Abstract
Wounding, apoptosis, or infection can trigger a proliferative response in neighboring cells to replace damaged tissue. Studies in Drosophila have implicated Jun N-terminal kinase (JNK)-dependent activation of Yorkie (Yki) as essential to regeneration-associated growth, as well as growth associated with neoplastic tumors. Yki is a transcriptional co-activator that is inhibited by Hippo signaling, a conserved pathway that regulates growth. We found identified a conserved mechanism by which JNK regulated Hippo signaling. Genetic studies in Drosophila identified Jub (also known as Ajuba LIM protein) as required for JNK-mediated activation of Yki, and showed that Jub contributed to wing regeneration after wounding and to tumor growth. Biochemical studies revealed that JNK promoted the phosphorylation of Ajuba family proteins in both Drosophila and mammalian cells. Binding studies in mammalian cells indicated that JNK increased binding between the Ajuba family proteins LIMD1 or WTIP and LATS1, a kinase within the Hippo pathway that inhibits the Yki homolog YAP. Moreover, JNK promoted binding of LIMD1 and LATS1 through direct phosphorylation of LIMD1. These results identify Ajuba family proteins as a conserved link between JNK and Hippo signaling, and imply that JNK increases Yki and YAP activity by promoting the binding of Ajuba family proteins to Warts and LATS.
Introduction
Many forms of tissue damage, including wounding, apoptosis or infection, can trigger a proliferative response in neighboring cells to replace damaged tissue (1, 2). This regenerative growth requires activation of the c-Jun N-terminal kinase (JNK) signaling pathway (2–4). JNK is a stress-activated kinase, which is stimulated by diverse signals such as wounding, irradiation, or oxidation, and which induces diverse biological responses, including cytoskeleton modulation, apoptosis, and cell proliferation, leading to modulation of morphogenesis, inflammation, regeneration, and tumorigenesis (4, 5). Induction of apoptosis enables tissues to get rid of stressed or damaged cells, and is a frequent response to JNK activation. Nonetheless, JNK activity is also indispensable in some contexts for maintaining tissue homeostasis by triggering compensatory cell proliferation or stem cell activation in response to injury (3, 4, 6, 7). Moreover, in some contexts JNK-promoted growth can promote tumorigenesis. For example, avoidance of cell competition or activation of the Ras oncogene in Drosophila enables cellular insults associated with JNK activation and apoptosis to instead trigger JNK-dependent tumorigenesis (8–14), and JNK activation has also been associated with tumorigenesis in mammals (4, 15, 16).
JNK can influence several signaling pathways, some of which have been implicated in JNK-promoted cell proliferation (17). One essential response for JNK-promoted proliferation in several contexts is activation of Yorkie (Yki in Drosophila, YAP in vertebrates) (18, 19). Yki is a transcriptional co-activator controlled by Hippo signaling, a conserved pathway that regulates growth during development, regeneration, and oncogenesis (18, 20). Within the Hippo pathway (Fig 1A), Yki and YAP are inhibited by the kinase Warts (Wts in Drosophila and LATS in vertebrates), which suppress Yki and YAP activity by keeping it in cytoplasm. Several factors that regulate Wts have been identified, including the kinase Hippo (Hpo in Drosophila and MST in vertebrates) which gives the pathway its name. Activation of Yki and YAP leads to tissue overgrowth and tumor formation, whereas loss of Yki and YAP impairs growth and can lead to apoptosis (18, 20). JNK-dependent activation of Yki is required for regenerative growth in multiple Drosophila tissues, including larval imaginal discs and adult intestines (8, 21–23), and is also required for growth associated with certain neoplastic tumor suppressors (8, 9, 14). However, the mechanism by which JNK signaling promotes Yki activation has been unknown. Here, we have employed a combination of genetic and biochemical approaches to elucidate a molecular mechanism linking JNK activity to Yki regulation. Moreover, we establish that JNK can promote YAP activity in mammalian cells, and that it does so through a conserved molecular mechanism.
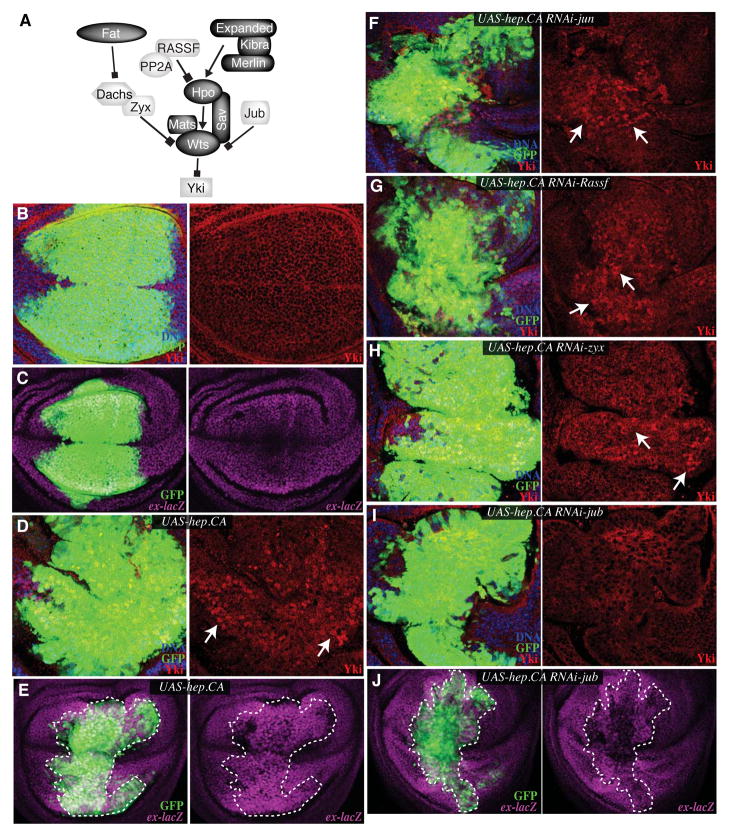
Fig. 1. Jnk activation of Yki in Drosophila wing discs requires Jub.
A) Simplified schematic of the Drosophila Hpo pathway. Proteins that inhibit Yki activity are indicated by dark shading and proteins that promote Yki activity are indicated by light shading. B–J) Wing discs stained for ex-lacZ (magenta) or Yki (red) and DNA (Hoechst, blue) with the salPE-Gal4 expression domain identified by expression from UAS-GFP (green). The right part of each panel shows a single channel from the stain to the left. White dashed lines outline the salPE-Gal4 expressing domain. Arrows point to examples of nuclear Yki. Discs are from animals with DroncI29 mutation, salPE-Gal4 and UAS-GFP transgenes, and B) control, C) ex-lacZ, D) UAS-hep.CA, E) ex-lacZ UAS-hep.CA, F) UAS-hep.CA UAS-RNAi-jun, G) UAS-hep.CA UAS-RNAi-Rassf, H) UAS-hep.CA UAS-RNAi-zyx, I) UAS-hep.CA UAS-RNAi-jub, J) ex-lacZ UAS-hep.CA UAS-RNAi-jub. Images are representative of at least 8 animals per genotype.
Results
JNK regulation of Drosophila Hippo signaling requires Ajuba LIM protein
To investigate the mechanism by which JNK regulates Hippo signaling, we took advantage of genetic approaches available in Drosophila. Expression of an activated form of the JNK kinase hemipterous (Hep.CA) in Drosophila wing discs, under the control of a Gal4 line expressed in the center of the wing (sal.PE-Gal4), results in strong Yki activation (8). Strong JNK activation normally promotes apoptosis; to reduce the apoptosis associated with Hep.CA expression, these flies also carry a mutation in the initiator caspase Dronc (DroncI29)( 24). Yki activation was reflected in these experiments both by the nuclear accumulation of Yki protein, and by the increased activity of a reporter for Yki’s transcriptional activity, ex-lacZ (Fig. 1B–E, S1H); this Yki activation was visible both in Hep.CA-expressing and neighboring cells. The biological effects of JNK activation are achieved through phosphorylation of target proteins (17), one of which is the transcription factor AP-1, a heterodimer of Fos and Jun proteins. RNAi directed against Drosophila Jun did not suppress the ability of Hep.CA expression to promote Yki activation (Figs 1F, S1A, S1H), suggesting that alternate targets of JNK were involved in mediating regulation of Yki activity. Although induction of Wingless and Decapentaplegic can contribute to proliferation in response to JNK activation (10, 25), prior studies suggest that these pathways do not contribute to autonomous activation of Yki (8, 26).
Because JNK promotes Yki activity, we introduced transgenes that should reduce Yki activity into flies with Hep.CA expression to examine the epistatic relationship between JNK and Hippo pathway components. The activation of Yki induced by Hep.CA expression was suppressed by activating the Hippo pathway through overexpressing Hpo or Wts, suggesting that Jnk promotes Yki activity at or upstream of Hpo and Wts (Fig. S1B–E, S1H) (8). Autonomous Yki activation was reduced by Hpo or Wts over-expression, although non-autonomous Yki activation was not completely blocked. We then examined whether depletion of components of the Hippo pathway that normally promote Yki activity could suppress Jnk-mediated Yki activation (Fig. 1A). Ras association family member (Rassf) interacts with a phosphatase complex and antagonizes Hippo activation (27, 28). RNAi directed against Rassf failed to suppress Yki activation (Figs 1G, S1F, S1H). Zyxin (Zyx) is a LIM domain protein that acts upstream of Wts within the Fat branch of the Hippo pathway (29); Zyx RNAi also failed to prevent activation of Yki by Jnk (Figs 1H, S1G, S1H). This lack of requirement for Zyx is consistent with observations that two other genes within the Fat branch of the Hippo pathway, dachs and fat, are not required for Yki activation induced by expression of Eiger (22). Ajuba LIM protein (Jub) is a LIM domain protein that interacts with both Wts and the scaffolding protein Salvador (Sav) (30). Knockdown of Jub by RNAi reduced the autonomous activation of Yki by Hep.CA (Fig 1I,J, S1H). Thus, JNK activation of Yki requires Jub, but not Zyx or Rassf.
Jub is required during wing regeneration and tumor growth
Regenerating wings are sensitive to reductions in Yki activity. For example, yki is normally recessive, because loss of one copy of yki has no discernible effects on normal wing growth, but heterozygosity for yki impairs wing regeneration after genetic ablation of the developing larval wing (8, 22). If Jub is normally important for Jnk-mediated Yki activation in vivo, then, given the essential role of Jnk in regeneration, regenerating wings might also exhibit sensitivity to Jub abundance. Indeed, heterozygosity for jub normally has no effect on wing growth or rates of development (Fig S2); however, loss of one copy of jub reduced the growth of regenerating wings (Fig. 2A–F).
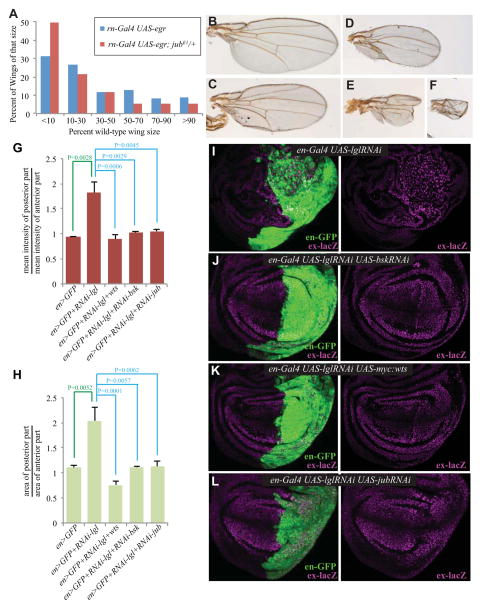
Fig. 2. Jub is required for Drosophila wing regeneration and for neoplastic tumor growth in wing discs with lgl knock-down.
A) Distribution of adult wing sizes in rn-Gal4 UAS-egr tubGal80ts (N=78) or rn-Gal4 UAS-egr tubGal80ts jubE1/+ (N=105) flies after larval wing ablation and recovery. B–F) Representative wings of 100% (B), 70% (C), 50% (D), 30% (E) and 10% (F) wild-type size. G) Quantification of the ratios of mean intensity of ex-lacZ expression in posterior to anterior compartments for the indicated genotypes (N=3 discs per genotype). H) Quantification of the ratios of area of the posterior to anterior compartments for the indicated genotypes (N=3 discs per genotype). Error bars indicate standard error, and P values less than 0.05 are shown. I–L) Wing discs stained for ex-lacz (β-gal, magenta) and with the posterior marked by expression of en-Gal4 UAS-GFP (green). The right part of each panel shows the ex-lacZ only stain from the image to the left. Discs are from animals with ex-lacZ, en-Gal4 and UAS-GFP transgenes and I) UAS-lglRNAi, J) UAS-lglRNAi UAS-bskRNAi, K) UAS-lglRNAi UAS-myc:wts, L) UAS-lglRNAi UAS-jubRNAi.
Loss of Lethal giant larvae (Lgl) in wing discs causes disruption of apical-basal cell polarity and formation of neoplastic tumors. These tumors are associated with activation of both Jnk and Yki, which are required for the associated over-proliferation (Figs 2G–K)(8, 9, 31). Because Jnk promotes Yki activation in Lgl-depleted cells (Fig 2G, I, J)(8), we used this as an independent model to confirm the requirement for Jub in Jnk-mediated Yki activation in vivo. Indeed, jub RNAi in Lgl-depleted cells suppressed both Yki activation and tissue overgrowth (Fig 2G, H, L).
JNK regulation of Hippo signaling is conserved in mammalian cells
Because both Jnk and Hippo signaling are conserved from Drosophila to humans, and JNK-triggered cell proliferation has also been implicated in repair of tissue damage and tumor growth in mammals (4, 6, 15, 16), we investigated whether JNK regulation of Yki is conserved. Basal JNK activity is required for cell proliferation in mammalian cell lines (32). When we treated the human mammary epithelial cell line MCF10A with the JNK inhibitor SP600125 (33), phosphorylation of the mammalian Yki homolog YAP on a key regulatory site, Ser127, was increased (Fig. 3A). Treatment of cultured cells with the JNK activator Anisomycin (34) significantly decreased phosphorylation of Ser127 of YAP (Fig 3A), an effect that was reversed by SP600125 (Fig. 3A). Characterization of the phosphorylation of the JNK substrate c-Jun (Fig. S3A) confirmed the expected effects of these treatments on JNK activity, and the same conditions were used in all drug treatment experiments. Phosphorylation of Ser127 in YAP by the kinase LATS is a key step in Hippo signaling, which promotes cytoplasmic localization of YAP through interaction with 14-3-3 proteins (20). Conversely, loss of phosphorylation at Ser127 activates YAP by increasing its nuclear localization. Thus, these effects suggest that JNK can promote YAP activation in mammalian cells, just as it can promote Yki activation in Drosophila. This notion was further supported by assaying expression of the YAP target gene CTGF, which encodes connective tissue growth factor. CTGF expression was reduced by Jnk inhibition and increased by Jnk activation (Fig 3B).
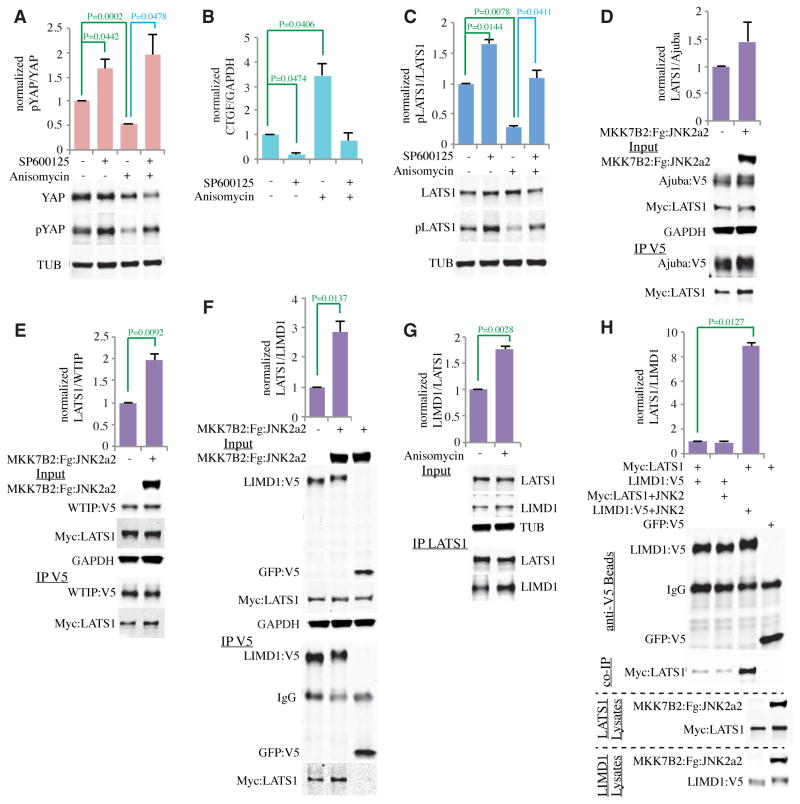
Fig. 3. JNK inhibits the Hippo pathway in mammalian cells and enhances LIMD1 and WTIP binding to LATS1.
A) Western blots on lysates of MCF10A cells treated with DMSO (control, indicated by −), SP600125, and/or Anisomycin as indicated (+), blotted using the indicated antisera. TUB is a loading control. Histograms show quantitation of the pYAP over YAP ratio from three biological replicates, normalized to the ratio in mock treated cells. B) Quantitation of CTGF mRNA abundance (N=3 biological replicates) by RT-PCR on MCF10A cells treated with DMSO (−), SP600125, and/or Anisomycin (+). GAPDH was used as an internal control. The CTGF over GAPDH ratio was normalized to the ratio in mock treated cells. C) Western blots on lysates of MCF10A cells treated with DMSO (−), SP600125, and/or Anisomycin (+), blotted using the indicated antisera. TUB is a loading control. Histograms show quantitation of the pLATS1 over LATS1 ratio from three biological replicates, normalized to the ratio in mock treated cells. D–F) Co-immunoprecipitation experiments from HEK293 cells co-transfected with Myc:LATS1 and Ajuba:V5 (D), WTIP:V5 (E), or LIMD1:V5 (F), in the presence or absence of a plasmid expressing activated-JNK2, as indicated. Blots marked “input” show relative amounts of the indicated proteins in cell lysates. Blots marked “IP V5” show relative amounts of protein precipitated by anti-V5 beads. Histograms show average ratio of LATS1/Ajuba family proteins from three biological replicates, normalized to the ratio in controls. G) Co-immunoprecipitation experiments from MCF10A cells treated or not with Anisomycin. Blots marked “Input” show relative amounts of endogenous LATS1 and LIMD1 in cell lysates. Blots marked “IP LATS1” show relative amounts of protein immunoprecipitated with anti-LATS1. Histogram shows average ratio of LIMD1/LATS1 from three biological replicates, normalized to the ratio in controls. H) In vitro binding experiments comparing the influence of JNK2 activation on LATS1 and LIMD1. Blot of anti-V5 beads shows amounts of LIMD1 or GFP (control) on beads, co-IP shows amounts of Myc:LATS1 precipitated by these beads. LIMD1 Lysates shows the relative amounts of LIMD1:V5 and JNK2 fusion protein in the lysates applied to V5 beads for purification, and LATS1 Lysates shows Myc:LATS1 and JNK2 fusion protein in the lysates added to beads. Histogram shows average ratio of LATS1/LIMD1 from three biological replicates, normalized to the ratio in controls. In all histograms, error bars indicate standard error, and P values less than 0.05 are shown.
The ability of JNK to reduce phosphorylation of Ser127 in YAP implies that Hippo signaling is being inhibited, because most upstream components of Hippo signaling ultimately impinge on LATS, the mammalian homologues of Drosophila Wts. LATS is activated by phosphorylation, and one key regulatory site in LATS1 is Thr1079, which is phosphorylated by the MST family of kinases (35, 36), the mammalian homologues of Drosophila Hpo. In MCF10A cells, LATS phosphorylation on Thr1079 was increased by treatment with SP600125 and decreased by treatment with Anisomycin (Fig 3C). These results suggest that JNK activity inhibits phosphorylation of LATS1 by MST. We extended these studies by examining the influence of two distinct JNK isoforms on Hippo signaling in HEK293 cells transfected with plasmids expressing the JNK kinase MKK7 fused with either JNK1 (MKK7B2:FLAG:JNK1) or JNK2 (MKK7B2:FLAG:JNK2), which results in constitutive activation of JNK (37). The JNK activity of the transfected fusion proteins was confirmed by phosphorylation of JNK (Fig. S3B). Phosphorylation of both Ser127 in endogenous YAP and Thr1079 in endogenous LATS1 was reduced when activated JNK1 or JNK2 were expressed in HEK293 cells (Fig S3C,D). Altogether, our results establish that JNK signaling regulates Hippo signaling in mammalian cells, and impinges upon the pathway at or upstream of the phosphorylation and activation of LATS, which is consistent with our genetic experiments in Drosophila.
JNK increases the binding of LIMD1 and WTIP to LATS1
Because genetic studies implicated Jub as essential to JNK-mediated regulation of Yki, we considered the possibility that JNK might influence the activity of Ajuba family proteins. Ajuba proteins can bind to both Wts and Sav in Drosophila cells, and their homologues LATS and WW45 in mammalian cells (30). The ability of Ajuba family proteins to promote Yki and YAP activity implies that they inhibit Wts and LATS activity through this binding (30). Thus we examined whether JNK could influence the binding between Ajuba family proteins and LATS through co-precipitation experiments in cultured cells. There are three mammalian Ajuba family proteins: Ajuba, LIM domain-containing protein 1 (LIMD1), and Wilms tumor protein 1-interacting protein (WTIP). Expression of constitutively activated-JNK significantly increased binding of LIMD1 and WTIP, but not that of Ajuba, to LATS1 (Fig. 3D,E,F). For LIMD1, we also confirmed that binding between endogenous LIMD1 and endogenous LATS1 was increased in MCF10A cells upon JNK activation by Anisomycin treatment (Fig. 3G). Thus, JNK activation increases binding between LIMD1 or WTIP and LATS1, which could in principle account for the decreased LATS activity associated with JNK activation.
To identify the protein that is targeted by JNK activation, we affinity-purified V5-tagged LIMD1 from HEK293 cells co-transfected or not with plasmids expressing activated-JNK, and then mixed purified LIMD1 with lysates either from cells expressing LATS1, or from cells expressing LATS1 and activated JNK. Co-transfection of activated JNK2 with LIMD1 resulted in a robust (nine-fold) increase in LIMD1 binding to LATS1 (Fig. 3H). Conversely, co-expression of constitutively-activated JNK2 with LATS1 did not increase binding between LIMD1 and LATS1 (Fig. 3H). Thus, the enhanced binding between LIMD1 and LATS1 is due to an influence of JNK2 on LIMD1 rather than on LATS1. Co-expression of constitutively activated JNK1 with LIMD1 gave a similar increase in binding to LATS1, confirming that either JNK protein can increase LATS1-LIMD1 binding. (Fig S3E). Similar experiments established that JNK also increased WTIP-LATS1 binding through an effect on WTIP (Fig S3F).
We also examined the influence of JNK activation on binding between Ajuba family proteins and WW45. However, the binding between Ajuba, LIMD1, or WTIP and WW45 was unaffected by JNK activation (Fig S3G, H, I).
JNK induces phosphorylation of Ajuba family proteins
Activation of JNK reduced the mobility of LIMD1 (Figs 3, S3), suggesting that it induces a post-translational modification. To examine whether Ajuba family proteins could be subject to JNK-promoted phosphorylation, lysates from HEK293 cells expressing an epitope-tagged Ajuba family protein, along with activated forms of JNK1 or JNK2 or negative controls, were analyzed by standard SDS-PAGE gradient gels and Phos-tag gels, which contain a phosphate-binding moiety that specifically retards the mobility of phosphorylated proteins (38, 39). Activation of JNK resulted in efficient phosphorylation of LIMD1, visible as a clear mobility shift of most protein on both standard gels and Phos-tag gels (Fig. 4A). For WTIP, a fraction of the protein was phosphorylated, based on the mobility shift observed on both standard and Phos-tag gels (Fig. 4A), although the phosphorylation profile of a substantial fraction of the protein was not altered. Ajuba was the least affected, because Phos-tag gels did not identify any new species with decreased mobility (namely, increased phosphorylation), although there was a modest shift in the proportions of faster and slower migrating isoforms on Phos-tag gels (Fig. 4A). The extent of phosphorylation of Ajuba family proteins by JNK thus correlated with the degree of increased binding to LATS1. Similar analysis for Drosophila Jub indicated that activation of Basket (Bsk), the Drosophila homolog of JNK, induced phosphorylation of Jub in S2 cells (Fig. S4A). To investigate whether the phosphorylation of Ajuba family proteins was direct, we also performed in vitro kinase assays, using LIMD1 purified from HEK293 cells or Jub purified from S2 cells, and a commercially-available active JNK. These experiments confirmed that Jub and LIMD1 could be directly phosphorylated by JNK in vitro (Fig 4B and S4B).
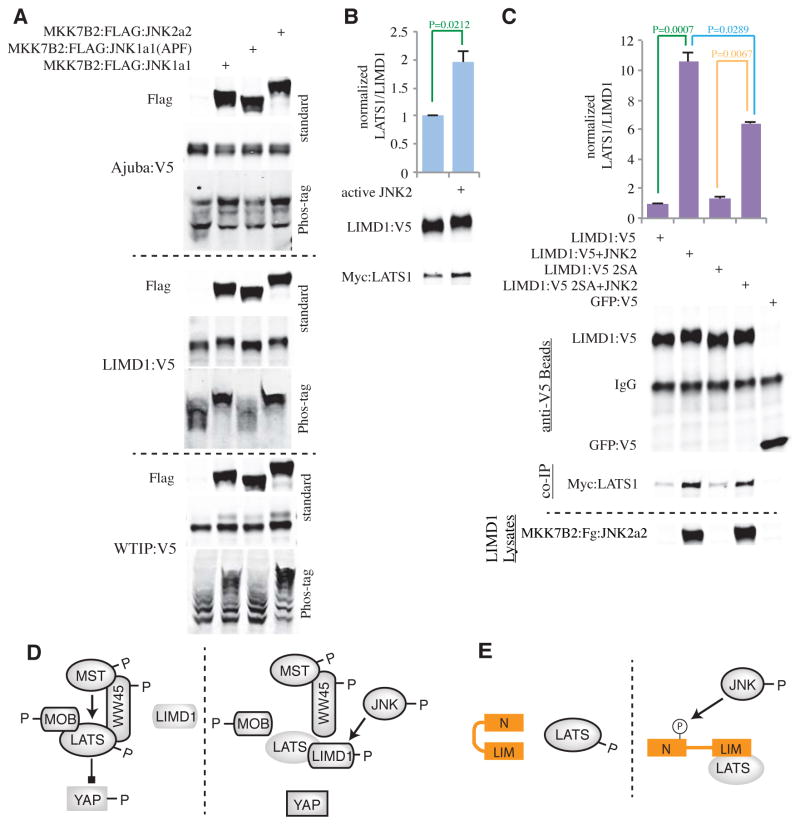
Fig. 4. JNK induces phosphorylation of Ajuba family proteins to increase binding to LATS1.
A) Western blots on lysates of HEK293 cells co-transfected with Ajuba:V5, LIMD1:V5, or WTIP:V5 and, as indicated, MKK7B2:FLAG:JNK1a1 (activated JNK1), MKK7B2:FLAG:JNK1a1(APF) (inactive JNK1), or MKK7B2:FLAG:JNK2a2 (activated JNK2). The expression of transfected JNK constructs is shown in Flag blots. For each Ajuba family protein, the upper blot shows a standard gel, and the lower blot shows a Phos-tag gel. Blots are representative of 3 biological replicates. B) In vitro binding of LIMD1 to LATS1 after in vitro phosphorylation of LIMD1 by active JNK2. The upper blot shows amount of LIMD1 on beads, and the lower blot shows Myc:LATS1 bound to the beads. The histogram shows the average LATS1/LIMD1 ratio from three biological replicates, normalized to the ratio without JNK2 phosphorylation. C) In vitro binding assays comparing wild-type LIMD1 and LIMD12SA mutant binding to LATS1. Anti-V5 beads blot shows LIMD1, LIMD12SA or GFP (control) protein on beads. Co-IP shows Myc:LATS1 bound to wild-type or mutant LIMD1. Expression of constitutively active JNK2 is shown in LIMD1 Lysates blot. The histogram shows the average LATS1/LIMD1 ratio from three biological replicates, normalized to the ratio in wild-type LIMD1 control. Error bars in B,C show standard error. P values less than 0.05 are indicated. D) Model illustrating influence of JNK on Hippo signaling, active proteins are outlined in black, P indicates phosphorylation. In the absence of JNK activation, MST (Hpo), MOB (Mats), and WW45 (Sav) can activate LATS (Wts), which then represses YAP (Yki) by phosphorylating it. When JNK is active, it promotes phosphorylation of Ajuba family proteins (LIMD1, WTIP, or Jub), which then bind more strongly to LATS. This binding inhibits LATS phosphorylation and consequent activation of LATS, possibly by occluding the phosphorylation site, or by inhibiting binding of WW45 or MOB. E) Model illustrating the proposed conformational change of Ajuba family proteins caused by JNK that enhances LATS binding. Without JNK activation, Ajuba family proteins stay as a ‘closed’ form and cannot be accessed by LATS (left). When JNK is activated, the N-terminus of Ajuba family proteins get phosphorylated which result in exposure of C-terminus to LATS (right).
Direct JNK phosphorylation of LIMD1 increases LIMD1-LATS1 binding
To investigate whether JNK enhances LIMD1-LATS1 binding directly through phosphorylating LIMD1, we incubated purified LIMD1 phosphorylated in vitro by JNK with cell lysates containing Myc-tagged LATS1. LIMD1-LATS1 binding was significantly increased by JNK-mediated phosphorylation of LIMD1 in vitro (Fig 4B).
We then identified candidate JNK phosphorylation sites on V5-tagged LIMD1 purified from cells with or without JNK2 activation by using mass spectrometry (LC-MS/MS). Eleven sites had increased phosphorylation in the presence of JNK activation (Fig S4C), nine of which conform to the minimal JNK site consensus (serine or threonine followed by proline). Of these, Ser272, Ser277, Ser421, and Ser424 have been reported to be phosphorylated in cells (40). A mutant version of LIMD1 with Ser272 and Ser277 changed to alanine (LIMD12SA) did not show a significant difference in binding to LATS1 without JNK activation, but with JNK activation, LATS1 binding was significantly but not completely reduced compared to wild-type LIMD1 (Fig. 4C). Thus, JNK phosphorylation of Ser272 and Ser277 accounts for roughly 40% of the JNK-dependent increase in LIMD1-LATS1 binding. A LIMD14SA mutant (in which Ser272, Ser277, Ser421, and Ser424 were changed to alanine) behaved similarly to LIMD12SA in these experiments (Fig. S4D). We also constructed a LIMD18A mutant (containing the mutations S187A, S197A, S211A, S255A, S272A, S277A, T294A, and S384A), and these mutations significantly reduced, but did not eliminate the increased LIMD1-LATS1 binding caused by JNK activation (Fig S4E).
Ser272 and Ser277 are within the N-terminal half of LIMD1, but Ajuba family proteins are reported to bind LATS proteins through their LIM domains (41), which are in the C-terminal half. To further investigate how JNK influences LIMD1-LATS1 binding, we assayed the influence of JNK on binding of a C-terminal LIMD1 polypeptide comprising the three LIM domains to LATS1. This polypeptide bound LATS1, but this binding was not affected by JNK activation (Fig S4F). This observation implies that the ability of JNK to increase the binding of LATS1 to the C-terminal half of LIMD1 requires JNK phosphorylation sites in the N-terminal half of LIMD1.
Discussion
JNK signaling has been implicated in proliferative responses to tissue damage during regeneration, compensatory cell proliferation, and tumorigenesis. In many cases, these proliferative responses depend upon activation of Yki, but mechanisms by which JNK activation promotes Yki activation have been unknown. Here, we have combined genetic and biochemical approaches to identify and characterize a molecular mechanism that links JNK to Yki regulation. Moreover, we have discovered that the ability of JNK to activate YAP is conserved in mammalian cells. Considering the important roles for both JNK and YAP activity in regeneration and tumorigenesis, the discovery that they can be linked in mammalian cells as they are in Drosophila suggests that a JNK-YAP link could also contribute to tumorigenesis and proliferative responses to tissue damage in mammals. JNK signaling also has pro-apoptotic activity, and the factors that control the balance between apoptotic and proliferative responses have remained unknown. Our identification of a key role for Ajuba family proteins and their regulation of Yki and YAP in the proliferative response provides a basis for further investigations of Ajuba family proteins as potential contributors to the divergent responses to JNK activation in different contexts.
Our results support a model in which JNK promotes Yki and YAP activity by phosphorylating Ajuba family LIM proteins and increasing their binding to Wts and LATS proteins, thereby preventing their activation by Hpo and MST (Fig. 4D). Although we have not yet identified the sites that completely account for the influence of JNK on LIMD1-LATS1 binding, our results show that the influence of JNK is mediated through an effect that ultimately impinges on LIMD1 rather than on LATS1, and that this effect could be at least partially recapitulated by in vitro phosphorylation of LIMD1 by JNK, and partially blocked by preventing phosphorylation of two Ser residues in the N-terminus. Thus, although we do not exclude the possibility of additional mechanisms, at least part of the effect of JNK can be ascribed to direct phosphorylation of the N-terminus. Because the C-terminus is the LATS1 binding region, these observations suggest a model in which phosphorylation of LIMD1 promotes formation of an “open” conformation in which the LIM domains are more accessible (Fig 4E). Intriguingly, direct evidence for a similar mechanism has been obtained for a related LIM-domain protein, Zyxin: phosphorylation of sites in the N-terminus of Zyxin reduces interaction of the N-terminus with the C-terminal LIM domains, and enhances the ability of the LIM domains to associate with other binding partners (42, 43). Our results also indicate that the responsiveness to JNK varies amongst the three mammalian family members, with LIMD1 being the most responsive and Ajuba the least responsive. Considering the requirement for jub in the regulation of Yki by JNK in Drosophila, it is noteworthy that amongst the three mammalian Ajuba family proteins, LIMD1 is the most closely related to Drosophila Jub, whereas Ajuba is the most divergent (30).
EGFR-Ras-ERK signaling has been linked to Yki activation (44). ERK can also connect to Hippo signaling through phosphorylation of Ajuba family proteins. Thus, these combined studies implicate Ajuba family proteins as a key regulatory node within the Hippo pathway for cross-regulation by other signaling pathways. The biochemical mechanisms are distinct: JNK promotes both LIMD1 and WTIP binding to LATS1 whereas ERK only promotes WTIP binding to LATS1, JNK promotes binding to LATS1, whereas ERK promotes binding to both LATS1 and WW45 or Sav, and JNK acts through sites in the N-terminus, whereas ERK acts through a site within the C-terminal LIM domains of WTIP (44). Nonetheless, there is a general conceptual similarity, in which phosphorylation influences the ability of Ajuba family proteins to bind to partners within the Hippo pathway, which might in all cases stem from a phosphorylation-induced conformational change. The observation that both pathways impinge upon Ajuba family proteins is particularly intriguing in light of the synergy between Ras and JNK activation in promoting tumorigenesis (9, 11–14), which might thus be at least partially explained by their impinging upon a shared biochemical mechanism for Yki and YAP regulation.
Materials and Methods
Fly stocks
The fly stocks used were as follows: salPE-Gal4 UAS-GFP UAS-hep.CA /CyOGFP; UAS-dcr2 DroncI29/TM6BGal80, ex-lacz salPE-Gal4 UAS-GFP UAS-hep.CA /CyOGFP; UAS-dcr2 DroncI29/TM6BGal80, ex-lacz en-Gal4 UAS-GFP/CyO; UAS-dcr2/TM6B, UAS-lglRNAi (vdrc 51249), UAS-bskRNAi (vdrc 104569), UAS-dRASSFRNAi (vdrc 110203), UAS-jubRNAi (vdrc 38442), UAS-zyxinRNAi (vdrc 104169), UAS-myc:wts.2, UAS-hpo, rn-Gal4 UAS-egr Gal80ts/TM6BGal80(45), and jubE1/FM7 (46).
Plasmids
V5-tagged human Ajuba, LIMD1, and LIMD1-C were generated by PCR using Ajuba or LIMD1 cDNA (Open Biosystems) as templates and inserting into pCDNA3.1-V5:His B vector (Life Technologies). Other plasmids used in this paper includes pCDNA3-MKK7B2:flag:Jnk1a1 (Addgene 19731), pCDNA3-MKK7B2:flag:Jnk1a1(APF) (Addgene 19730), pCDNA3-MKK7B2:flag:Jnk2a2(Addgene 19727), pCDNA3-myc:lats1 (35), pCDNA3-GFP:V5, pCDNA3-WTIP:V5, pUAST-3Xflag:jub (44). MKK7B2:flag:Jnk1a1(APF) is a kinase dead form of JNK1 fused with MKK7, in which the activation motif Thr1959-Pro-Tyr1965 is replaced with Ala-Pro-Phe (37). LIMD1:V5 mutants were made using Quickchange lightning multi site-directed mutagenesis kit (Agilent Technologies).
Cell culture, transfection and treatment
Drosophila S2 cells were cultured in Schneider’s Drosophila medium (Life Technologies) supplemented with 10% FBS (Sigma) and Antibiotic-Antimycotic (Life Technologies) at 25°C. HEK293 cells were cultured in DMEM medium (Life Technologies) supplemented with 10% FBS and Antibiotic-Antimycotic, and MCF10A cells were cultured in DMEM/F12 medium (Life Technologies) supplemented with 5% horse serum, 20μg/ml EGF, 10μg/ml insulin, 0.1μg/ml chloratoxin, 0.5μg/ml hydrocortisone and Antibiotic-Antimycotic at 37°C and 5% CO2. S2 cells were transfected with Cellfectin II (Life Technologies), and HEK293 and MCF10A cells were transfected with Lipofectamine 2000 (Life Technologies) according to manufacturer’s protocols, and harvested 24h after transfection. 50μM SP600125 (Santa Cruz Biotechnology) and/or 50ng/ml Anisomycin (Abcam) were applied to MCF10A cells for 4h after 24h serum starvation, for co-treatments cells were pretreated with DMSO (−) or 50μM SP600125 for 1h, followed by treatment with 50ng/ml Anisomycin and/or 50μM SP600125 for 4h.
Immunoblotting and immunoprecipitation
Cells were lysed in lysis buffer (50mM Tris·HCl pH7.4, 150mM NaCl, 1% Triton X-100, 0.1% CHAPS, 0.1% NP-40, 1mM EDTA, 5% glycerol) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Calbiochem). Protein samples were applied to 4–15% gradient gels (Bio-rad). For immunoprecipitation, protein samples were incubated with mouse anti-V5 agarose affinity gel (Sigma) overnight or rabbit anti-Lats1 (1:150, Cell signaling Technology) overnight followed by incubation with protein G sepharose (GE Healthcare) for 1h at 4°C. Antibodies used for immunoblotting include rabbit anti-Lats1 (1:2000, Cell Signaling Technology), rabbit anti-phospho-Lats1(T1079) (1:2000, Cell Signaling Technology), rabbit anti-phospho-Yap(S127) (1:4000, Cell Signaling Technology), rabbit anti-Yap (1:2000, Epitomics), rabbit anti-phospho-c-Jun (S73) (1:1000, Cell Signaling Technology), rabbit anti-Myc (1:2000, Santa Cruz Biotechnology), mouse anti-V5 (1:10000, Life Technologies), rabbit anti-LIMD1 (1:2000, Bethyl Laboratories). Blots were visualized and quantified using fluorescent-conjugated secondary antibodies (Li-Cor Biosciences) and Odyssey Imaging System (Li-Cor Biosciences).
Statistical Analysis
Statistical significance was determined using paired two-tailed t test for two sample comparisons or ANOVA for multiple samples analysis, after logarithm transformation of normalized or ratio values, with P<0.05 set as the criteria for significance. The Tukey test was used to derive adjusted P values for multiple comparisons. Error bars on figure panels show standard error of the mean.
Phos-tag gel
For Phos-tag gel, cells were lysed in 50mM Tris·HCl pH7.5, 150mM NaCl, 1% Triton X-100, 0.1% NP-40. Lysates containing transfected Drosophila Jub or human LIMD1 were applied to 6% SDS-PAGE containing 25μM Phos-tag Acrylamide AAL-107 (NARD Institute) and 50μM MnCl2. Lysates containing transfected human Ajuba or WTIP were applied to 8% SDS-PAGE containing 25μM Phos-tag Acrylamide AAL-107 and 50μM MnCl2.
In vitro kinase assay
Flag tagged Jub was expressed in S2 cells in 6-well plates and purified using EZviewRed Anti-flag M2 Affinity Gel (Sigma). After washing with lysis buffer, beads with purified proteins were added to kinase buffer (50mM Tris·HCl pH7.5, 1mM DTT) supplemented with protease inhibitor cocktail, phosphatase inhibitor cocktail, and magnesium/ATP cocktail (1:5, Millipore). For each reaction, 500ng JNK1a1 (to phosphorylate Drosophila Jub) or JNK2a2 (to phosphorylate LIMD1, Millipore) was added. The mixture was incubated at 15°C for 1h.
In vitro binding assay
V5-tagged LIMD1, LIMD1-C, WTIP, or GFP were transfected into HEK293 cells with or without MKK7B2:FLAG:Jnk2a2. Cells were lysed in lysis buffer (50mM Tris·HCl pH7.4, 150mM NaCl, 1% Triton X-100, 0.1% CHAPS, 0.1% NP-40, 1mM EDTA, 5% glycerol) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Calbiochem). Lysates were incubated with mouse anti-V5 agarose (Sigma) for 3h at 4°C, then washed with lysis buffer 4 times. The beads with V5-tagged proteins were incubated with cell lysates containing Myc-tagged Lats1 with or without MKK7B2:FLAG:Jnk2a2 or MKK7B2:FLAG:Jnk1a1 overnight at 4°C. Beads were then washed with lysis buffer 6 times, and applied to SDS-PAGE.
Mass spectrometry
V5-tagged LIMD1 was transfected into HEK293 cells with or without MKK7B2:FLAG:Jnk2a2. After protein extraction, LIMD1:V5 was purified using anti-V5 agarose (Sigma), and applied to 4–15% gradient gel. Gels were stained using Gelcode blue stain reagent (Pierce). Bands were cut from gel and analyzed by the Biological Mass Spectrometry Facility of the UMDNJ- Rutgers for LC-MS/MS analysis.
Quantitative RT-PCR
RNA was extracted from MCF10A cells treated with different drugs using Trizol reagent (Life Technologies). SuperScript III reverse transcriptase (Life Technologies) was used for reverse transcription. Quantitative PCR was conducted using QuantiTect SYBR green PCR kit (Qiagen).
Immunostaining
Inverted anterior part of Drosophila larvae were fixed in 4% PFA for 20min at room temperature, then washed with PBS containing 1% BSA and 0.1% Triton X-100 and blocked by 5% Donkey serum. Antibodies used for immunostaining include rabbit anti-Yki (1:400), mouse anti-β-gal (1:400, DHSB). The intensity of ex-lacZ staining and compartment area were quantified using Image J software, comparing GFP-expressing to non-GFP expressing cells within the wing pouch.
Wing regeneration experiment
Larvae were raised at 18°C for 8 days after egg laying then transferred to 29°C for 40h to ablate the developing larval wing by inducing pro-apoptotic gene expression through inactivation of Gal80ts. After ablation, larvae were shifted back to 18°C and maintained at 18°C until eclosion. Adult wings were mounted in Gary’s magic mountant and photographed using ProgRes Mac Capture Pro software. Wing sizes were quantified using ImageJ software.
Supplementary Material
Fig S1. Epistasis between JNK and Hippo pathway components in regulation of Yki
Fig S2 Heterozygosity for jub does not affect the rate of development or adult wing size without ablation
Fig S3 JNK regulates Hippo signaling and enhances LIMD1 and WTIP binding to LATS1
Fig S4. Phosphorylation of Ajuba family proteins by JNK and mapping of phosphorylation sites in LIMD1.
Acknowledgments
We thank the Developmental Studies Hybridoma Bank and the Bloomington stock center for antibodies and Drosophila stocks, and Xin Zhao for statistical consulting.
Funding: This research was supported by Human Frontiers Science Program grant RGP0016/2010 and the Howard Hughes Medical Institute.
Footnotes
Author contributions: GS performed the experiments. GS and KI conceived the experiments and wrote the manuscript.
Data availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) through the PRIDE partner repository with the dataset identifier PXD000384 and DOI 10.6019/PXD000384.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Developmental cell. 2011 Jul 19;21:172. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends in Cell Biology. 2008 Oct 01;18:467. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergantinos C, Corominas M, Serras F. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development. 2010 Apr;137:1169. doi: 10.1242/dev.045559. [DOI] [PubMed] [Google Scholar]
- 4.Chen F. JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer research. 2012 Feb 15;72:379. doi: 10.1158/0008-5472.CAN-11-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogoyevitch MA, Ngoei KR, Zhao TT, Yeap YY, Ng DC. c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochimica et biophysica acta. 2010 Mar;1804:463. doi: 10.1016/j.bbapap.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Iimuro Y, Fujimoto J. TLRs, NF-kappaB, JNK, and Liver Regeneration. Gastroenterology research and practice. 2010;2010 doi: 10.1155/2010/598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasaki J, Shibata N, Sakurai T, Agata K, Umesono Y. Role of c-Jun N-terminal kinase activation in blastema formation during planarian regeneration. Development, growth & differentiation. 2011 Apr;53:389. doi: 10.1111/j.1440-169X.2011.01254.x. [DOI] [PubMed] [Google Scholar]
- 8.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Developmental biology. 2011 Feb 1;350:139. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menendez J, Perez-Garijo A, Calleja M, Morata G. A tumor-suppressing mechanism in Drosophila involving cell competition and the Hippo pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010 Aug 17;107:14651. doi: 10.1073/pnas.1009376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Garijo A, Shlevkov E, Morata G. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development (Cambridge, England) 2009 Mar 25; doi: 10.1242/dev.034017. [DOI] [PubMed] [Google Scholar]
- 11.Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Current biology: CB. 2006 Jun 6;16:1139. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 12.Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. The EMBO Journal. 2006 Nov 15;25:5294. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leong GR, Goulding KR, Amin N, Richardson HE, Brumby AM. scribble mutants promote aPKC and JNK-dependent epithelial neoplasia independently of Crumbs. BMC Biology. 2009;7:62. doi: 10.1186/1741-7007-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohsawa S, et al. Mitochondrial defect drives non-autonomous tumour progression through Hippo signalling in Drosophila. Nature. 2012 Oct 25;490:547. doi: 10.1038/nature11452. [DOI] [PubMed] [Google Scholar]
- 15.Zhang JY, Selim MA. The role of the c-Jun N-terminal Kinase signaling pathway in skin cancer. American journal of cancer research. 2012;2:691. [PMC free article] [PubMed] [Google Scholar]
- 16.Hui L, Zatloukal K, Scheuch H, Stepniak E, Wagner EF. Proliferation of human HCC cells and chemically induced mouse liver cancers requires JNK1-dependent p21 downregulation. The Journal of clinical investigation. 2008 Dec;118:3943. doi: 10.1172/JCI37156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogoyevitch MA, Kobe B. Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiology and molecular biology reviews: MMBR. 2006 Dec;70:1061. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Developmental dynamics: an official publication of the American Association of Anatomists. 2012 Jan;241:3. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irvine KD. Integration of intercellular signaling through the Hippo pathway. Seminars in cell & developmental biology. 2012 Sep;23:812. doi: 10.1016/j.semcdb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010 Oct 19;19:491. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Current biology: CB. 2010 Sep 14;20:1580. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Developmental biology. 2011 Feb 15;350:255. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- 23.Shaw RL, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010 Dec;137:4147. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development (Cambridge, England) 2005 Jun;132:2125. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Developmental cell. 2004 Oct;7:491. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Developmental cell. 2008 Aug;15:309. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Molecular Cell. 2010 Aug 27;39:521. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Polesello C, Huelsmann S, Brown NH, Tapon N. The Drosophila RASSF homolog antagonizes the hippo pathway. Current biology: CB. 2006 Dec 19;16:2459. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rauskolb C, Pan G, Reddy BV, Oh H, Irvine KD. Zyxin links fat signaling to the hippo pathway. PLoS biology. 2011 Jun;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Current biology: CB. 2010 Apr 13;20:657. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Current biology: CB. 2010 Apr 13;20:573. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 32.Du L, et al. Inhibition of cell proliferation and cell cycle progression by specific inhibition of basal JNK activity: evidence that mitotic Bcl-2 phosphorylation is JNK-independent. The Journal of biological chemistry. 2004 Mar 19;279:11957. doi: 10.1074/jbc.M304935200. [DOI] [PubMed] [Google Scholar]
- 33.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proceedings of the National Academy of Sciences of the United States of America. 2001 Nov 20;98:13681. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barancik M, Htun P, Schaper W. Okadaic acid and anisomycin are protective and stimulate the SAPK/JNK pathway. Journal of cardiovascular pharmacology. 1999 Aug;34:182. doi: 10.1097/00005344-199908000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Chan EH, et al. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005 Mar 17;24:2076. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 36.Praskova M, Xia F, Avruch J. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Current biology: CB. 2008 Mar 11;18:311. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei K, et al. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Molecular and cellular biology. 2002 Jul;22:4929. doi: 10.1128/MCB.22.13.4929-4942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006 Apr;5:749. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008 Mar;135:1081. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2008 Aug 5;105:10762. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS letters. 2006 Feb 6;580:782. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- 42.Call GS, et al. Zyxin phosphorylation at serine 142 modulates the zyxin head-tail interaction to alter cell-cell adhesion. Biochemical and biophysical research communications. 2011 Feb 21;404:780. doi: 10.1016/j.bbrc.2010.12.058. [DOI] [PubMed] [Google Scholar]
- 43.Hirota T, et al. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol. 2000 May 29;149:1073. doi: 10.1083/jcb.149.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy BVVG, Irvine KD. Regulation of Hippo Signaling by EGFR-MAPK Signaling through Ajuba Family Proteins. Developmental cell. 2013 Apr 11;24:459. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Developmental cell. 2009 Jun;16:797. doi: 10.1016/j.devcel.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabino D, Brown NH, Basto R. Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. Journal of cell science. 2011 Apr 1;124:1156. doi: 10.1242/jcs.076711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Epistasis between JNK and Hippo pathway components in regulation of Yki
Fig S2 Heterozygosity for jub does not affect the rate of development or adult wing size without ablation
Fig S3 JNK regulates Hippo signaling and enhances LIMD1 and WTIP binding to LATS1
Fig S4. Phosphorylation of Ajuba family proteins by JNK and mapping of phosphorylation sites in LIMD1.