μm stripes/100 μm spacings, and 100 μm stripes/100 μm spacings was verified using high-resolution optical micrographs and ImageJ analysis. PNIPAM microgels existed as monolayers within stripes and spacings, as revealed by atomic force microscopy (AFM). Upon cell seeding on PNIPAM micropatterned substrates, NIH3T3 fibroblast cells preferentially adhered within spacings to form cell patterns. Three days after cell seeding, cells proliferated to form confluent cell layers. The thermoresponsiveness of the underlying PNIPAM microgels was then utilized to recover fibroblast cell sheets from substrates simply by lowering the temperature, without disrupting the underlying PNIPAM microgel patterns. Harvested cell sheets similar to these have been used for multiple tissue engineering applications. Also, this simple, low cost, template-free dip coating technique can be utilized to micropattern multifunctional PNIPAM microgels, generating complex stimuli-responsive substrates to study cell-material interactions and allow drug delivery to cells in a spatially and temporally-controlled manners.
Introduction
A multitude of approaches have been employed to pattern cells on substrates at a variety of length scales. Cell patterning allows control over cell morphology, alignment, and even cell functions via interaction with underlying biochemical and/or topographical cues.1, 2 Cell patterning methodologies include, but are not limited to, microcontact printing (soft lithography),3 photochemistry,4 inkjet printing,5 laser bioprinting,6 and photolithography.7 These approaches require costly templates and frequent retooling with expensive optics and radiation sources to generate cell patterns. It is therefore desirable to devise template-free high-throughput cell patterning techniques. Controlled cell adhesion on substrates can then lead to the formation of patterned tissues for a myriad of tissue engineering applications. Herein, we achieve controlled cell adhesion by first patterning thermoresponsive PNIPAM microgels via a simple, low cost, template-free dip coating methodology and then using the patterned substrates to control cell adhesion at a variety of length scales.
Poly(N-isopropylacrylamide) (PNIPAM) is a well-known, often-utilized thermoresponsive material, which exhibits a lower critical solution temperature (LCST) of ~31 °C in water.8 Below the LCST, PNIPAM-water interactions are favorable resulting in fully extended soluble PNIPAM chains. However, above the phase transition temperature, PNIPAM is hydrophobic and interactions between PNIPAM chains dominate, expelling water, and leading to phase separation. This transition results in insolubility of the linear PNIPAM.9, 10 The thermoresponsive nature of PNIPAM has enabled PNIPAM-based materials to act as substrates for harvesting cell sheets in an enzyme-free manner.11-14 Intact tissue-like cell sheets with preserved extracellular matrix (ECM)15 can be used individually or can be layered/rolled to create tissues of larger sizes or with defined lamellar organizations, an approach known as “cell sheet engineering”. Specifically, cell sheet engineering has been reported using PNIPAM coated substrates such as tissue culture polystyrene,16-18 micropatterned polystyrene,11 micropatterned poly(dimethyl siloxane) (PDMS),14 porous cell culture membranes,19 and treated glass/silicon wafers.13, 20 Two recent reviews discuss in greater detail the various approaches for cell sheet engineering.21, 22
PNIPAM can be crosslinked to form temperature-responsive hydrogels with volume phase transition temperatures (VPTT) between 31-32 °C.23, 24 PNIPAM hydrogels in colloidal forms are referred to as “microgels” (diameters in the nm - μm range) and are biocompatible.25 Because of their small size and high surface area-to-volume ratios, PNIPAM microgels exhibit faster responses towards temperature changes as compared to bulk PNIPAM hydrogels.26 Moreover, PNIPAM microgels can be synthesized through dispersion polymerization in an aqueous solution, where particle sizes can be controlled to generate narrow particle size distributions.9, 27 Due to their temperature responsiveness, PNIPAM microgels have found widespread biomedical applications, including bio-separations,28 drug delivery,29, 30 and biosensor applications31.
PNIPAM microgels can be conveniently fabricated into monolayers on substrates simply by spin coating32 or solvent evaporation (air drying).33, 34 For example, Kawaguchi et al. reported assembly of ordered two-dimensional PNIPAM microgel arrays by air-drying the microgels on polystyrene (PS) substrates.34 Recently, Schmidt et al. demonstrated the successful adhesion and temperature-responsive detachment of cells on PNIPAM surfaces generated by air-drying microgels.35 In addition to air-drying, unpatterned PNIPAM microgel monolayers have also been deposited on positively-charged silicon wafers simply by immersing substrates in the particle dispersion.36
The methods for microgel monolayer formation are extremely convenient but are uncontrollable with respect to the patterns generated. In fact, forming ordered arrays of nano/microparticles of defined dimensions on solid substrates without utilizing a lithography-based templating strategy was not possible until Ghosh et al. generated “stripe/spacing” patterns through dip coating of polystyrene particles onto gold-coated silicon wafers.37 The stripe/spacing patterns of varying dimensions were formed simply by withdrawing gold-coated silicon wafers from polystyrene microparticle suspensions at different withdrawal speeds.37 Watanabe et al. further determined that the stripe width via this method could be controlled by varying particle concentrations and withdrawal rates, unlike the distance between the stripes (spacings), which depended on stripe thickness and the surface tension of solvent used to suspend microparticles.38 It was also observed that evaporation rate did not affect stripe width and/or spacing, but it did affect the growth rate of stripe-patterned films.38 In a recent study, particle stripe patterns with controllable periodicity were successfully generated by altering the particle dispersion liquid levels used to deposit the particles.39
To the best of our knowledge, dip coating of hydrogel-based particles (of PNIPAM or any other polymer) has not been utilized to form patterns. Here we report a template-free micropatterning (generation of stripe/spacing) of PNIPAM microgels on polystyrene (PS) substrate via dip coating methodologies. By varying substrate withdrawal speeds from PNIPAM microgel dispersions, we controlled the dimensions of the PNIPAM micropatterns. We demonstrated that the resulting stripe/spacing microstructure could be employed for controlling cell adhesion. We also utilized the thermoresponsiveness of PNIPAM coated surfaces to demonstrate enzyme-free cell detachment simply by lowering the temperature below the VPTT of the PNIPAM microgels. Our developed methodology is simple and low cost and does not require lithographically-generated photomasks to form micropatterns. Also, this template-free technique can be utilized to pattern multifunctional PNIPAM microgels to form spatially complex stimuli-responsive substrates for biosensing and drug-delivery applications. Additionally, we demonstrate that dip coated PNIPAM microgel substrates can be employed to rapidly recover cell sheets of desired dimensions for applications in tissue engineering.
Experimental Section
Materials
N-isopropylacrylamide (NIPAM, 97%), N, N'-methylenebisacrylamide (BIS, 99%), potassium persulfate (KPS, 99%), styrene (>99%), sodium styrene sulfonate (>90%), Triton X-100, and hydrogen peroxide solution (30%) were all purchased from Sigma-Aldrich. Sodium dodecyl sulfate (SDS, 99%) was purchased from J.T. Baker. Sulfuric acid (98%) was obtained from Mallinckrodt Chemical, Ethanol (ACS grade) from BDH chemicals, Bovine Serum Albumin (BSA) from Sigma Life Sciences, and Fetal Bovine Serum (FBS) was purchased from Atlanta Biologicals. High glucose Dulbecco's Modified Eagle's Medium (HG-DMEM, HyClone) was purchased from Fisher Thermoscientific, Dulbecco's phosphate buffered saline (DPBS) from Gibco/Life Technologies, and tissue culture T75 flasks were purchased from Grenier bio-one. Antibiotic-antimycotic solution (100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL amphotericin B) was purchased from Cellgro. DAPI (4′,6-diamidino-2-phenylindole), TRITC-conjugated phalloidin, FITC-conjugated secondary antibody, and anti-Vinculin antibody were purchased from Millipore. All reagents were used as received, and distilled deionized water was used in all experiments.
Particle synthesis and characterization
PNIPAM microgels were synthesized using free-radical dispersion polymerization.40 In a 100 ml round bottom flask equipped with a magnetic stir bar, 0.76 g of NIPAM, 0.032 g of BIS and 0.32 ml of 1% (w/w) SDS were dissolved in 50 mL of deionized water. The solution was purged with argon for 40 min and then heated to 60 °C with continuous stirring. Polymerization was initiated by addition of 0.5 ml of aqueous 0.123 M KPS. The reaction was carried out for 5 hr and then allowed to cool to room temperature. The resulting particles were washed in deionized water by repetitive centrifugation (13,000 rpm, 2 hours, 15 °C) at least four times.
Polystyrene (PS) microspheres were synthesized by emulsion polymerization. In a 100 ml round bottom flask equipped with a magnetic stir bar, 10 g of styrene and 0.01 g of sodium styrene sulfonate were dissolved in 70 ml of deionized water. The solution was purged with argon for 50 min and then heated to 70 °C under vigorous stirring.
Polymerization was initiated by adding 20 ml of aqueous solution of 9.25 mM KPS. The reaction was carried out for 18 hours at 70 °C. The resulting particles were washed in deionized water by repetitive centrifugation (4,000 rpm, 10 min, 15 °C) at least four times. Particle size was measured using dynamic light scattering (Brookhaven Instruments model 90 Plus) on particles dispersed in deionized water.
Micropattern fabrication and characterization
Non-coated 75 mm × 25 mm glass slides (VWR) were cleaned by soaking in freshly prepared Piranha solution (98% sulfuric acid: 30% hydrogen peroxide = 3:1 (v/v)) for 10 min. The slides were rinsed with deionized water twice and sonicated in ethanol for an additional 10 min. After rinsing with ethanol, the slides were dried at 60 °C. PS substrates were cut as 75 mm × 25 mm rectangles from regular, non-coated PS petri dishes (Becton, Dickinson and Company). The cut substrates were cleaned by sonication in ethanol for 10 min, rinsed with ethanol and then dried at 60 °C. The cleaned substrates were used immediately for patterning particles by dip coating.
Purified PNIPAM microgels or PS microspheres were dispersed in deionized water at a concentration of 0.02 wt% and then filtered with a 0.1 μm pore size membrane. The particle dispersion was maintained at 40 °C in an oil bath during the process. For micropatterning PNIPAM microgels or PS microspheres, the substrate to be patterned was attached to a programmable syringe pump (NE-1000, New Era), as shown in Scheme 1A. The substrate was immersed into a PNIPAM microgel dispersion, and after temperature stabilization the substrate was withdrawn upward at 90 μm/min or 50.9 mm/min continuously, or by alternating the two speeds depending on the experiment. The setup was kept in a plastic chamber to prevent interference from dust and airflow. To fabricate the surfaces coated with PNIPAM microgels at different densities, 10 μl of PNIPAM microgel dispersion (2 × 10-2, 2 × 10-3 or 2 × 10-4 wt% of PNIPAM particles in deionized water) was dropped on a horizontal cleaned substrate and air-dried at room temperature.
Scheme 1.
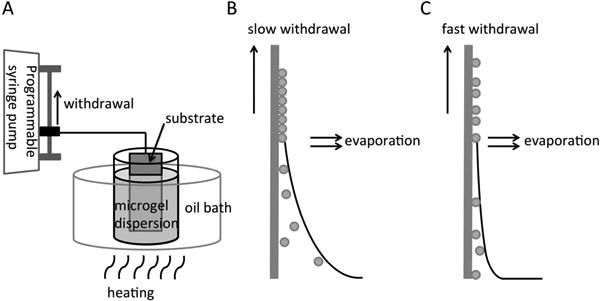
(A) The setup for the dip coating process employed for patterning PNIPAM microgels on substrates. The substrate was vertically immersed in a particle dispersion heated to 40 °C, and then withdrawn by a programmable syringe pump. (B) Particles deposited with slow withdrawal speeds (50.9 mm/min), and (C) Particles deposited with fast withdrawal speeds (90 μm/min). The sketches in (B) and (C) were adapted from the work of Dimitrov and Nagayama, and Watanabe et al.38, 41
The coated substrates were observed with a differential interference contrast (DIC) microscope (BX51, Olympus) equipped with a digital camera. The images were analyzed with ImageJ software to calculate the average dimension of the micropatterns. Statistically significant results between results were determined using ANOVA using Kruskal-Wallis approximation with a significant level of p<0.05. For higher magnifications, the samples were sputter-coated with gold and then observed with a focused ion beam scanning electron microscope (FIB-SEM, Zeiss Auriga). The surface topography of substrates dip coated with PNIPAM microgels was analyzed by an atomic force microscope (AFM) equipped with a heating unit (MFP-3D, Asylum). AFM imaging and analysis were performed and processed using the MFP-3D software in IgorPro (WaveMetrics Inc., Lake Oswego, OR). Gold-coated silicon nitride cantilevers (TR400PB, Olympus) with a force constant = 0.09 N/m and resonance frequency = 32 kHz were used to image the samples in air under ambient conditions, or in the cell culture medium heated to 37 °C. Samples were adhered to the Asylum Closed Fluid Cell or Bioheater with vacuum grease for imaging. A 10 μm × 10 μm area was scanned for each image at a scan rate of 0.5 Hz.
Cell seeding and adhesion on patterned substrates
NIH3T3 fibroblasts were obtained from ATCC and maintained in high glucose Dulbecco's modified Eagle's medium (HG-DMEM, Hyclone) supplemented with 10% FBS and 1% antibiotic-antimycotic solution (culture media) at 37 °C in humidified atmosphere containing 5% CO2. For cell seeding, NIH3T3 fibroblasts were trypsinized, collected via centrifugation (1500 rpm, 5 min, 25 °C), resuspended in 5 ml of culture media, and counted with a hemocytometer. Patterned substrates with PNIPAM microgels or PS microspheres (control) were seeded with fibroblasts at 12,000 cells/cm2 in 12-well plates. Substrates were incubated at 37 °C in humidified atmosphere of 5% CO2. After 24 hr, 48 hr, and 72 hr incubations (depending on the study), cell adhesion and behavior was observed with an inverted optical microscope (Nikon Eclipse Ti 2000) using 10×Nikon Plan Fluor objective (0.30 NA). For cell detachment studies, PNIPAM-coated substrates in 12-well plates were washed with culture media at 25 °C, and cell behavior was observed using an inverted optical microscope. Images were analyzed using NIS-Elements AR and ImageJ software.
Fluorescent staining of aligned cells on PNIPAM-coated substrate
The actin cytoskeleton, cell nuclei, and focal adhesion of fibroblasts seeded on patterned PNIPAM substrates were fluorescently stained using the Actin Cytoskeleton and Focal Adhesion Staining kit (Millipore). Adherent NIH3T3 fibroblasts on PNIPAM-coated substrates were rinsed with DPBS (1×) and fixed with 4% paraformaldehyde in DPBS (Gibco/Life Technologies) for 15 min at 25 °C. After rinsing with wash buffer (DPBS containing 0.05% Tween-20), cells were permeabilized with 0.1% Triton X-100 in DPBS for 5 min. Cells were then blocked with 1% BSA in DPBS for 30 min at room temperature. After which cells were incubated for 1 hr with Anti-Vinculin (primary) antibody (purified clone 7F9 diluted in blocking solution at room temperature (0.01 mg/mL in BSA-DPBS). Cells were washed thrice with wash buffer and incubated with secondary antibody, i.e., FITC-conjugated secondary antibody at 0.02 mg/mL in wash buffer) for 60 min along with TRITC-conjugated Phalloidin (0.006 mg/mL). Through this staining protocol, fibroblasts F-actin were detected using TRITC-conjugated phalloidin and focal contacts were revealed using anti-Vinculin monoclonal antibody followed by incubation with FITC-conjugated secondary antibody. For nuclei staining, cells were treated with DAPI (4′,6-diamidino-2-phenylindole) at 0.001 mg/mL for 5 min at room temperature, followed by extensive washing with wash buffer. Finally, fluorescent images of stained cells were obtained by an inverted fluorescent microscope using 20× Nikon Plan S Fluor objective (0.45 NA) (for high magnification imaging) and image was processed with NIS Elements AR software. Monochrome images of TRITC-conjugated Phalloidin, Anti-Vinculin and DAPI were overlaid and displayed in pseudocolor for all fixed cell samples.
Simultaneous cell-PNIPAM microgel imaging using SEM
Cells adherent on PNIPAM microgel patterned substrates 24 hour after cell seeding were fixed with 4% paraformaldehyde in DPBS. After fixing, cell substrates were sputter-coated with gold (Denton Vacuum Desk II) and imaged using Zeiss Auriga CrossBeam SEM-FIB instrument. Imaging was performed and processed using the SmartSEM software (Zeiss).
Water contact angle measurements
Static water contact angle was measured using captive bubble method16 with a VCA Optima XE system (AST Products, Inc., MA). Samples were immersed in deionized water in a glass chamber, and the contact angle was measured after placing an air bubble onto the sample surfaces. Measurements were conducted at ambient temperature (25 °C) or 37 °C.
Results and Discussion
Synthesis and temperature-responsiveness of PNIPAM microgels
The temperature-dependent size of PNIPAM microgels was analyzed using Dynamic Light Scattering (DLS) (See Supporting Information, Figure S1). The polydispersity of formed particles was consistently ~0.005 at 25 °C, indicating the narrow particle size distributions achieved using this method. The particles displayed temperature dependent size, with an average diameter of ~450 nm at 25 °C decreasing to ~200 nm at 40 °C as water is expelled from the particles. The VPTT of PNIPAM microgels was 31.3 °C, as determined by the inflection point of the Boltzmann sigmoid curve fit of the data (see Supporting Information, Figure S1). The VPTT is similar to the VPTT of PNIPAM hydrogels (31-32 °C) reported by Schild et al.23
Micropatterning of PNIPAM microgels on PS substrates
Dip coating is a well-established method for depositing particles onto substrates.37, 38, 41 Particles deposit through convective currents that form by evaporation from a rising meniscus.41 The convective currents carry particles into the region of the meniscus, and as the meniscus moves down the substrate, either by a falling liquid level or by lifting the substrate, a coating of particles is deposited onto the surface of the substrate. For dip coating to be effective, the meniscus must have a small contact angle with the substrate. As a result, aqueous particle suspensions have not been previously reported to coat effectively onto hydrophobic substrates, such as unmodified polystyrene. As a proof of concept, PS microspheres were dip coated on glass and PS substrates using the methodology described in Scheme 1. PS microspheres were successfully deposited on hydrophilic glass substrates but not on hydrophobic PS substrates (see Supporting Information, Figure S2). This is likely due to the high contact angle of PS (> 84°),42, 43 which results in little or no rising meniscus at the contact line when the PS substrate is dipped into aqueous solutions. A nearly flat meniscus prevents convective current formation that concentrates particles at the contact line. Surprisingly, PNIPAM microparticles were effectively deposited on both hydrophilic glass and hydrophobic PS using the same method (See Supporting Information Figure S2).
PNIPAM microgel dispersions were heated to 40 °C before dip coating, therefore microgels were in their collapsed state (diameter = 200 nm). Substrates were attached to a programmable pump then immersed in dispersions and vertically withdrawn at defined speeds. Substrates withdrawn at slower rates resulted in more particle deposition on substrates due to longer residence times, allowing for convection of particles to the interface (Scheme 1B). Alternatively, substrates withdrawn at higher speeds resulted in thinner meniscuses, resulting in lower densities of particle deposition on substrates (Scheme 1C).37, 38 To create surface micropatterns, substrates were withdrawn from particle dispersions at two alternating speeds, 90 μm/min and 50.9 mm/min. Upon vertical substrate withdrawal at these two alternating speeds, PNIPAM microgels assembled as striped patterns on substrates (Figure 1). The areas coated at slow withdrawal speeds, exhibiting dense particle concentrations, are referred to as “stripes” and the areas coated at fast substrate vertical withdrawal speeds with sparse particle deposition are termed “spacings”. By controlling the withdrawal distances, three sets of stripe/spacing micropatterns were achieved: 50 μm stripe/50 μm spacing (Figure 1A, 1D), 50 μm stripe/100 μm spacing (Figure 1B, 1E) and 100 μm stripe/100 μm spacing (Figure 1C, 1F).
Figure 1.
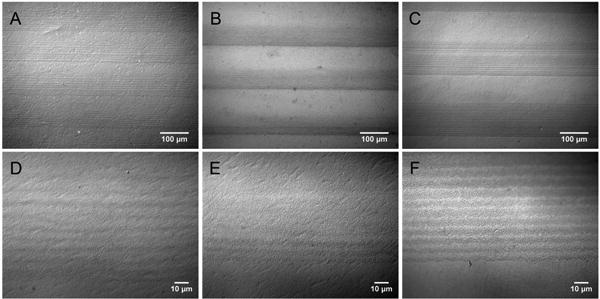
Light microscope images of PS substrates micropatterned with PNIPAM microgels via dip coating method at various resolutions. Substrates were coated via alternating speeds to create the micropatterns of 50 μm stripes/50 μm spacings at (A, D), 50 μm stripes/100 μm spacings (B, E) and 100 μm stripes/100 μm spacings (C, F). Samples were imaged under the magnification of 200X (A-C) and 1000X (D-F).
PNIPAM microgel deposition on PS substrates is shown in Figure 1. While PNIPAM microgels deposit on hydrophobic PS substrates via dip coating, hydrophobic PS microspheres do not (See Supporting Information Figure S2). This may be due to differences in stiffnesses of PNIPAM microgels and PS microparticles. Upon adsorption onto the PS surface, PNIPAM microgels deform substantially.44 Particle flattening results in greater contact areas between particles and the surface.44 Therefore, the increased contact area between PNIPAM microgels and PS substrate as well as the flexibility of PNIPAM chains within the microgel leads to increased PNIPAM microgel-substrate interactions due to Van der Waals and hydrophobic interactions.45 Unlike the PNIPAM microgels, PS particles are non-deformable, composed of polymer chains with restricted mobility due to the highly crosslinked, glassy characteristic of the polymer, leading to far less Van der Waals and hydrophobic bonding. Our observed results are supported by the recent work by Sorrell et al. where deformability of microgels was found to affect microgel-based thin film composition.46 In fact, the deformability of PNIPAM microgels enables these particles to be deposited onto a wide range of substrates that would otherwise be impossible to coat with rigid particles such as PS or silica. The PS substrate was chosen for the remainder of studies as it is commonly used in cell culture experiments.
To further examine the stripes/spacing patterns of PNIPAM microgels, optical microscope images (Figure 1) were analyzed using ImageJ software. Line profiles (perpendicular to the stripes) were used to define stripes as regions where the image had low average gray values (darker), and spacings as regions where images had high average gray values (brighter). Edges were selected manually as the points where the gray value changed inflects and changes at least 25 units within line profiles (See Supporting Information Figure S3). The size distribution of the stripes and spacings for different micropatterned substrates, i.e, 50 μm stripes/50 μm spacings, 50 μm stripes/100 μm spacings, and 100 μm stripes/100 μm spacings are shown in Figure 2. For the patterns with 50 μm stripes/100 μm spacings and 100 μm stripes/100 μm spacings, the stripe/spacing width distribution was found to be consistent with expected values as set in the withdrawal speed profiles (Figure 2A). This was not true for 50 μm stripes/50 μm spacings micropatterns of PNIPAM microgels, where the observed average stripe width (average stripes = 65 ± 9 μm) was larger than 50 μm, while the observed spacing was lower than 50 μm (average spacing = 45.4 ± 8.2 μm). These inconsistent results could be due to insufficient disruption of meniscuses at the 50 μm spacings (between stripes). The width of the spacings may not be sufficient to precisely break the meniscus when the withdrawal speed changes. As a result, PNIPAM microgel deposition within stripes does not terminate immediately when the substrate withdrawal speed increases to form the spacing.
Figure 2.
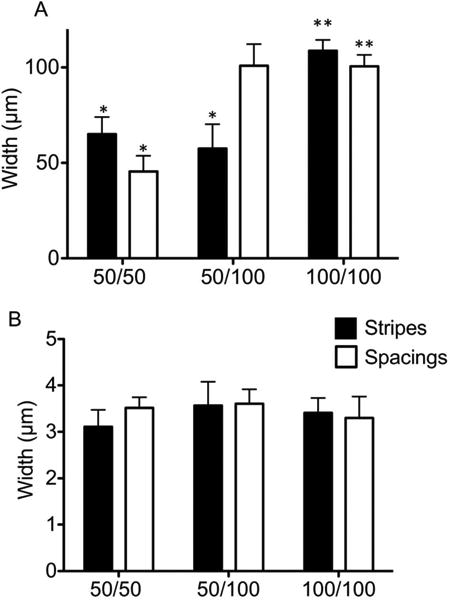
Histograms of (A) patterned PNIPAM microgels within 50 μm stripes/50 μm spacings, 50 μm stripes/100 μm spacings, and 100 μm stripes/100 μm spacings patterned samples and (B) stripe-width and spacings of spontaneously-formed patterns within stripe regions on substrates with 50 μm stripes/50 μm spacings, 50 μm stripes/100 μm spacings, and 100 μm stripes/100 μm spacings. * p<0.05 vs hypothetical value 50.0 calculated using one-sample t-test and ** p<0.05 vs hypothetical value 100.0 calculated using one-sample t-test. n=19, error bars represent standard deviation.
Within the patterns, fluctuations of gray values within the stripes were observed, indicating the existence of smaller stripes within the striped-patterned areas (Figure 1D-F). Images from Figure 1A-C were examined for these fluctuations both within the 50 μm and 100 μm stripes. The widths of these small stripes and the distances in between these stripes were 3-4 μm (Figure 2B) as analyzed by ImageJ (see Supporting Information, Figure S3). As substrates are withdrawn vertically at constant rates within the stripe regions, smaller stripes are formed spontaneously. This phenomenon is not uncommon when using dip coating methodologies for patterning microspheres such as silica or polystyrene on wettable surfaces. The mechanism for the stripes formation is the gravity-driven “stick-slip” motion of the contact line,37, 38 similar to the well known “coffee ring” phenomenon observed when a suspension is dried on a horizontal substrate.47, 48
Stability of PNIPAM microgel patterns upon temperature cycling
The stability of PNIPAM microgel patterns deposited on PS substrates was examined after thermal cycling. The PNIPAM-coated substrates with 50 μm stripes/50 μm spacings were immersed in cell culture media. Images (Figure 3) show that PNIPAM micropatterns remained unperturbed even after 3 days at 37 °C. In addition, the patterns were exposed to three temperature cycles (cooled to 25 °C for 1 hr every 24 hr) with no alteration in patterns. This observation confirms that PNIPAM microgels deposited on PS by the simple dip coating method were robust and did not release in tissue culture media even upon cycling the temperature above and below the VPTT of PNIPAM microgels.
Figure 3.
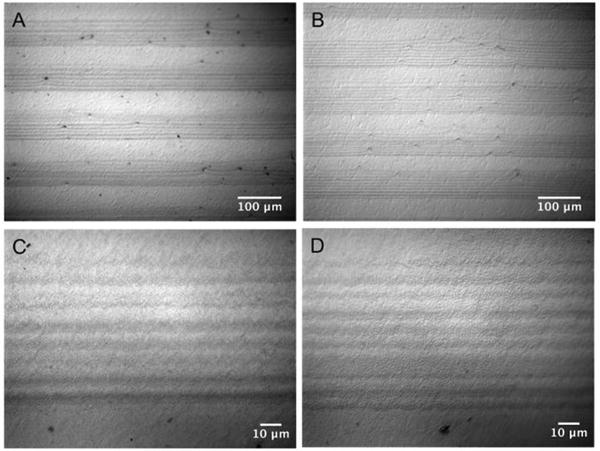
DIC microscope images of PS substrates coated with PNIPAM micropatterns of 50 μm stripes/50 μm spacings immersed in culture medium at 37 °C for 3 days (A, C) or after 3 temperature cycles of 23 hr at 37 °C/1 hr at 25 °C (B, D).
Topographical examination of PNIPAM micropatterns
PNIPAM micropatterned substrates were further examined using SEM and AFM to determine if PNIPAM patterns exist as monolayers or as multilayers, and also if there are topographical differences between stripe and spacing regions. SEM images (Figure 4A) showed densely packed particles in stripes and sparsely deposited particles in spacings. The thickness of PNIPAM micropatterns was further investigated by AFM (Figure 4B, C). AFM images revealed that particles deposited as a single layer all through the substrate, and the height of particles in both dense (stripes) regions and sparse (spacings) regions were very similar (Figure 4B, C). The height of particles on dry substrates at room temperature was ~80 nm as revealed by AFM, and the height increased to 120 nm when substrates were tested after incubation in culture medium at 37 °C due to swelling of the PNIPAM microgels (see Supporting Information, Figure S4 and Figure S5).
Figure 4.
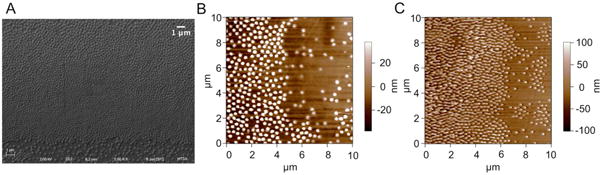
Images of substrates coated with PNIPAM of 50 μm stripes/50 μm spacings by (A) SEM, (B) AFM in air, and (C) AFM in cell culture media at 37 °C.
Cell seeding on patterned substrates
NIH3T3 fibroblast cell adhesion and alignment were examined on PNIPAM micropatterns. Fibroblasts have been previously cultured on micropatterned surfaces grafted with PNIPAM to obtain harvestable cell sheets or tissue constructs.14, 49, 50 Fibroblasts adhered to the PNIPAM-coated substrates generating cell patterns replicative of the stripes/spacings (Figure 5A-C) as observed in brightfield images. The thinnest cell stripes were formed on the substrate with 50 μm stripes/50 μm spacings micropatterns (Figure 5A), and most of the elongated fibroblasts aligned along the direction of the PNIPAM micropatterns. However, cell alignment was lost when the dimensions of PNIPAM micropatterns were increased to 100 μm stripes/100 μm spacings micropatterns or 50 μm stripes/100 μm spacings micropatterns (Figure 5B-C). A closer examination of this behavior using fluorescent staining of F-actin (Phalloidin, red), nuclei (DAPI, blue), and focal contacts (Vinculin, green) illustrated that the adhered fibroblasts were randomly aligned on 100 μm stripes/100 μm spacings micropatterns (Figure 5D) as well as 50 μm stripes/100 μm spacings micropatterns. This might be the result of reduced spatial constraints presented by wider stripe/spacing micropatterns so the fibroblasts can attach and spread freely within the striped micropatterns.
Figure 5.
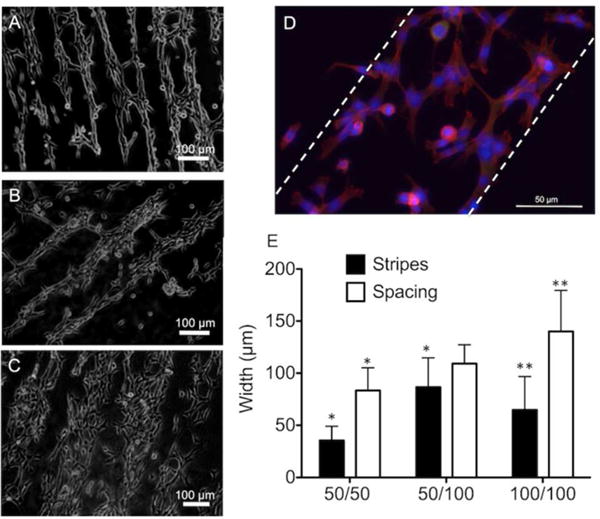
Representative optical micrographs of NIH3T3 fibroblasts patterns on micropatterned PS substrates with (A) 50 μm stripes/50 μm spacings, (B) 50 μm stripes/100 μm spacings, and (C) 100 μm stripes/100 μm spacings at 24 hr after seeding. The data presented herein is compiled from at least five different images captured on three independent days of experimentation. (D) Representative fluorescent images of adherent NIH3T3 fibroblasts on substrates with PNIPAM micropatterns of 100 μm stripes/100 μm spacings. Dashed lines indicate cell pattern. Actin and nuclei were stained with AlexaFluor568-phalloidin (red) and DAPI (blue), respectively. Cellular focal adhesions were stained with anti-vinculin fluorescein (green). (E) Histogram of cells adhered on patterned PNIPAM substrates with stripe widths and spacings of 50 μm stripes/50 μm spacings, 50 μm stripes/100 μm spacings, and 100 μm stripes/100 μm spacings. * p<0.05 versus hypothetical value 50.0 calculated using one-sample t-test and ** p<0.05 versus hypothetical value 100.0 calculated using one-sample t-test. n=13, error bars represent standard deviation.
Cellular morphologies on the three different PNIPAM patterned substrates (50 μm stripes/50 μm spacings (Figure 5A), 50 μm stripes/100 μm spacings (Figure 5B), and 100 μm stripes/100 μm spacing (Figure 5C)) were further characterized 24 hours after seeding. As plotted in Figure 5E, widths of cell stripes and spacings for different micropatterned substrates exhibited broad distributions. Also, the widths of PNIPAM microgel patterns and resulting cell stripes and cell spacings did not completely replicate each other. For example, the micropatterns employing 50 μm stripes/50 μm spacing, the stripes were 65.0 ± 9.0 μm in width (Figure 2A) but cell stripes on these substrates had an average width of 35.3 ± 13.8 μm (Figure 5E). Similarly, average PNIPAM spacings for 50 μm stripes/50 μm spacing samples were 45.5 ± 8.2 μm (Figure 2A), but the cell spacing corresponded to 83.4 ± 21.3 μm. To more closely examine these inconsistencies, fibroblast preference for stripes or spacings on PNIPAM microgels were further investigated. PNIPAM micropatterns with different stripe/spacing ratios were created on single PS substrates. The width of PNIPAM stripes was increased from 100 μm to 350 μm while keeping the spacing constant at 50 μm (Figure 6A-B). Upon cell seeding, it was observed that the entire PS substrates were uniformly patterned with cell stripes of 50 μm widths. These 50 μm wide stripes were separated by increasing distances of 100 μm to 350 μm (Figure 6C-D). Thus, cells preferentially adhered onto the spacings between PNIPAM stripes rather than on the stripes. A few cells were observed on PNIPAM stripes but these cells exhibited rounded morphologies, indicative of poor attachment onto the stripes. To further confirm that the spacings of the stripes/spacings pattern were the preferred cell adhesive substrate, PNIPAM micropatterned substrates were imaged 24 hours after seeding via high-resolution SEM. SEM can scan and image much smaller areas as compared to typical microscopy, but it generates high-resolution images detailing both cell structure as well as PNIPAM microgels in a single frame. Using SEM it was observed that fibroblasts did adhere to spacings, as cells were clearly observed to be surrounded by densely packed PNIPAM microgels (Figure 6E-F).
Figure 6.
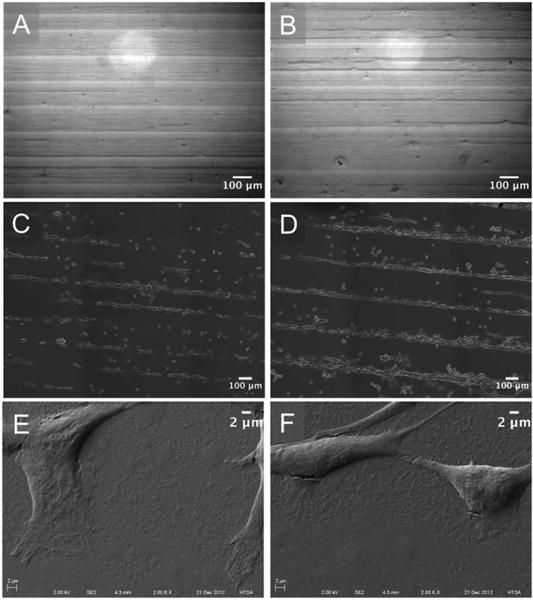
DIC microscope images of PNIPAM micropatterns on PS substrates (A-B) and the optical microscope images of NIH3T3 fibroblasts adhered on substrates after cell seeding (C-D) with PNIPAM micropatterns of 50 μm spacings with 100μm (A, C) or 200 μm stripes (B, D). (E-F) SEM images of fixed NIH3T3 cells on PNIPAM micropatterned substrates with100 μm stripes/100 μm spacings.
Our observations highlight that, in addition to chemical cues, a variety of material properties play roles in controlling cell-material interactions. In the current study, cells are able to sense differences between densely packed and sparsely distributed PNIPAM microgels patterned onto PS surfaces, and prefer to adhere to the substrate with sparsely deposited PNIPAM particles. This preferential cell adhesion on spacings could be due to the following reasons: differences in (a) topography, (b) hydrophobicity, or (c) PNIPAM density. Our data verifies that the stripes and spacings are compositionally similar, i.e. both regions are covered with monolayers of PNIPAM microgels (as seen by AFM images of patterned substrates) (Figure 4B,C) but at different densities. Height differences between ridges and grooves of the microgroove patterns, which have been shown previously to affect cell adhesion and patterning,14 are not significantly different between the two PNIPAM microgel regions. In order to investigate if a difference in hydrophobicity is responsible for the observed differences in cell adhesion, PNIPAM microgel-coated PS substrates were further characterized using contact angle measurements (Table 1). Importantly, the dimension of patterned PNIPAM microgels is in the sub-micrometer range, while the measured contact angles are representative of macroscopic surface characteristics of the patterned substrates. Also, PNIPAM is hydrophilic compared to the PS substrates even at physiological temperatures (37 °C, T > VPTT). Substrates coated with PNIPAM microgels exhibited lower contact angles compared to the PS substrates although substrates coated with sparsely deposited PNIPAM particles (representative of “spacing” in PNIPAM micropatterned region) had a greater contact angle (60.3°) than the densely patterned PNIPAM areas (representative of “stripes” on PNIPAM micropatterned substrate, 39.6°). The relative similarities of contact angles between the stripes and spacings is not surprising because surface contact angle is not sensitive to micron-sized differences in the patterns due to the macroscopic nature of the analytical technique. For PNIPAM-coated substrates, contact angle did not change significantly as the temperature was cycled between 25 °C and 37 °C. Again, due to the macroscopic nature of contact angle measurements, small (micron-scaled) variations in hydrophilicity due to temperature-induced dehydration of deposited PNIPAM microgels are not quantifiable. In addition, it has been reported previously that cell detachment from PNIPAM surfaces occurs irrespective of changes in contact angle, similar to our observations.20
Table 1.
Physical characteristics of different polystyrene substrates with or without PNIPAM microgels and the corresponding cell attachment.
| substrate | contact angle measurement(degree)c,f | cell adhesion(cells/cm2)d,f | cell detachment e | |
|---|---|---|---|---|
| 25 °C | 37 °C | |||
| plain PS | 77.6 ± 1.9 | 69.6 ± 1.1 | (5.1 ± 4.3) × 10 3 | No |
| densely-packed PNIPAM microgels on PS a | 39.6 ± 3.7 | 41.6 ± 3.9! | (6.6 ± 4.5) × 10 3 | Yes |
| sparsely-distributed PNIPAM microgels on PS b | 60.3 ± 2.8 | 61.6 ± 1.4 | (20.7 ± 11.4) × 10 3* | Yes |
| 50 μm stripes/50 μm spacings | 45.5 ± 3.9 | 42.8± 4.0 | (29.2 ± 8.0) × 10 3* | Yes |
| 50 μm stripes/100 μm spacings | 38.8 ± 2.7 | 41.7 ± 3.0 | (12.9 ± 5.0) × 10 3* | Yes |
| 100 μm stripes/100 μm spacings | 43.7 ±1.1 | 39.7 ± 1.6 | (15.4 ± 3.2) ×10 3* | Yes!! |
PS substrate dip coated with PNIPAM microgels at the withdrawal rate of 90 μm/min.
PS substrate dip coated with PNIPAM microgels at the withdwaral rate of 50.9 mm/min.
Static water contact angle using the captive bubble method. Measurements were conducted at ambient temperature (24.5 °C) or 37 °C by heating the chamber with a heating strip.
Cells were seeded on samples at 12000 cells/cm2 and incubated at 37 °C for 24 hours. These substrates were imaged using DIC and cell adhesion per unit area was calculated from at least 10 different optical micrographs using 3 different substrates per condition.
Cell detachment was performed by washing the substrate with tissue culture media at 25 °C.
Data represents mean ± st dev.
p<0.05 vs plain PS.
To further investigate if PNIPAM density is controlling cell adhesion of NIH3T3 fibroblasts to spacings (i.e., sparse PNIPAM microgel distributions) rather than stripes (dense PNIPAM microgel distributions), different PNIPAM microgel suspensions were deposited (using substrate withdrawal rates of 50.9 mm/min or 90 μm/min) on PS substrates. Fibroblasts were then seeded on the PNIPAM-modified substrates. PNIPAM density was observed to substantially effect cell adhesion. On hydrophilic PS substrates coated with high densities of PNIPAM microgels (obtained via slow substrate withdrawal rates (50.9 mm/min) from PNIPAM microgel suspensions) (contact angle, 39.6°), very few cells attached (Table 1) but as the PNIPAM microgel concentrations were reduced further (obtained via substrate withdrawal rates of 90 μm/min), generating hydrophobic substrates (contact angle, 60.3°), greater fibroblast adhesion and spreading was observed. These results are similar to those reported by Xue et al. where the differences in reversible cellular adhesion were studied on substrates with different PNIPAM grafting densities.51 The same group also showed that differences in cell adhesion correlated directly with fibronectin adsorption onto grafted PNIPAM substrates.51
Surface hydrophobicity has been shown to control adsorption of a variety of proteins, which allow for cell adhesion through integrin-receptor binding.52 The cell-material interactions observed on dip coated micropatterned PNIPAM substrates here may be the result of differential protein adsorption of the stripes and spacings. Differences in PNIPAM densities on PS substrates affect surface hydrophobicity as measured by contact angle measurements (Table 1). The spacings in dip coated micropatterned substrates are relatively hydrophobic (contact angle = 60.3°, Table 1), which has been shown to positively affect cell adhesive protein adsorption available from the cell culture media.53, 54 In contrast, the “stripes” in dip coated micropatterned substrates are more hydrophilic (contact angle = 39.6°), resulting in less cell adhesive protein adsorption during cell seeding due to the presence of inhibitory water monolayers.55, 56 Therefore, surface hydrophobicity via PNIPAM density is likely to be responsible for generating cell micropatterns on dip coated PNIPAM microgel substrates, where preferential cell adhesion is observed only on spacings immediately after seeding.
Cell proliferation and temperature-responsive detachment
Fibroblasts adhered to dip coated PS substrates and form cell patterns for culture periods up to 24 hours, likely due to preferential protein adsorption. However, at time points longer than 24 hours, patterns were gradually lost (Figure 7A-C). Specifically, when seeded on micropatterned substrate composed of 100 μm stripes/100 μm spacings and imaged longitudinally at 24 hr, 48 hr, and 72 hr, fibroblast density increased over time and gaps that existed between cell stripes at 24 hours decreased as cells proliferated to cover the entire substrate. After 72 hr, cells formed confluent sheets on PNIPAM-coated substrates. Thus, while the fibroblasts adhered onto the spacings (with sparsely deposited PNIPAM microgels) initially, after extended culture times, cells were capable of adhering onto PNIPAM stripes (with densely packed PNIPAM microgels). The mechanism for altered cell adhesion over time is unclear. It is possible that attached fibroblasts secrete cell adhesive proteins in addition to the cohort available in cell culture media. These proteins may include vitronectin, laminin, collagen, and fibronectin,57 which have different binding affinities to the stripes and spacings and also may be secreted in different concentrations with temporal variations. The cell-produced proteins may, therefore, adsorb differentially to the micropatterned substrate areas, making the entire surface, not just the spacings, more amenable to cell adhesion, resulting in evolution of confluent cell sheets.
Figure 7.
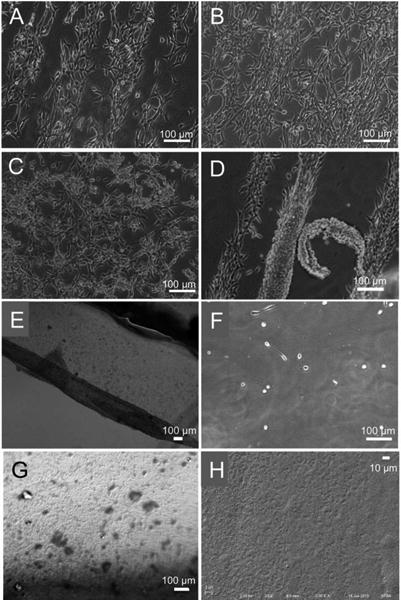
Representative optical images of adherent NIH3T3 fibroblasts on PS substrates coated with PNIPAM micropatterns of 100 μm stripes/100 μm spacings at 37 °C for 24 hr (A), 48 hr (B) and 72 hr (C), and detached fibroblasts upon lowering the temperature to 25 °C (D, E). On the substrate after cell detachment (F), the PNIPAM particles were observed under DIC microscope (G) and SEM (H).
To examine temperature-responsive detachment of the developed cell sheets, we washed patterned cell substrates with tissue culture medium at 25 °C. Patterned fibroblasts stripes started to detach within 3 minutes (Figure 7D) and all cells detached 5 minutes after lowering the temperature. Confluent cell layers established by 72 hours of cell seeding behaved similarly when washed with tissue culture medium at 25 °C, i.e., fibroblasts started to detach from the substrate as soon as the temperature was lowered (within 3 min) (Figure 7D). Complete detachment of cells implies that the fibroblasts could detach from both the spacings and stripes. Thus, both sparsely deposited as well as densely packed PNIPAM microgels imparted thermal responsiveness to the coated substrates that, in turn, facilitated complete cell sheet detachment. Cell detachment from dip coated PNIPAM substrates was found to be faster than most techniques used to form cell sheets via the thermoresponsiveness of PNIPAM, i.e. spin coating (12 min), UV crosslinking (30 min), sol-gel processes (2 hours), and electron beam polymerization (1 hour).58 This faster temperature response compared to spin-coated or air-dried PNIPAM polymers is likely due to the PNIPAM microgels providing much more rapid response to temperature fluctuations due to high surface area-to-volume ratios.24 We also investigated whether cells detached with patterned PNIPAM microgels upon temperature cycling. PNIPAM microgel patterned substrates seeded with fibroblasts (after 48 hours) were washed with tissue cultre medium at 25 °C. It should be noted that we confirmed via imaging that temperature cylcing between 25 °C and 37 °C does not detch PNIPM microgels from dip coated PNIPAM substrate (Figure 3). However, we also analyzed patterns after cell detachment using an optical microscope (Figure 7E) and a scanning electron microscope (Figure 7F). The PNIPAM microgels were retained on the surface even after temperature-responsive cell detachment.
To further support that PNIPAM microgels were responsible for cell detachment, unmodified substrates (PS substrates alone) and substrates coated with PNIPAM microgels at constant 90 μm/min and 50.9 mm/min withdrawal speeds, respectively, were also tested. Figure 8 shows that 24 hr after seeding, fibroblasts adhered to unmodified PS substrates sparsely, but did not cover the entire surface. However, on PNIPAM-coated substrates, cell density was significantly higher (Figure 8E, F). Note that on substrates coated at 90 μm/min withdrawal speeds, fibroblasts adhered onto spontaneously formed PNIPAM micropatterns. After lowering the temperature, cells attached to unmodified PS substrates remained adherent, but cells seeded onto PNIPAM substrates detached readily, due to the thermal responsiveness of the underlying PNIPAM patterns. Only a few non-adherent cells with rounded morphologies were observed on the PNIPAM-coated substrates after one temperature cycling. Thus, using this system, the thermal responsiveness of PNIPAM microgels was harnessed to form cell sheets in an enzyme-free manner.
Figure 8.
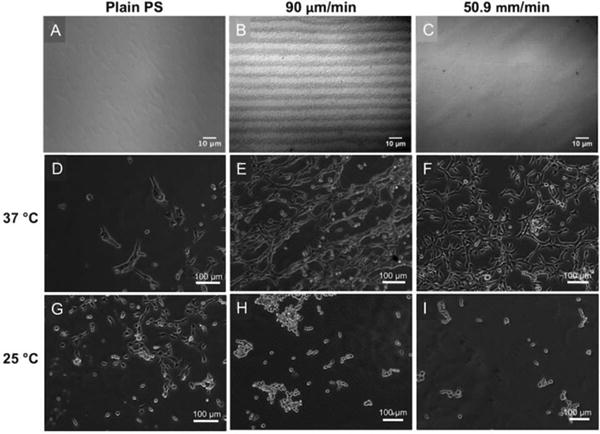
DIC microscopy images of the substrates (A-C) and representative optical microscope images of NIH3T3 fibroblast cells cultured on the substrates at 37 °C for 24 hr (D-F) and after lowering the temperature to 25 °C (G-I). Plain PS substrate (A, D, G) and substrates coated with PNIPAM microgels at constant speed of 90 μm/min (B, E, H) and 50.9 mm/min (C, F, I) were compared.
Conclusions
PNIPAM micropatterns were created on PS substrates using a simple and low cost approach. Through a rate-controlled dip coating process, PNIPAM microgels self-assembled into regularly spaced stripes with variable, yet controllable, widths and spacings. When fibroblasts were seeded on patterned substrates, cells preferentially adhered within spacings consisting of sparsely distributed PNIPAM microgels but not on densely packed PNIPAM stripes. Over three days, fibroblasts proliferated and eventually became confluent and detached as a single cell sheet upon temperature reduction, due to the thermoresponsiveness of PNIPAM, while the deposited PNIPAM micropatterns remained intact. The current work presents a controllable, template-free, low-cost micropatterning technique to pattern soft microparticles on both hydrophobic and hydrophilic substrates. PNIPAM micropatterns may prove useful for a multitude of applications. For example, surface-modified PNIPAM microgels (using chemical moieties such as acids, amines, or even full proteins) can also be micropatterned using this simple technique. These functional micropatterned PNIPAM substrates could be utilized for fundamental cell-material interaction studies where cell behaviors can be controlled spatially as well as temporally via the underlying PNIPAM microgels.45
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the National Institutes of Health (P30A1078498 and 5R01AI080770) and the University of Rochester for funding. We would also like to thank Graham Marsh and Dr. Richard E. Waugh at the University of Rochester for help with Atomic Force Microscopy. Also, authors wish to thank Erin Antonienko for help with data tabulation, and Xuefei Zhang for the assistance with image processing.
Abbreviations
- TRITC
Tetramethylrhodamine-5-(and 6)-isothiocyanate
- FITC
Fluorescein isothiocyanate
Footnotes
Notes: The authors declare no competing financial interest.
Supporting Information: Temperature dependence of PNIPAM particle size, images of PNIPAM and PS particles deposited on glass and PS substrate, image analysis for calculating the widths of the spontaneously formed PNIPAM stripes, AFM images and the cross section profiles, images of fibroblast cells on substrates coated with different densities of PNIPAM particles. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rydholm AE, Held NL, Benoit DS, Bowman CN, Anseth KS. Modifying Network Chemistry in Thiol-Acrylate Photopolymers Through Postpolymerization Functionalization to Control Cell-Material Interactions. J Biomed Mater Res A. 2008;86:23–30. doi: 10.1002/jbm.a.31526. [DOI] [PubMed] [Google Scholar]
- 2.Khire SV, Benoit DSW, Anseth KS, Bowman CN. Ultrathin Gradient Films Using Thiol-Ene Polymerizations. J Poly Sci. 2006;44:7027–7039. [Google Scholar]
- 3.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric Control of Cell Life and Death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 4.Barbucci R, Lamponi S, Magnani A, Pasqui D. Micropatterned Surfaces for the Control of Endothelial Cell Behaviour. Biomol Eng. 2002;19:161–170. doi: 10.1016/s1389-0344(02)00022-9. [DOI] [PubMed] [Google Scholar]
- 5.Gauvreau V, Laroche G. Micropattern Printing of Adhesion, Spreading, and Migration Peptides on Poly(tetrafluoroethylene) Films to Promote Endothelialization. Bioconj Chem. 2005;16:1088–1097. doi: 10.1021/bc049717s. [DOI] [PubMed] [Google Scholar]
- 6.Guillemot F, Souquet A, Catros S, Guillotin B, Lopez J, Faucon M, Pippenger B, Bareille R, Rémy M, Bellance S, Chabassier P, Fricain JC, Amédée J. High-Throughput Laser Printing of Cells and Biomaterials for Tissue Engineering. Acta Biomater. 2010;6:2494–2500. doi: 10.1016/j.actbio.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 7.Wilson K, Stancescu M, Das M, Rumsey J, Hickman J. Direct Patterning of Coplanar Polyethylene Glycol Alkylsilane Monolayers by Deep-Ultraviolet Photolithography as a General Method for High Fidelity, Long-term Cell Patterning and Culture. J Vac Sci Tech B. 2011;29:021020–021010. doi: 10.1116/1.3549127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heskins M, Guillet JE. Solution Properties of Poly(N-isopropylacrylamide) J Macromol Sci Part A. 1968;2:1441–1455. [Google Scholar]
- 9.Otake K, Inomata H, Konno M, Saito S. Thermal Analysis of the Volume Phase Transition with N-isopropylacrylamide Gels. Macromolecules. 1990;23:283–289. [Google Scholar]
- 10.Wu C. A Comparison Between the “Coil-to-Globule” Transition of Linear Chains and the “Volume Phase Transition” of Spherical Microgels. Polymer. 1998;39:4609–4619. [Google Scholar]
- 11.Isenberg BC, Tsuda Y, Williams C, Shimizu T, Yamato M, Okano T, Wong JY. A Thermoresponsive, Microtextured Substrate for Cell Sheet Engineering with Defined Structural Organization. Biomaterials. 2008;29:2565–2572. doi: 10.1016/j.biomaterials.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong B, Choi JS, Jeon S, Choi IS. The Control of Cell Adhesion and Detachment on Thin Films of Thermoresponsive Poly[(N-isopropylacrylamide)-r-((3-(methacryloylamino)propyl)-dimethyl(3-sulfopropyl)ammonium hydroxide)] Biomaterials. 2009;30:5514–5522. doi: 10.1016/j.biomaterials.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Patel NG, Cavicchia JP, Zhang G, Zhang Newby Bm. Rapid Cell Sheet Detachment Using Spin-Coated pNIPAAm Films Retained on Surfaces by an Aminopropyltriethoxysilane Network. Acta Biomater. 2012;8:2559–2567. doi: 10.1016/j.actbio.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 14.Tekin H, Ozaydin-Ince G, Tsinman T, Gleason KK, Langer R, Khademhosseini A, Demirel MC. Responsive Microgrooves for the Formation of Harvestable Tissue Constructs. Langmuir. 2011;27:5671–5679. doi: 10.1021/la200183x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Yamato M, Shimizu T, Sekine H, Ohashi K, Kanzaki M, Ohki T, Nishida K, Okano T. Reconstruction of Functional Tissues with Cell Sheet Engineering. Biomaterials. 2007;28:5033–5043. doi: 10.1016/j.biomaterials.2007.07.052. [DOI] [PubMed] [Google Scholar]
- 16.Akiyama Y, Kikuchi A, Yamato M, Okano T. Ultrathin Poly(N-isopropylacrylamide) Grafted Layer on Polystyrene Surfaces for Cell Adhesion/Detachment Control. Langmuir. 2004;20:5506–5511. doi: 10.1021/la036139f. [DOI] [PubMed] [Google Scholar]
- 17.Okano T, Yamada N, Sakai H, Sakurai Y. A Novel Recovery System for Cultured Cells Using Plasma-Rreated Polystyrene Dishes Grafted with poly(N-isopropylacrylamide) J Biomed Mater Res. 1993;27:1243–1251. doi: 10.1002/jbm.820271005. [DOI] [PubMed] [Google Scholar]
- 18.Yamada N, Okano T, Sakai H, Karikusa F, Sawasaki Y, Sakurai Y. Thermo-Responsive Polymeric Surfaces; Control of Attachment and Detachment of Cultured Cells. Die Makromolekulare Chemie. 1990;11:571–576. [Google Scholar]
- 19.Kwon OH, Kikuchi A, Yamato M, Sakurai Y, Okano T. Rapid Cell Sheet Detachment from Poly(N-isopropylacrylamide)-Grafted Porous Cell Culture Membranes. J Biomed Mater Res. 2000;50:82–89. doi: 10.1002/(sici)1097-4636(200004)50:1<82::aid-jbm12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi H, Nakayama M, Yamato M, Okano T. Controlled Chain Length and Graft Density of Thermoresponsive Polymer Brushes for Optimizing Cell Sheet Harvest. Biomacromolecules. 2010;11:1991–1999. doi: 10.1021/bm100342e. [DOI] [PubMed] [Google Scholar]
- 21.da Silva RMP, Mano JF, Reis RL. Smart Thermoresponsive Coatings and Surfaces for Tissue Engineering: Switching Cell-Material Boundaries. Trends Biotech. 2007;25:577–583. doi: 10.1016/j.tibtech.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Kumashiro Y, Yamato M, Okano T. Cell Attachment–Detachment Control on Temperature-Responsive Thin Surfaces for Novel Tissue Engineering. Ann Biomed Eng. 2010;38:1977–1988. doi: 10.1007/s10439-010-0035-1. [DOI] [PubMed] [Google Scholar]
- 23.Schild HG. Poly(N-isopropylacrylamide): Experiment, Theory and Application. Prog Polym Sci. 1992;17:163–249. [Google Scholar]
- 24.Pelton R. Temperature-Sensitive Aqueous Microgels. Adv Colloid Interface Sci. 2000;85:1–33. doi: 10.1016/s0001-8686(99)00023-8. [DOI] [PubMed] [Google Scholar]
- 25.Weng H, Zhou J, Tang L, Hu Z. Tissue Responses to Thermally-Responsive Hydrogel Nanoparticles. J Biomater Sci Polym Ed. 2004;15:1167–1180. doi: 10.1163/1568562041753106. [DOI] [PubMed] [Google Scholar]
- 26.Reese CE, Mikhonin AV, Kamenjicki M, Tikhonov A, Asher SA. Nanogel Nanosecond Photonic Crystal Optical Switching. J Am Chem Soc. 2004;126:1493–1496. doi: 10.1021/ja037118a. [DOI] [PubMed] [Google Scholar]
- 27.Kawasaki H, Sasaki S, Maeda H. Effect of Introduced Electric Charge on the Volume Phase Transition of N-isopropylacrylamide Gels. J Phys Chem B. 1997;101:4184–4187. [Google Scholar]
- 28.Kawaguchi H, Fujimoto K. Smart Latexes for Bioseparation. Bioseparation. 1998;7:253–258. doi: 10.1023/a:1008055211667. [DOI] [PubMed] [Google Scholar]
- 29.Lopez VC, Hadgraft J, Snowden MJ. The Use of Colloidal Microgels as a (Trans)dermal Drug Delivery System. Int J Pharm. 2005;292:137–147. doi: 10.1016/j.ijpharm.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 30.Oh JK, Drumright R, Siegwart DJ, Matyjaszewski K. The Development of Microgels/Nanogels for Drug Delivery Applications. Prog Polym Sci. 2008;33:448–477. [Google Scholar]
- Kim J, Nayak S, Lyon LA. Bioresponsive Hydrogel Microlenses. J Am Chem Soc. 2005;127:9588–9592. doi: 10.1021/ja0519076. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt S, Motschmann H, Hellweg T, von Klitzing R. Thermoresponsive Surfaces by Spin-Coating of PNIPAM-co-PAA Microgels: A Combined AFM and Ellipsometry Study. Polymer. 2008;49:749–756. [Google Scholar]
- 33.Tsuji S, Kawaguchi H. Colored thin films prepared from hydrogel microspheres. Langmuir. 2005;21:8439–8442. doi: 10.1021/la050271t. [DOI] [PubMed] [Google Scholar]
- 34.Tsuji S, Kawaguchi H. Self-Assembly of poly(N-isopropylacrylamide)-Carrying Microspheres Into Two-Dimensional Colloidal Arrays. Langmuir. 2005;21:2434–2437. doi: 10.1021/la047477z. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt S, Zeiser M, Hellweg T, Duschl C, Fery A, Möhwald H. Adhesion and Mechanical Properties of PNIPAM Microgel Films and Their Potential Use as Switchable Cell Culture Substrates. Adv Funct Mater. 2010;20:3235–3243. [Google Scholar]
- 36.Nerapusri V, Keddie JL, Vincent B, Bushnak IA. Swelling and Deswelling of Adsorbed Microgel Monolayers Triggered by Changes in Temperature, pH, and Electrolyte Concentration. Langmuir. 2006;22:5036–5041. doi: 10.1021/la060252z. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh M, Fan F, Stebe KJ. Spontaneous Pattern Formation by Dip Coating of Colloidal Suspensions on Homogeneous Surfaces. Langmuir. 2007;23:2180–2183. doi: 10.1021/la062150e. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe S, Inukai K, Mizuta S, Miyahara MT. Mechanism for Stripe Pattern Formation on Hydrophilic Surfaces by Using Convective Self-Assembly. Langmuir. 2009;25:7287–7295. doi: 10.1021/la900315h. [DOI] [PubMed] [Google Scholar]
- 39.Mino Y, Watanabe S, Miyahara MT. Colloidal Stripe Pattern with Controlled Periodicity by Convective Self-Assembly with Liquid-Level Manipulation. ACS Applied Materials & Interfaces. 2012;4:3184–3190. doi: 10.1021/am300526g. [DOI] [PubMed] [Google Scholar]
- 40.Garcia A, Marquez M, Cai T, Rosario R, Hu Z, Gust D, Hayes M, Vail SA, Park CD. Photo-, Thermally, and pH-Responsive Microgels†. Langmuir. 2006;23:224–229. doi: 10.1021/la061632n. [DOI] [PubMed] [Google Scholar]
- 41.Dimitrov AS, Nagayama K. Continuous Convective Assembling of Fine Particles Into Two-Dimensional Arrays on Solid Surfaces. Langmuir. 1996;12:1303–1311. [Google Scholar]
- 42.Dann JR. Forces Involved in the Adhesive Process: I. Critical Surface Tensions of Polymeric Solids as Determined with Polar Liquids. J Colloid Interface Sci. 1970;32:302–320. [Google Scholar]
- 43.Omenyi SN, Neumann AW, van Oss CJ. Attraction and Repulsion of Solid Particles by Solidification Fronts I. Thermodynamic Effects. J App Phys. 1981;52:789–795. [Google Scholar]
- 44.Hofl S, Zitzler L, Hellweg T, Herminghaus S, Mugele F. Volume Phase Transition of “Smart” Microgels in Bulk Solution and Adsorbed at an Interface: A Combined AFM, Dynamic Light, and Small Angle Neutron Scattering Study. Polymer. 2007;48:245–254. [Google Scholar]
- 45.Guan Y, Zhang Y. Microgels for Biomedical Applications: From Dispersed Particles to 3D Assembly. Soft Matter. 2011;7:6375–6384. [Google Scholar]
- 46.Sorrell CD, Lyon LA. Deformation Controlled Assembly of Binary Microgel Thin Films. Langmuir. 2008;24:7216–7222. doi: 10.1021/la800092q. [DOI] [PubMed] [Google Scholar]
- 47.Dushkin CD, Yoshimura H, Nagayama K. Nucleation and Growth of Two-Dimensional Colloidal Crystals. Chem Phys Lett. 1993;204:455–460. [Google Scholar]
- 48.Deegan RD, Bakajin O, Dupont TF, Huber G, Nagel SR, Witten TA. Capillary Flow as the Cause of Ring Stains from Dried Liquid Drops. Nature. 1997;389:827–829. [Google Scholar]
- 49.Takahashi H, Nakayama M, Itoga K, Yamato M, Okano T. Micropatterned Thermoresponsive Polymer Brush Surfaces for Fabricating Cell Sheets with Well-Controlled Crientational Structures. Biomacromolecules. 2011;12:1414–1418. doi: 10.1021/bm2000956. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi H, Nakayama M, Shimizu T, Yamato M, Okano T. Anisotropic Cell Sheets for Constructing Three-Dimensional Tissue with Well-Organized Cell Orientation. Biomaterials. 2011;32:8830–8838. doi: 10.1016/j.biomaterials.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Xue C, Choi BC, Choi S, Braun PV, Leckband DE. Protein Adsorption Modes Determine Reversible Cell Attachment on Poly(N-isopropyl acrylamide) Brushes. Adv Funct Mater. 2012;22:2394–2401. [Google Scholar]
- 52.Horbett Thomas A. Biomaterials: Interfacial Phenomena and Applications. Vol. 199. American Chemical Society; Washington D.C: 1982. Protein Adsorption on Biomaterials; pp. 233–244. [Google Scholar]
- 53.Xu FJ, Zhong SP, Yung LY, Kang ET, Neoh KG. Surface-Active and Stimuli-Responsive Polymer--si(100) Hybrids from Surface-Initiated Atom Transfer Radical Polymerization for Control of Cell Adhesion. Biomacromolecules. 2004;5:2392–2403. doi: 10.1021/bm049675a. [DOI] [PubMed] [Google Scholar]
- 54.Mizutani A, Kikuchi A, Yamato M, Kanazawa H, Okano T. Preparation of Thermoresponsive Polymer Brush Surfaces and Their Interaction with Cells. Biomaterials. 2008;29:2073–2081. doi: 10.1016/j.biomaterials.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Zhu Y, Li B, Gao C. Fabrication of Thermoresponsive Polymer Gradients for Study of Cell Adhesion and Detachment. Langmuir. 2008;24:13632–13639. doi: 10.1021/la802556e. [DOI] [PubMed] [Google Scholar]
- 56.Matsuno A, Fujimaki T, Mizutani A, Ide F, Tanaka H, Asano S, Miyawaki S, Uno T, Tanaka J, Nakaguchi H, Sasaki M, Murakami M, Yamazaki K, Ishida Y. Disappearance of Gadolinium Enhancement in a Chemoresistant Astrocytoma of the Tectum After High-Dose Interferon Beta. Tumori. 2008;94:853–855. doi: 10.1177/030089160809400614. [DOI] [PubMed] [Google Scholar]
- 57.Morita H, Yasumitsu H, Watanabe Y, Miyazaki K, Umeda M. Characterization of Cell-Adhesive Proteins Secreted by Cell Lines Growing in Protein-and Lipid-Free Synthetic Medium: Mouse L.P3 Cells Secrete a Procollagen Molecule with Potent Cell-Attachment Activity. Cell Struct Funct. 1993;18:61–72. doi: 10.1247/csf.18.61. [DOI] [PubMed] [Google Scholar]
- 58.Nash ME, Carroll WM, Velasco D, Gomez J, Gorelov AV, Elezov D, Gallardo A, Rochev YA, Elvira C. Synthesis and Characterization of a Novel Thermoresponsive Copolymer Series and Their Application in Cell and Cell Sheet Regeneration. J Biomater Sci Polym Ed. 2013;24:253–268. doi: 10.1080/09205063.2012.690274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


