Abstract
Effectively and accurately assessing total microbial community diversity is one of the primary challenges in modern microbial ecology. This is particularly true with regard to the detection and characterization of unculturable populations and those present only in low abundance. We report a novel strategy, GC fractionation combined with denaturing gradient gel electrophoresis (GC-DGGE), which combines mechanistically different community analysis approaches to enhance assessment of microbial community diversity and detection of minority populations of microbes. This approach employs GC fractionation as an initial step to reduce the complexity of the community in each fraction. This reduced complexity facilitates subsequent detection of diversity in individual fractions. DGGE analysis of individual fractions revealed bands that were undetected or only poorly represented when total bacterial community DNA was analyzed. Also, directed cloning and sequencing of individual bands from DGGE lanes corresponding to individual G+C fractions allowed detection of numerous phylotypes that were not recovered using a traditional random cloning and sequencing approach.
One of the primary challenges in modern microbial ecology is effectively and accurately assessing total microbial diversity, particularly with regard to detection of unculturable and fastidious bacterial species and those present in low abundance (i.e., minority populations). A common thread described in numerous published studies and textbooks regarding microbial community diversity is that, in most environments, only 0.1 to 1.0% of bacteria detected by direct microscopic enumeration can be recovered on even the most general of laboratory media. As a result, microbial ecologists generally have the opinion that the vast majority of microbial diversity remains uncharacterized due to this gap between culturable and direct estimates of microbial biomass and diversity. This concern has spurred the development of a number of molecular approaches for studying microbial communities, often based on analysis of nucleic acids directly extracted from environmental samples, that attempt to bridge this gap. These molecular community analysis approaches can be organized into two general classes: compilation-based analyses that combine individual bits of data to obtain a sense of community structure, and total community analyses that characterize the whole community in a single analysis.
Compilation-based strategies often involve a random (“shotgun”) approach, wherein related functional or ribosomal gene sequences from individual community members are PCR amplified and cloned from total community DNA for phylogenetic analysis or comparison to existing databases. Such techniques have proven very powerful and have been widely applied, generating much information on microbial diversity in a variety of systems, especially where ecologically relevant, but as-yet-uncultured, microbial community members are concerned (e.g., see references 5, 7, 8, 13, 15, 25, 28, 34, 36, 37, and 43-45). However, it is becoming clear that compilation-based approaches, which typically analyze 100 to 300 randomly obtained individual sequences (9), are limited in their ability to accurately detect total diversity where communities are complex. Thus, in microbial communities comprised of hundreds to thousands of individual taxa (e.g., soils or the gastrointestinal [GI] tract), individual taxa present in lower abundance (i.e., minority populations) will go undetected.
Some recent studies of microbial diversity have taken a theoretical approach by estimating total community diversity based on mathematical extrapolation from a partial analysis of the total community (6, 9, 10, 20, 25, 26, 41). These approaches, however, provide no specific information regarding the identity of minority populations, since their presence is only inferred and no clones are actually obtained and analyzed.
By contrast, total community analyses typically attempt to capture a sense of total community structure or diversity through a single, more direct analysis of total community DNA. A number of different approaches have been developed, including monitoring DNA reannealing kinetics (40, 42), restriction analysis of PCR amplicons from community DNA (10, 14, 24, 39), denaturing gradient gel electrophoresis (DGGE) of community amplicons (12, 27, 40), and fractionation of total community DNA based on G+C content (17). While these approaches typically probe the entire community, including minority populations, by direct analysis of total community DNA, they generally do not provide high-resolution identification of the populations present and do not focus on minority populations.
The limitations described above suggest that novel approaches are required to more comprehensively assess microbial diversity and enable detection and characterization of taxa that are present in low abundance yet perform important functions in the community. The present study combines two mechanistically different community analysis methods (GC fractionation and DGGE; GC-DGGE) with phylogenetic analysis of DNA sequences to obtain information on minority populations in the GI tract that were not detected by a typical random cloning survey of the same community.
DGGE-based approaches have been widely embraced to provide rapid, comparative analyses of apparent diversity of microbial communities in a variety of environments. Further, individual bands of interest can be excised from the gel for cloning or direct sequence analysis (32). However, because this approach relies on PCR amplification with its potential biases (21, 31, 38) and on visualization of resultant PCR products on gels, it is not quantitative and also likely underestimates true diversity in complex communities where taxa present only in low abundance go undetected.
GC fractionation of total community DNA (17) is independent of PCR amplification and thus provides a sense of relative abundance of bacterial populations, though only at low resolution. The output from this approach is a fractionated profile of the entire community that indicates relative abundance of DNA as a function of G+C content and inferential information regarding the taxa comprising the community. This technique has been successfully employed to study and compare microbial community structures in a variety of environments, including soils and sediments (16, 17), bioreactors (18), and GI tracts of insects and animals (1-3, 33). In addition, this technique physically fractionates total community DNA into aliquots that represent different G+C contents. These highly purified fractions are of high molecular weight and thus are suitable for additional molecular manipulations, including PCR amplification, DGGE analysis, and cloning.
Since each approach to microbial community analysis has its own inherent strengths and limitations, we reasoned that combining mechanistically different approaches should afford better resolution, provide more information, and thus increase the ability to accurately detect and assess total community diversity, including minority populations. We and others have previously shown how GC fractionation combined with 16S rRNA gene sequencing provides a useful method for the directed detection of bacterial populations of interest in the GI tract of humans and animals (2, 4) and in a volcanic soil (30). Herein we report a study that combines GC fractionation, DGGE analysis, and directed cloning and sequencing to assess microbial community diversity and specifically detect minority populations in the community.
This combined approach (GC-DGGE) overcomes both the primary limitation of GC fractionation, low resolution that does not indicate the number or identity of different taxa in a particular G+C fraction, and also the primary limitation of DGGE, the inability to detect populations present in low abundance, to better assess total community diversity. Initial fractionation of total community DNA based on G+C content effectively reduces the complexity of the community DNA mixture being analyzed such that the total diversity within each fraction can be more effectively assessed. The GC-DGGE approach also enables detection of taxa that are present in low abundance, since their DNA is localized into one or a few fractions and thus effectively purified away from the bulk of total community DNA. Additionally, by cloning and sequencing DGGE bands from individual fractions, we can gain insight into the identity of specific taxa of interest.
MATERIALS AND METHODS
Recovery of enterome DNA from chicken digesta.
For this study, a “super-pool” DNA sample representing the cecal enterome (chromosomal DNA of the total bacterial community of the cecum) of broiler chickens worldwide was created by combining equal amounts of enterome DNA extracted from the cecal digesta of approximately 500 individual birds from the United States, United Kingdom, Australia, France, and Finland. Bacterial community DNA was recovered from each animal essentially as described previously (3), except that the Tris-EDTA buffer contained 50 mM EDTA. We have previously shown that this method recovers >95% of bacteria from the cecal matrix and affords essentially quantitative lysis (3). Thus, the super-pool DNA sample represented the full diversity of microbial communities present in chicken ceca from several countries worldwide.
GC fractionation of bacterial community DNA.
Purified enterome super-pool DNA was subjected to equilibrium density centrifugation for fractionation based on G+C content essentially as described previously (17), except that the cesium chloride solution was buffered with 5 mM Tris (pH 8.0). This approach fractionates the genomic DNA of the component taxa of the community as a function of its characteristic G+C content. This separation is based on differential density imposed by the AT-dependent DNA-binding dye bis-benzimidazole (17). Following ultracentrifugation, a Brandel model SYR-94 syringe pump (Brandel, Inc., Gaithersburg, Md.) was used to pass the formed gradients through an ISCO UA-5 UV absorbance detector (ISCO, Inc., Lincoln, Nebr.) set to 280 nm (to minimize background absorbance due to the cesium chloride gradient) and then to a fraction collector. The G+C content represented by each gradient fraction was determined by linear regression analysis (r2 > 0.99) of data obtained from control gradients containing standard DNA samples of known G+C composition as described elsewhere (17).
PCR amplification of 16S rRNA gene sequences.
Fractions from the region of the gradient containing DNA were subsequently desalted using PD-10 columns (Amersham Pharmacia Biotech, Piscataway, N.J.) and the manufacturer's recommended protocol. bis-Benzimidazole is presumably also removed in this process or at least does not interfere with subsequent PCRs. Partial 16S sequences of rRNA genes representing the organisms in the super-pool sample and from individual gradient fractions were amplified for DGGE analysis and cloning by PCR as described previously (1, 19). For direct random cloning from super-pool bacterial community DNA, primers 536f (5′-CAGCMGCCGCGGTAATWC-3′) and 907r (5′-CCGTCAATTCMTTTRAGTTT-3′) were used. To amplify sequences from individual G+C fractions for DGGE analysis, a 40-base GC clamp was added to the 536f primer to generate primer 536fC (11) and used in conjunction with the 907r primer. PCR conditions were as previously described (1).
These primers were derived from generally conserved sequences that have previously been shown to be present in the greatest percentage of eubacteria (and also Archaea and Eucarya) (35) and were predicted to capture >75% of all eubacterial sequences based on those present in the database at the time of that study (>10,000). However, greater recovery of total diversity was expected in the present study because primers 536f, 536fC, and 907r include degenerate bases at the positions having mismatches with known rRNA sequences in the Schmalenberger study. These degeneracies overcome inefficient binding of primers with mismatches by including appropriate primer variations and were thus expected to help minimize PCR bias in the present study.
DGGE analysis.
DGGE was performed to compare the banding patterns obtained from individual fractions obtained by GC profiling to the pattern obtained from total cecal enterome DNA. Procedures and conditions for DGGE were essentially as described previously (11), except that 750 ng of PCR amplicons from each G+C fraction and from the unfractionated super-pool sample was loaded into each lane. Following staining with SYBR Green I (BioWhittaker Molecular Applications, Rockland, Maine) using the manufacturer's recommendations, gel banding patterns were visualized and captured using a Bio-Rad Gel Doc 1000 and Molecular Analyst software (Bio-Rad Laboratories, Hercules, Calif.). GelCompar version 4.0 software (Applied Maths, Kortrijk, Belgium) was then used to normalize the positions of sample bands (i.e., remove “smiling”) based on band positions in multiple known marker lanes to facilitate comparison of sample band patterns.
Cloning, sequencing, and phylogenetic analysis.
Amplified bacterial 16S rRNA PCR products (∼400 bp) from super-pool DNA or from bands excised from the DGGE gel were subsequently cloned into the EcoRVsite of the pT7Blue-3 plasmid vector using the Perfectly Blunt cloning kit (Novagen, Madison, Wis.). Plasmid clones were identified based on blue-white screening and then grown overnight in Luria-Bertani medium amended with ampicillin (300 μg/ml) and tetracycline (15 μg/ml). Plasmid DNA was subsequently purified using Qiagen mini-prep kits (Qiagen, Valencia, Calif.) according to the manufacturer's specifications. The insert size of individual clones was confirmed by restriction fragment analysis using EcoRI.
Selected DGGE bands were excised from the gel, cloned, and sequenced as described in detail previously (11). To ensure that individual clones corresponded to the bands of interest, purified plasmid DNA from putative clones was used as template for PCR using the 536fC and 907r primers, and the products were analyzed via DGGE alongside PCR products from the super-pool DNA.
All confirmed clones from the super-pool survey and from DGGE bands were subjected to double-stranded DNA sequence analysis (MWG Biotech, High Point, N.C.) and sequence comparison to determine the best match to known sequences using the Ribosomal Database Project II (RDP II) website (http://www.cme.msu.edu/RDP/html/index.html) (23). Screening for potential chimeric products was performed using the Chimera Check software at the RDP II site, and potential chimeras were not considered further. The species number designations and phylotype number designations given below in the tables are arbitrary and are based on a best-match analysis that included the sequences from this work, such that any two clones given the same species number or phylotype number are related to each other at an Sab of≥0.95.
Nucleotide sequence accession number.
Sequences obtained by random cloning from unfractionated super-pool DNA were deposited in GenBank under accession numbers AY574393 to AY574431, while sequences obtained from excised bands in the DGGE gel were deposited under accession numbers AY574432 to AY574568.
RESULTS AND DISCUSSION
DGGE analysis of total community DNA and individual GC fractions.
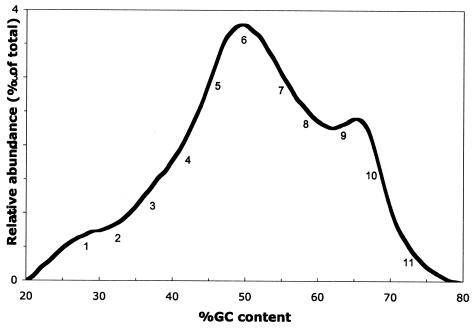
The GC fractionation approach generates a profile of the structure of the total bacterial community as a plot or histogram of relative abundance of DNA versus percent G+C content of genomic DNA (Fig. 1). This approach has previously been shown to accurately and reproducibly fractionate total microbial community DNA, producing an average standard deviation across six replicate cecal community profiles of 5.0% (1). The super-pool community analyzed in this study was comprised of microbial taxa having genomic G+C contents ranging between 20 and 80%, with the most abundant taxa having G+C contents in the 45 to 55% and the 63 to 70% G+C ranges. The GC profile presented in Fig. 1 is generally similar to those obtained in prior studies of cecal microbial community DNA from individual chickens or smaller groups of birds that compared microbial communities based on position in the intestine (3) and feed composition independent of geographical location (1, 3). The super-pool community DNA in the present study, however, should better represent the diversity of taxa present in broiler chickens worldwide, since it is comprised of pooled DNA from approximately 500 birds geographically distributed over much of the world.
FIG. 1.
GC profile of pooled cecal community DNA (super-pool DNA) from ∼500 individual birds from the United States, United Kingdom, Australia, France, and Finland. Numbers indicate locations of individual fractions subjected to DGGE analysis.
In the present study we extended the utility and capabilities of the GC profiling approach by combining it with DGGE analysis and phylogenetic analysis of 16S rRNA gene sequences. To accomplish this, individual fractions representing subsets of the total microbial community (Fig. 1) were subjected to DGGE analysis and compared to the DGGE patterns obtained from unfractionated total community DNA (Fig. 2). The DGGE patterns from the individual fractions displayed a general progression from bands in the upper region of the gel towards bands in the lower region of the gel. This was not unexpected, since fraction 1 comprises the lowest genomic G+C content, which increases progressively with fraction number, as do the concentrations of denaturants in the DGGE gel. Since G+C content is expected to be a major factor governing Tm and, thus, denaturation of DNA, the general trend towards increased migration into the denaturant gradient with increasing G+C content was anticipated. This pattern of migration was reproducible, as an essentially identical pattern was obtained from a replicate DGGE analysis that included separate PCRs from these same fractions (data not shown). Other researchers have noted that some, but not all, degenerate primers can produce multiple bands from a single template (22). In the present study, there was no evidence of the clusters of tightly spaced bands that are indicative of that phenomenon. Further, all primer sequences were removed from the consensus DNA sequences prior to analysis, so that all phylogenetic information came from regions internal to the primers and thus did not reflect minor degeneracies in the primers themselves.
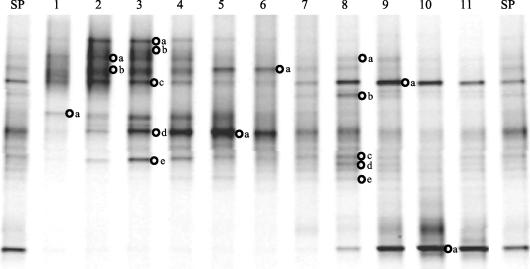
FIG. 2.
DGGE analysis of partial 16S rRNA gene sequences from super-pool DNA and individual gradient fractions (image normalized to remove smiling, using GelCompar software). SP indicates DGGE patterns from unfractionated super-pool DNA, and lane numbers indicate DGGE patterns from individual GC gradient fractions (Fig. 1). The lettered circles indicate individual bands excised from the gel for cloning and DNA sequence analysis.
Perhaps more importantly, in several cases specific bands that were not apparent or were only weakly visible in DGGE lanes corresponding to the total super-pool sample were noticeably enriched in specific fractions (Fig. 2). Examples include lane 1, band a; lane 3, bands a, b, and e; and lane 8, bands b, c, and d. Presumably, the basis for this enrichment is that the reduced complexity and increased relative abundance of templates in the individual fractions compared to the total super-pool DNA allowed templates that were present in low abundance or poorly amplified in the total community to be amplified more effectively. It is also possible that the limited range of G+C content in each fraction facilitated more-even amplification of template mixtures having similar G+C content. This enhanced amplification facilitated visualization and subsequent cloning and characterization of the taxa corresponding to these bands. Thus, the GC-DGGE approach is well-suited to allow visualization and recovery of sequences from minority populations of bacteria whose presence would otherwise go undetected in DGGE analysis of the corresponding unfractionated community DNA.
Sequencing and phylogenetic analysis of DGGE bands from individual fractions.
Individual DGGE bands of interest were excised from the gel, cloned, sequenced, and subjected to phylogenetic analysis to determine the closest known relatives to these cecal inhabitants. The criteria for selection of bands (Fig. 2) were that (i) they represent examples of bands that have migrated similarly in widely separated individual fractions such that they appear as a single band in the super-pool lanes of Fig. 2 (e.g., lane 3, band c, and lane 9, band a; lane 3, band d, and lane 5, band a); (ii) they represent bands in high abundance in the community (e.g., lane 5, band a; lane 9, band a; lane 10, band a); or (iii) they were undetected or in very low relative abundance in the DGGE pattern from the total super-pool sample (e.g., lane 1, band a; lane 3, bands a and b; lane 6, band a; lane 8, band b). For several of the excised bands, multiple clones were generated to assess the degree to which single DGGE bands harbored heterogeneous mixtures of rRNA gene sequences from more than one population.
In all, 17 different bands were excised from the DGGE gel, from which 39 independent clones were generated and subjected to DNA sequence analysis. Based on prior experience comparing partial 16S sequences from type strains to newly cloned sequences (2) we made the following assignments: when an Sab score of a cloned sequence was ≥0.95 in relation to a type strain of a known species, the cloned sequence was assigned to that species. When the Sab score of a cloned sequence was greater than 0.70 but less than 0.95 in relation to a known sequence, the clone sequence was assigned to that genus. Where the Sab score of a cloned sequence was less than 0.70 in relation to any known sequence, that clone was labeled an unidentified phylotype. Based on the criteria described above, five of the clones were assigned to the species level: Bifidobacterium saeculare (two clones), Lactobacillus crispatus (two clones), and Lactobacillus reuteri (one clone) (Table 1). An additional 17 clones were assigned to genera including Atopobium, Bacteroides, Butyrivibrio, Clostridium, Eubacterium, Fusobacterium, Pectinstud, and Ruminococcus. The remaining 16 of the clones represented 13 different unidentified phylotypes (Table 1). It is interesting that the majority (13 of 14) of unknown phylotypes were recovered from fractions 1 to 3, which represent regions in the total community profile where the relative abundance of DNA, and hence organisms in the community, is low (Fig. 1). This likely represents a manifestation of the aforementioned phenomenon where, based on the generally used random cloning approaches for phylogenetic surveys, the majority of sequences obtained and deposited in the databases to date come from the most abundant organisms in the community.
TABLE 1.
Best-match identification of phylotypes of clones from excised DGGE bands
| Identificationa | Location and clone no.b |
|---|---|
| Species level | |
| B. saeculare | 10-a-1, 10-a-2 |
| L. crispatus | 5-a-1, 5-a-2 |
| L. reuteri | 8-e-1 |
| Genus level | |
| Atopobium sp. 1 | 10-a-3 |
| Bacteroides sp. 4 | 8-a-1, 8-a-2 |
| Butyrivibrio sp. 3 | 8-d-1 |
| Clostridium sp. 1 | 8-b-1, 8-b-2 |
| Eubacterium sp. 9 | 8-e-2 |
| Eubacterium sp. 8 | 5-a-3 |
| Fusobacterium sp. 1 | 9-a-1, 9-a-2, 9-a-3 |
| Pectinstud sp. 1 | 3-e-1, 8-c-2 |
| Ruminococcus sp. 1 | 8-d-3 |
| Ruminococcus sp. 27 | 8-e-3 |
| Ruminococcus sp. 4 | 8-c-1, 8-c-3 |
| Ruminococcus sp. 26 | 8-d-2 |
| Unidentified phylotypes | |
| Unidentified phylotypes 22 | 1-a-1 |
| Unidentified phylotype 33 | 3-d-2, 3-d-1 |
| Unidentified phylotype 25 | 2-b-1, 3-c-1 |
| Unidentified phylotype 31 | 3-c-2 |
| Unidentified phylotype 23 | 2-a-1, 2-a-3 |
| Unidentified phylotype 24 | 2-a-2 |
| Unidentified phylotype 32 | 3-c-3 |
| Unidentified phylotype 26 | 2-b-2 |
| Unidentified phylotype 30 | 3-b-1 |
| Unidentified phylotype 28 | 3-a-1 |
| Unidentified phylotype 27 | 2-b-3 |
| Unidentified phylotype 34 | 6-a-1 |
| Unidentified phylotype 29 | 3-a-2 |
Species-level identification, Sab ≥ 0.95; genus-level identification, Sab between 0.70 and 0.95; unidentified phylotype, Sab < 0.70.
Band clones indicates lane number, band letter, and clone number (refer to Fig. 2).
It is also worth noting that in 9 of the 13 cases where multiple clones were made from a given excised band, more than one phylotype was detected from that band. In fact, in four cases where three clones were obtained from a single band, all three represented different phylotypes (Table 1). This indicates that sequences from different microbial species are comigrating and that diversity estimates based solely on counting the number of bands in DGGE lanes are probably conservative. This is perhaps not surprising, since molecules with different primary sequence might have similar denaturation kinetics due to having similar G+C content or different localized domains of ready denaturation or higher stability that, due to the cooperativity of the denaturation process, control the overall migration properties of the molecule in the DGGE gel. Further, minor variations in the length of these partial 16S rRNA gene amplicons might facilitate comigration of individual amplicons having different G+C contents or primary sequences.
Two sets of bands in Fig. 2 (lane 3, band c and lane 9, band a; and lane 3, band d and lane 5, band a) are from regions that have intense bands in the unfractionated super-pool sample and also have bands in essentially all of the G+C fractions. These represent striking examples of where a single DGGE band from total community DNA can harbor multiple phylotypes, as evidenced by the various identities of the clones obtained from these bands (Table 1). However, GC fractionation generally separated these phylotypes to different fractions.
Random cloning of unfractionated super-pool DNA.
A random cloning-based 16S rRNA phylogenetic survey was performed on the same super-pool DNA sample that was analyzed by GC-DGGE to compare the relative efficiencies of the two approaches for detecting less-abundant members of the bacterial community. In all, 136 randomly selected, confirmed clones were analyzed. This general approach and the number of clones studied are typical in size and scope to numerous other phylogenetic surveys of microbial communities from a variety of environments as observed by Dunbar et al. (9). As in many, perhaps most, reports of phylogenetic analysis of 16S rRNA gene sequences obtained from environmental samples, the majority of sequences obtained did not exactly match any known organisms in the RDP II database. Indeed, only 1 sequence of the 136 analyzed was an exact match (Sab = 1.00) to a known organism, Lactobacillus salivarius subsp. salicinius (strain H0268 ATCC 11742T) (Table 2).
TABLE 2.
Best-match identification of phylotypes from shotgun cloning of unfractionated super-pool DNAa
| Identification | n | Identification | n | |
|---|---|---|---|---|
| Species level | ||||
| Bacteroides fragilis | 3 | |||
| Bacteroides merdae | 1 | |||
| Bifidobacterium saeculare | 10 | |||
| Enterococcus mundti | 1 | |||
| Escherichia coli | 1 | |||
| Klebsiella pneumoniae | 1 | |||
| Lactobacillus crispatus | 10 | |||
| Lactobacillus gasseri | 2 | |||
| Lactobacillus pontis | 1 | |||
| Lactobacillus salivarius | 1 | |||
| Streptococcus bovis ATCC 27960 | 6 | |||
| Genus level | ||||
| Bacteroides sp. 1 | 1 | |||
| Bacteroides sp. 2 | 1 | |||
| Bacteroides sp. 3 | 1 | |||
| Bacteroides sp. str. BV-1 | 1 | |||
| Bifidobacterium sp. 1 | 1 | |||
| Butyrivibrio sp. 1 | 1 | |||
| Butyrivibrio sp. 2 | 1 | |||
| Clostridium sp. 1 | 5 | |||
| Clostridium sp. 2 | 1 | |||
| Clostridium sp. 3 | 1 | |||
| Clostridium sp. 4 | 1 | |||
| Clostridium sp. 5 | 1 | |||
| Clostridium sp. 6 | 1 | |||
| Clostridium sp. 7 | 1 | |||
| Clostridium sp. 8 | 1 | |||
| Clostridium sp. 9 | 1 | |||
| Clostridium sp. 10 | 1 | |||
| Clostridium sp. 11 | 1 | |||
| Eubacterium sp. 1 | 2 | |||
| Eubacterium sp. 2 | 1 | |||
| Eubacterium sp. 3 | 1 | |||
| Eubacterium sp. 4 | 1 | |||
| Eubacterium sp. 5 | 1 | |||
| Eubacterium sp. 6 | 1 | |||
| Eubacterium sp. 7 | 2 | |||
| Fusobacterium sp. 1 | 6 | |||
| Fusobacterium sp. 2 | 1 | |||
| Fusobacterium sp. 3 | 1 | |||
| Ruminococcus sp. 1 | 4 | |||
| Ruminococcus sp. 2 | 2 | |||
| Ruminococcus sp. 3 | 2 | |||
| Ruminococcus sp. 4 | 2 | |||
| Ruminococcus sp. 5 | 1 | |||
| Ruminococcus sp. 6 | 1 | |||
| Ruminococcus sp. 7 | 1 | |||
| Ruminococcus sp. 8 | 1 | |||
| Ruminococcus sp. 9 | 1 | |||
| Ruminococcus sp. 10 | 1 | |||
| Ruminococcus sp. 11 | 1 | |||
| Ruminococcus sp. 12 | 1 | |||
| Ruminococcus sp. 13 | 1 | |||
| Ruminococcus sp. 14 | 1 | |||
| Ruminococcus sp. 15 | 1 | |||
| Ruminococcus sp. 16 | 1 | |||
| Ruminococcus sp. 17 | 1 | |||
| Ruminococcus sp. 18 | 1 | |||
| Ruminococcus sp. 19 | 1 | |||
| Ruminococcus sp. 20 | 1 | |||
| Ruminococcus sp. 21 | 1 | |||
| Ruminococcus sp. 22 | 1 | |||
| Ruminococcus sp. 23 | 1 | |||
| Ruminococcus sp. 24 | 1 | |||
| Ruminococcus sp. 25 | 1 | |||
| Sporobacter sp. 1 | 1 | |||
| Sporobacter sp. 2 | 1 | |||
| Unidentified phylotypes | ||||
| Unidentified phylotype 1 | 5 | |||
| Unidentified phylotype 2 | 3 | |||
| Unidentified phylotype 3 | 2 | |||
| Unidentified phylotype 4 | 1 | |||
| Unidentified phylotype 5 | 1 | |||
| Unidentified phylotype 6 | 1 | |||
| Unidentified phylotype 7 | 1 | |||
| Unidentified phylotype 8 | 1 | |||
| Unidentified phylotype 9 | 1 | |||
| Unidentified phylotype 10 | 1 | |||
| Unidentified phylotype 11 | 1 | |||
| Unidentified phylotype 12 | 1 | |||
| Unidentified phylotype 13 | 1 | |||
| Unidentified phylotype 14 | 1 | |||
| Unidentified phylotype 15 | 1 | |||
| Unidentified phylotype 16 | 1 | |||
| Unidentified phylotype 17 | 1 | |||
| Unidentified phylotype 18 | 1 | |||
| Unidentified phylotype 19 | 1 | |||
| Unidentified phylotype 20 | 1 | |||
| Unidentified phylotype 21 | 1 |
Criteria for identification were as described for Table 1. n indicates the number of clones obtained for that phylotype.
The 37 clone sequences that had Sab scores of ≥0.95 represented Bacteroides fragilis (3 clones), Bacteroides merdae (1 clone), B. saeculare (10 clones), Enterococcus mundti (1 clone), Escherichia coli (1 clone), Klebsiella pneumoniae (1 clone), L. crispatus (10 clones), Lactobacillus gasseri (2 clones), Lactobacillus pontis (1 clone), L. salivarius (1 clone), and Streptococcus bovis (6 clones) (Table 2). Seventy-one of the sequences produced Sab scores between 0.70 and 0.95, often representing several different phylotypes within a given genus. For example, there were 4 clones representing 4 different phylotypes within the genus Bacteroides, 15 clones representing 11 different phylotypes within the genus Clostridium, and 31 clones representing 25 different phylotypes within the genus Ruminococcus (Table 2). The remaining 28 sequences produced Sab scores below 0.70 and were distributed into 21 different unidentified phylotypes (Table 2). These findings are generally consistent with prior reports on chicken cecal microflora composition using phylogenetic approaches by this and other groups (1, 3, 14, 29). Collectively, these data indicate that there are many as-yet-unknown microbes inhabiting the chicken GI tract, at least in the context of phylogenetic characterization.
The most abundant species, based on frequency of cloning, were B. saeculare (7% of total) and L. crispatus (7% of total). At the genus level, the most abundant microbes were Ruminococcus (23% of total), Clostridium (11% of total), Lactobacillus (10% of total), and Bifidobacterium (8% of total). It is important to note that there were 87 unique phylotypes represented in this set of 136 clones, and the number of representatives of individual phylotypes in this study ranged between 1 (for 71 phylotypes) and 10 (for 2 phylotypes). Based on simple probability estimates and ignoring any potential cloning bias, a given phylotype would need to represent at least 2.2% of the total to be detected in this study with 95% confidence and at least 3.4% of the total to be detected with 99% confidence. Since more than half of the phylotypes detected in this survey were represented only once (0.7% of total), it can be assumed that there were many more phylotypes present in low abundance that went undetected. If we theoretically consider a system where there are 200 different phylotypes with relative abundances normally distributed across an order of magnitude from 0.1 to 1% of the total, 2,000 sequences (=10 times the number of phylotypes) would be required to detect all 200 phylotypes with 95% confidence. This is likely a conservative estimate, since others have noted that complex microbial communities often exhibit a log-normal distribution and thus contain a higher proportion of rare populations (6, 9). Alternatively, an approach that can dissect or fractionate the complexity of the entire community, allow directed recovery of unique phylotypes, and facilitate detection of populations present in low abundance could be employed to enhance detection of the diversity present in the community. Preliminary fractionation of total community DNA based on G+C content, as described here, represents one such approach.
In the context of the present study, it is most important to compare the suites of clones obtained by random cloning and the combined GC-DGGE approach. While it is difficult to apply statistical approaches to directly compare these disparate data sets, one from random sampling and the other from directed sampling, chi-square analysis indicated that the distribution of clones in the data sets obtained by the two approaches was very different (P < 0.01). The results were also quite striking in a qualitative sense. Where DGGE bands were selected for cloning because they were abundantly represented in the unfractionated super-pool sample (e.g., lane 5, band a; lane 9, band a; lane 10, band a), the corresponding phylotypes were well-represented in the random cloning survey (Table 3). Where DGGE bands from individual fractions were of intermediate abundance in the lanes from the unfractionated super-pool sample (e.g., lane 2, bands a and b; lane 3, band e; lane 8, bands b, c and d), the corresponding phylotypes from DNA fractions representing high relative abundance in the total community (e.g., fraction 8) were generally detected by the random cloning approach, whereas those from fractions of low relative abundance (lanes 2 and 3) were not (Table 3). In cases where DGGE bands were selected because they were undetected or in very low abundance in the super-pool lanes (e.g., lane 1, band a; lane 3, bands a and b; lane 6, band a), the corresponding phylotypes were not recovered by the random cloning approach. Overall, 22 of the 28 phylotypes recovered from excised DGGE bands (79%) were not detected by the random cloning approach. If one excludes clones from bands selected because they were abundant in the unfractionated super-pool sample (lane 5, band a; lane 9, band a; lane 10, band a), 20 of the 23 different phylotypes recovered from the remaining targeted DGGE bands (87%) were not represented in the pool of phylotypes recovered by random cloning.
TABLE 3.
Overlapping phylotype clones between random cloning and GC-DGGE approaches
| Best-match identification | n (random)a | Band clone(s) |
|---|---|---|
| B. saeculare | 10 | 10-a-1, 10-a-2 |
| L. crispatus | 10 | 5-b-1, 5-b-2 |
| Clostridium sp. 1 | 5 | 8-b-1, 8-b-2 |
| Fusobacterium sp. 1 | 6 | 9-a-1, 9-a-2, 9-a-3 |
| Ruminococcus sp. 1 | 2 | 8-d-3 |
| Ruminococcus sp. 4 | 1 | 8-c-1, 8-c-3 |
n (random) indicates the number of times the phylotype was encountered by random cloning from unfractionated super-pool DNA.
Collectively, these data demonstrate that the GC-DGGE approach allows directed detection, recovery, and analysis of phylotypes that are not, or are only poorly, represented when total bacterial community DNA is analyzed directly. Further, although an analysis of PCR bias was not an objective of this study, the data presented herein indicate that the PCR-based, random cloning approach taken with the super-pool DNA was relatively free of PCR bias, since sequences from abundant fractions were readily detected while those from fractions of low abundance were detected only rarely or not at all.
The power of combined approaches.
In this paper we report a novel strategy, GC-DGGE, which combines mechanistically different microbial community analysis approaches to facilitate enhanced assessment of microbial community diversity and detection of minority populations of microbes. The underlying basis for this strategy is to employ GC fractionation as an initial step to divide total community DNA into fractions based on G+C content, thereby effectively reducing the complexity of the community in each fraction. This reduced complexity facilitates detection of diversity based on DGGE analysis and directed cloning and sequencing of individual bands from DGGE lanes corresponding to individual fractions. This combined approach (GC-DGGE) overcomes both the primary limitation of GC fractionation, low resolution that does not indicate the number or identity of different taxa in a particular G+C fraction, and also the primary limitation of DGGE, the inability to detect populations present in low abundance, to allow enhanced detection of total microbial community diversity.
Acknowledgments
We gratefully acknowledge Harri Makivuokko for invaluable assistance with sample preparation and GC fractionation and Linda Schimmelpfennig for excellent technical assistance in cloning, sequencing, and data analysis. We also offer our sincere gratitude to Bruno Pot of Applied Maths for invaluable help and advice with image analysis of DGGE gels.
This work was funded in part by the National Technology Agency of Finland, Tekes.
REFERENCES
- 1.Apajalahti, J. H., A. Kettunen, M. R. Bedford, and W. E. Holben. 2001. Percent G+C profiling accurately reveals diet-related differences in the gastrointestinal microbial community of broiler chickens. Appl. Environ. Microbiol. 67:5656-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apajalahti, J. H., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apajalahti, J. H., L. K. Sarkilahti, B. R. Maki, J. P. Heikkinen, P. H. Nurminen, and W. E. Holben. 1998. Effective recovery of bacterial DNA and percent-guanine-plus-cytosine-based analysis of community structure in the gastrointestinal tract of broiler chickens. Appl. Environ. Microbiol. 64:4084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apajalahti, J. H. A., A. Kettunen, P. H. Nurminen, H. Jatila, and W. E. Holben. 2003. Selective plating underestimates abundance and shows differential recovery of bifidobacterial species from human feces. Appl. Environ. Microbiol. 69:5731-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong, E. F., D. G. Franks, and A. L. Alldredge. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924-934. [Google Scholar]
- 8.Devereux, R., and G. W. Mundfrom. 1994. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl. Environ. Microbiol. 60:3437-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunbar, J., S. M. Barns, L. O. Ticknor, and C. R. Kuske. 2002. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 68:3035-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feris, K. P., P. W. Ramsey, C. Frazar, M. C. Rillig, J. E. Gannon, and W. E. Holben. 2003. Structure and seasonal dynamics of hyporheic zone microbial communities in free-stone rivers of the western United States. Microb. Ecol. 10.1007/s00248-002-0100-x. [DOI] [PubMed]
- 12.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1993. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl. Environ. Microbiol. 59:1294-1302. (Erratum, 61:4517, 1995.) [DOI] [PMC free article] [PubMed]
- 14.Gong, J., R. J. Forster, H. Yu, J. R. Chambers, P. M. Sabour, R. Wheatcroft, and S. Chen. 2002. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 208:1-7. [DOI] [PubMed] [Google Scholar]
- 15.Gray, J. P., and R. P. Herwig. 1996. Phylogenetic analysis of the bacterial communities in marine sediments. Appl. Environ. Microbiol. 62:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gsell, T. C., W. E. Holben, and R. M. Ventullo. 1997. Characterization of the sediment bacterial community in groundwater discharge zones of an alkaline fen: a seasonal study. Appl. Environ. Microbiol. 63:3111-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holben, W. E., and D. Harris. 1995. DNA-based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 18.Holben, W. E., K. Noto, T. Sumino, and Y. Suwa. 1998. Molecular analysis of bacterial communities in a three-compartment granular activated sludge system indicates community-level control by incompatible nitrification processes. Appl. Environ. Microbiol. 64:2528-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holben, W. E., P. Williams, M. A. Gilbert, M. Saarinen, L. K. Sarkilahti, and J. H. Apajalahti. 2002. Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 44:175-185. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishii, K., and M. Fukui. 2001. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 67:3753-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. J. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyzer, G., E. C. de Wall, and A. J. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neefs, J.-M., Y. V. de Peer, L. Hendriks, and R. De Wachter. 1990. Compilation of small ribosomal RNA sequences. Nucleic Acids Res. 18:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Netherwood, T., H. J. Gilbert, D. S. Parker, and A. G. O'Donnell. 1999. Probiotics shown to change bacterial community structure in the avian gastrointestinal tract. Appl. Environ. Microbiol. 65:5134-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanguinetti, C. J., E. Dias Netto, and A. J. G. Simpson. 1994. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. BioTechniques 17:915-919. [PubMed] [Google Scholar]
- 33.Santo Domingo, J. W., M. G. Kaufman, M. J. Klug, W. E. Holben, D. Harris, and J. M. Tiedje. 1998. Influence of diet on the structure and function of the bacterial hindgut community of crickets. Mol. Ecol. 7:761-767. [Google Scholar]
- 34.Schleper, C., W. Holben, and H. P. Klenk. 1997. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl. Environ. Microbiol. 63:321-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmalenberger, A., F. Schwieger, and C. C. Tebbe. 2001. Effect of primers hybridizing to different evolutionarily conserved regions of the small-subunit rRNA gene in PCR-based microbial community analyses and genetic profiling. Appl. Environ. Microbiol. 67:3557-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt, T. M., E. F. deLong, and N. R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173:4371-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stackebrandt, E., W. Liesack, and B. M. Goebel. 1993. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 7:232-236. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki, M., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takai, K., D. P. Moser, M. DeFlaun, T. C. Onstott, and J. K. Fredrickson. 2001. Archaeal diversity in waters from deep South African gold mines. Appl. Environ. Microbiol. 67:5750-5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torsvik, V., F. L. Daae, R. A. Sandaa, and L. Ovreas. 1998. Novel techniques for analysing microbial diversity in natural and perturbed environments. J. Biotechnol. 64:53-62. [DOI] [PubMed] [Google Scholar]
- 41.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity-magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 42.Torsvik, V., K. Salte, R. Sorheim, and J. Goksoyr. 1990. Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl. Environ. Microbiol. 56:776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda, T., Y. Suga, and T. Matsuguchi. 1995. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur. J. Soil Sci. 46:415-421. [Google Scholar]
- 44.Ward, D. M., M. J. Ferris, S. C. Nold, and M. M. Bateson. 1998. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 62:1353-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]