Abstract
Gibberella zeae (asexual state Fusarium graminearum) is a major causal agent of wheat head blight and maize ear rot in North America and is responsible for contamination of grain with deoxynivalenol and related trichothecene mycotoxins. To identify additional trichothecene biosynthetic genes, cDNA libraries were prepared from fungal cultures under trichothecene-inducing conditions in culture and in planta. A gene designated LH1 that was highly expressed under these conditions exhibited only moderate (59%) similarity to known trichothecene biosynthetic cytochrome P450s. To determine the function of LH1, gene disruptants were produced and assessed for trichothecene production. Gene disruptants no longer produced 15-acetyldeoxynivalenol, which is oxygenated at carbon 7 (C-7) and C-8, but rather accumulated calonectrin and 3-deacetylcalonectrin, which are not oxygenated at either C-7 or C-8. These results indicate that gene LH1 encodes a cytochrome P450 responsible for oxygenation at one or both of these positions. Despite the relatively low level of DNA and amino acid sequence similarity between the two genes, LH1 from G. zeae is the probable homologue of Tri1, which encodes a cytochrome P450 required for C-8 oxygenation in F. sporotrichioides.
Trichothecenes are potent inhibitors of protein synthesis and virulence factors in wheat head scab (4). Many of the genes required for trichothecene biosynthesis are clustered in a 26-kb region in both the Fusarium sporotrichioides and F. graminearum genomes (2, 11). Sequence differences have been identified in the gene clusters between T-2 toxin-producing strains of F. sporotrichioides and deoxynivalenol (DON)-producing strains of F. graminearum that can account for their different trichothecene phenotypes (3, 12, 12, 13). For example, the genes responsible for C-4 oxygenation and acetylation, Tri13 and Tri7, respectively, are missing or inactive in the F. graminearum DON-producing strains but are present in T-2 toxin-producing strains of F. sporotrichioides and nivalenol-producing strains of F. graminearum. Not all trichothecene biosynthetic genes are in the cluster, and three additional trichothecene-related genes have been identified that are physically separated from the main cluster. Tri101, which encodes trichothecene C-3 acetyltransferase (10, 15), is located on linkage group 3, while the main cluster is located on linkage group 1 (9). With the release of the F. graminearum genomic sequence (www.genome.wi.mit.edu), Tri101 has been located on contig 1.321 whereas the main cluster is on contig 1.159. A second small group of at least two trichothecene-related genes that encode the C-8 oxygenase (Tri1) and the C-8 acyltransferase (Tri16) in F. sporotrichioides also is known (18, 21). These researchers could not identify a homologue of Tri1 in F. graminearum by Southern analysis.
A putative homologue of Tri1 in F. graminearum, LH1, was found when we examined an expressed sequence tag (EST) database derived from a variety of F. graminearum expression libraries. In order to determine the function of this gene, disruption mutants were made and analyzed. This is the first characterization of a possible multifunctional oxygenase required for trichothecene biosynthesis.
MATERIALS AND METHODS
Strains and growth conditions.
Originally collected from maize caryopsis in Ottawa, Ontario, Canada, by G. A. Neish in 1981, F. graminearum wild-type strain DAOM180378 was provided to us by C. Babcock of the Canadian Collection of Fungal Cultures (CCFC/DAOM), Agriculture & Agri-Food Canada, Ottawa. The F. graminearum wild-type strain, Z-3639 (NRRL 29214; USDA/ARS Culture Collection, Peoria, Ill), was isolated from scabby wheat by R. Bowden (Kansas State University) (1) and was maintained on V-8 juice agar (26) slants. F. graminearum LH1D3, LH1D41, LH1D44, LH1D47, LH1D59, LH1D65, and LH1D72 (available from the Fungal Genetics Stock Center [FGSC9497 to FGSC9503]) contain disrupted sequences of LH1, as described below. Transgenic strains GzTri3D-11 (FGSC9488) and GzTri11D-12 (FGSC9496) were constructed from Z-3639 with disrupted sequences of Tri3 (17) and Tri11 (16), respectively, as described for F. sporotrichioides. All fungal cultures were initially grown on V-8 juice agar plates under an alternating 12-h, 25°C light-12-h, 22°C dark cycle. Fungal cultures were maintained on V-8 slants. Spores or agar plugs of fungal cultures were stored long term at −80°C in 10% glycerol.
Culture conditions and cDNA library construction.
The Fg03 cDNA library was prepared from a fungal culture (strain DAOM180378) grown under conditions designed to promote trichothecene production, based on a protocol of Miller and Blackwell (19). A 3.5-ml macerated suspension of potato dextrose agar (Difco Laboratories, Detroit, Mich.)-grown mycelium in distilled water was used to inoculate 50 ml of first-stage medium (3 g of NH4Cl, 2 g of MgSO4 · 7H2O, 0.2 g of FeSO4 · 7H2O, 2 g of KH2PO4, 2 g of peptone, 2 g of yeast extract, 2 g of malt extract, and 20 g of glucose in 1 liter of distilled water) in a 250-ml flask and grown at 28°C in the dark (with shaking at ∼250 rpm) for 3 days. The culture was ground in a Waring blender and concentrated by centrifugation (20 min, 1,000 × g, 4°C), and 50% of the supernatant was removed. A 3.5-ml aliquot of the concentrated culture was used to inoculate 50 ml of second-stage production medium (modified MYRO) [1 g of (NH4)2HPO4, 3 g of KH2PO4, 0.2 g of MgSO4 · 7H2O, 5 g of NaCl, 40 g of sucrose, and 10 g of glycerol in 1 liter of distilled water] in 250-ml flasks and grown under the same conditions as first-stage medium for 8 days. The mean DON concentration of the cultures was determined by competitive direct enzyme-linked immunosorbent assay (25) to be 165 μg/ml (courtesy of M. Savard, Agriculture & Agri-Food Canada, Ottawa). The culture was harvested and partially dried by filtration through Whatman 1MM filter paper on a Büchner funnel and stored at −70°C until use.
The Fg05 cDNA library was prepared with RNA isolated from mycelium (strain DAOM180378) cultured on cornmeal. Cornmeal powder (25 g) was hydrated with 12 ml of water in a 250-ml flask and inoculated with mycelium washed from two confluent V-8 agar plates. The culture was grown in the dark at 25°C without agitation and harvested after 11 days. The mean DON concentration was determined by competitive direct enzyme-linked immunosorbent assay to be 46 μg/ml.
Total fungal RNA was isolated by using the standard TRIzol method (Invitrogen, Carlsbad, Calif.), and poly(A) RNA was isolated by using the PolyATract system (Promega, Madison, Wis.). In order to construct a cDNA library with a more complete representation of mRNAs, the Fg03 library was prepared by using a modified subtractive suppression hybridization (SSH) (5) protocol that reduces the bias toward the more abundant mRNA species. This normalization protocol involved allowing the mRNA to hybridize to itself, and as the more abundant mRNA species would hybridize more easily to each other, they would form a dimer that would be eliminated from the PCR amplification step that followed. Consequently, the mRNA population is normalized so that the more abundant mRNA species are decreased to a level similar to that of the less abundant species. The protocol was a modified version of the PCR-Select cDNA subtraction kit (Clontech Laboratories, Inc., Palo Alto, Calif.) whereby cDNA was not digested with RsaI prior to hybridizations and no subtraction with an alternate mRNA population was attempted. The Fg05 cDNA library was not normalized and was constructed by using the GeneRacer kit (Invitrogen). For both Fg03 and Fg05 cDNA libraries, cDNA was cloned undirected into the pGEM-T vector (Promega) and transformed into SURE 2 cells (Stratagene, La Jolla, Calif.).
Sequencing and gene analysis.
Single-pass 5′- or 3′-end sequencing of random cDNA clones was conducted with a LI-COR 4200 sequencing system (LI-COR, Lincoln, Nebr.) with an average read length of 619 nucleotides. Redundant ESTs were grouped into contigs with the Seqman II assembly program (DNASTAR Inc., Madison, Wis.) with a minimum match of 90% over a length of 30 bp. Bioinformatic analyses were performed with the Lasergene software package (DNASTAR, Inc.). BLAST searches were done with the TBLASTN program (http://www.ncbi.nlm.nih.gov/BLAST) by using the default parameters.
Infected wheat heads.
Wheat heads (cv. Roblin) at mid-anthesis stage were sprayed with a conidial suspension of F. graminearum (105 spores/ml). Following inoculation, plants were kept for 2 days at high humidity in a dew chamber (15°C walls, 20°C water, 30°C air) without light. The infected plants were then kept for 2 days under misting conditions with a 10-s mist every 20 min at 25°C. The resulting fungal mycelial tissue was sampled and stored at −70°C.
Solid rich medium.
Mycelia were grown on V-8 agar plates for 7 days at 28°C with 14 h of light to obtain a culture grown on rich medium. Mycelia were harvested from plates with 0.6 M MgSO4, ground under liquid N2, and stored at −70°C until further use.
Stripped wheat leaves.
Leaves (100 g) from 39-day-old wheat plants were soaked in oxalate buffer at 95°C for 48 h. The leaves were placed on Miracloth (Calbiochem, La Jolla, Calif.), rinsed with water, and ground in liquid N2. The ground tissue was resuspended twice in methanol, separated through filtration, and suspended in chloroform with stirring overnight at room temperature. The ground leaf tissue was rinsed two times with ethanol and three times with water, and excess water was absorbed by Whatman filter paper. An aliquot of the ground leaf tissue (5 g) was autoclaved and added to 500 ml of 1× synthetic low-nutrient agar (20), with 1 g of sodium acetate per liter added, to form the medium. This medium was inoculated with two agar plugs of F. graminearum DAOM180378 and shaken at 200 rpm at 28°C for 6 days. The culture was harvested and partially dried by vacuum filtration through sterile Miracloth (Calbiochem) and stored at −70°C until use.
Northern analyses.
Total RNA was isolated by the standard TRIzol method (Invitrogen) from the (trichothecene-producing and cornmeal) cultures described above.
Total RNA (10 μg) was transferred to Magnacharge membrane (Osmonics Inc., Westborough, Mass.) in accordance with the manufacturer's instructions. A 650-bp EcoRI fragment from cDNA clone Fg03_09a11 was random prime labeled with the Prime-a-Gene Labeling System (Promega) and [32P]dCTP (Amersham Biosciences, Piscataway, N.J.) for use as a hybridization probe. Essentially, hybridizations were done in 50% formamide buffer overnight at 42°C. Final washes were done with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate at 65°C. The blot was exposed to Kodak (Rochester, N.Y.) BioMax MS film with a Kodak BioMax TranScreen-HE intensifying screen.
Gene disruption and transformations.
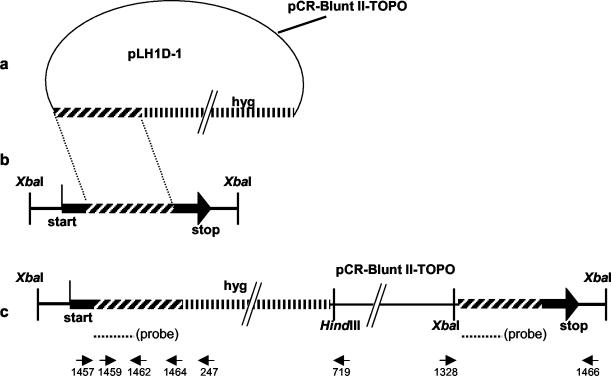
To make the disruption vector for F. graminearum LH1, the gene (GenBank AY339129) was amplified from strain Z-3639 with Pfu polymerase (Stratagene) by following the manufacturer's recommendations (95°C for 2 min, 95°C for 30 s, 52°C for 30 s, and 72°C for 2 min for 30 cycles, followed by 72°C for 10 min) with primers 1459 and 1464 (Table 1), the latter containing an AscI site (underlined), with a PTC-100 thermocycler (MJ Research, Watertown, Mass.). The resulting 1,281-bp fragment was purified from an agarose gel (UltraClean; MoBio, Solana Beach, Calif.) and cloned into pCR-Blunt II-TOPO (Invitrogen). This plasmid was cut with AscI, and an AscI fragment containing the promoter 1-hygB selectable marker (30) was inserted. This insertion formed a disruption vector (pLH1D-1) consisting of a truncated portion of the coding region lacking 268 bp from the 5′ end and 202 bp from the 3′ end of LH1, followed by the hygromycin selectable marker.
TABLE 1.
DNA primers used in this study
| Primer | Sequence 5′→3′ | Gene |
|---|---|---|
| 247 | GGTCAACATGATGTCAGG | hyg |
| 719 | GACGTTGTAAAACGACGGCCAGTG | Plasmid |
| 1113 | ATGTTTCAATATTCCCTATGGC | FgTri11 |
| 1114 | TCATCTTGGGTCAAGGTAC | FgTri11 |
| 1199 | CGGATCATCTATCCTTGAG | FgTri11 |
| 1207 | GGAGGGCCTAGTTTAGG | FgTri11 |
| 1211 | CGTGCCCTGGGGACCAACAC | hyg |
| 1212 | CACAGCCGTGGTTGGTGGGG | hyg |
| 1275 | CAACCATCAAGATGAGCGC | FgTri3 |
| 1276 | CCTCGAGAAACCTATGTACC | FgTri3 |
| 1277 | GTTCTAATAACACAGAGC | FgTri3 |
| 1278 | GTCCTTGGAGTACCACCG | FgTri3 |
| 1328 | CACACAGGAAACAGCTATGACC | Plasmid |
| 1457 | GTCATGGCTCTCATCACCAG | FgLH1 |
| 1459 | GCCTCCCACTCTGGCCGATG | FgLH1 |
| 1462 | GTAGCAGTGCGTCTCTGAG | FgLH1 |
| 1464 | CATGGCGCGCCCAGGGCAGGCGTGTTCTCa | FgLH1 |
| 1466 | GTCAACAAGTGGCGAGATC | FgLH1 |
The underlined sequence is an AscI site.
The disruption vectors for FgTri11 and FgTri3 were made by inserting the promoter1-hygB selectable marker into the middle region of each gene, thereby disrupting the formation of an intact, functional protein. FgTri11 was amplified from strain Z-3639 with Pfu polymerase under the conditions described earlier, with primers 1113 and 1114 (Table 1). The 1.73-kb fragment was inserted into pT7-Blue3 with the Perfectly Blunt Cloning Kit (Novagen, EMDBiosciences). The newly created vector was cut with the restriction enzyme EcoRV at a unique site in the FgTri11 sequence, 724 bp from the start site and 1,014 bp from the stop site; treated with CIAP; and ligated with the SacI/XmnI fragment containing the promoter1-hygB marker from the vector pUCH4 (14). Transformation of fungal protoplasts was performed as previously described (22).
Disruption analysis.
Fungal transformants were analyzed with PCR and Southern techniques (24). To confirm the disruption of LH1, primers outside of the cloned sequence (1457 and 1466) were paired with internal ones (247, 719, and 1328) in order to detect true insertions versus ectopic insertions (Fig. 1). The same technique was used for confirmation of the disruption of FgTri11 and FgTri3 with primers specific to their sequences (Table 1).
FIG. 1.
Disruption of LH1 in F. graminearum. (a) Diagram of disruptant plasmid pLH1D-1. The hatched segment on the left represents a 1,281-bp fragment of LH1. The vertically lined segment on the right represents a promoter 1-hygB fragment. (b) Diagram of the Z-3639 genomic DNA containing the LH1 region. “Start” and “stop” are the ends of LH1. (c) Diagram of the LH1 region of a transformant resulting from a single crossover event. The binding position of the probe used for Southern analysis is indicated; arrows indicate the positions of primers. Primer sequences are given in Table 1.
In the Southern analysis, genomic DNAs of the F. graminearum wild-type and transformants strains were digested with XbaI and HindIII (New England Biolabs, Beverly, Mass.), blotted to NYtran SuperCharge membrane (Schleicher & Schuell, Keene, N.H.), and hybridized to a 32P-labeled probe (Prime-A-Gene; Promega) consisting of 443 bp of the coding region (overlapping exons 1 through 4) of LH1. Primers 1459 and 1462 (Table 1) were used to amplify the probe from the Z-3639 template. For analysis of FgTri11 disruptants, the genomic DNA was restricted with PvuII/NcoI, while BglII was used for FgTri3 disruptants.
Trichothecene toxin assays.
Mutant strains were maintained on slants of V-8 juice agar with hygromycin B (300 μg/ml). Liquid cultures were grown on GYEP medium (5% glucose, 0.1% yeast extract, 0.1% peptone; 20 ml in 50-ml Erlenmeyer flasks). Liquid cultures of F. graminearum were inoculated with a 20-mm mycelial plug cut from 1-week-old cultures grown on V-8 juice agar. All liquid cultures were grown at 28°C in the dark and shaken at 200 rpm. Liquid cultures of F. graminearum were harvested after 7 days and extracted with ethyl acetate by vortexing in a conical tube, and the concentrated extract was analyzed by gas chromatography (GC) as described previously (17). To isolate sufficient quantities of the compounds produced by the F. graminearum mutant strains for spectral analyses, putative mutant strains were grown on cornmeal cultures. Cornmeal cultures were prepared by inoculating cornmeal (25 g moistened with 11 ml of water) in 300-ml Erlenmeyer flasks with spores washed from V-8 juice agar plates. The inoculated cornmeal was incubated in the dark for 7 days at 28°C. Cultures were extracted overnight with 150 ml of ethyl acetate. Compound identifications were confirmed by GC-mass spectrometry (MS).
Feeding studies.
Liquid cultures of transgenic strains LH1D41, LH1D59, GzTri3D-11, and GzTri11D-12 were prepared as described above for Fg03 cDNA library construction. Second-stage production cultures (20 ml in 50-ml Erlenmeyer flasks) were amended with an acetone solution containing one of four 15-ADON precursors—isotrichodermin, 8-hydroxy-3-decalonectrin, 7,8-dihydroxycalonectrin, or calonectrin (250 μM final concentration). Isotrichodermin was isolated and purified from FsTri11D cultures (16). 7,8-Dihydroxycalonectrin and calonectrin were isolated and purified from GzTri8D mutant (14) strains, and 8-hydroxy-3-decalonectrin was isolated and purified from FsTri13D mutant strains (2). Aliquots (5 ml)were removed from amended cultures after 2 to 6 days, extracted with ethyl acetate (2 ml), and analyzed by GC-MS for conversion to 15-ADON.
Chemical analyses.
GC measurements were made by flame ionization detection with a Hewlett-Packard 5890 gas chromatograph fitted with a 30-m fused silica capillary column (DB1; 0.25 μm; J&W Scientific Co., Palo Alto, Calif.). For routine screening of the trichothecene toxin phenotype, the column was held at 120°C at injection, heated to 210°C at 15°C/min and held for 1 min, and then heated to 260°C at 5°C/min and held for 8 min. Low-resolution mass spectra were obtained by GC-MS with the same temperature program and a Hewlett-Packard 5891 mass-selective detector fitted with a DB-5-MS column (15 m by 0.25 mm [film thickness]).
Nucleotide sequence accession numbers.
The seven EST sequences representing LH1 have been deposited in the National Center for Biotechnology Information GenBank database under the following accession numbers: Fg03_05c04, CF075160; Fg03_05d01, CF075163; Fg03_08d01, CF075159; Fg03_09a03, CF075158; Fg03_09a11, CF075161; Fg03_09g12, CF075162; Fg05_04m07, CF120339.
RESULTS
We constructed an F. graminearum EST database derived from two cDNA libraries, Fg03 and Fg05, prepared from RNA produced by trichothecene-producing liquid cultures and solid substrate cultures, respectively. The analysis revealed numerous genes that were only found in the Fg03 and Fg05 libraries and not represented by previously released F. graminearum ESTs (28). Two sequence contigs were identified whose best BLAST match was a P450 monooxygenase from F. sporotrichioides (TRI1 protein, AAK77933). These two contigs contained six ESTs from the Fg03 library and one EST from the Fg05 library. Further sequencing of the seven clones representing the above ESTs confirmed that they were overlapping and were derived from a single gene, tentatively named LH1. A cDNA consensus sequence of these seven ESTs could potentially code for a 512-residue protein exhibiting 59% amino acid identity with FsTRI1.
Expression analysis of LH1.
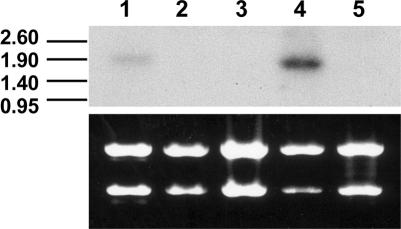
A 650-bp EcoRI fragment from the Fg03_09a11 EST clone was used as a probe to investigate the expression of LH1 under various growth conditions. Expression of a single transcript of approximately 1.9 kb was detected in the trichothecene-producing liquid culture and, at a lower level, in Fusarium-infected wheat heads (Fig. 2).
FIG. 2.
Northern blot analysis of LH1 with the 32P-labeled gene-specific Fg03_09a11 probe hybridized with total RNA isolated from F. graminearum grown under the following conditions: lane 1, infected wheat heads; lane 2, stripped wheat leaves in liquid culture; lane 3, solid rich medium; lane 4, trichothecene-producing liquid culture; lane 5, solid cornmeal substrate. The blot was exposed to film for 5 h at −70°C. The corresponding ethidium bromide-stained gel (showing strong 25S and 18S rRNA bands) prior to blotting is shown under the hybridization panel. Sizes of markers are indicated in kilobases on the left.
Disruption of LH1 in F. graminearum.
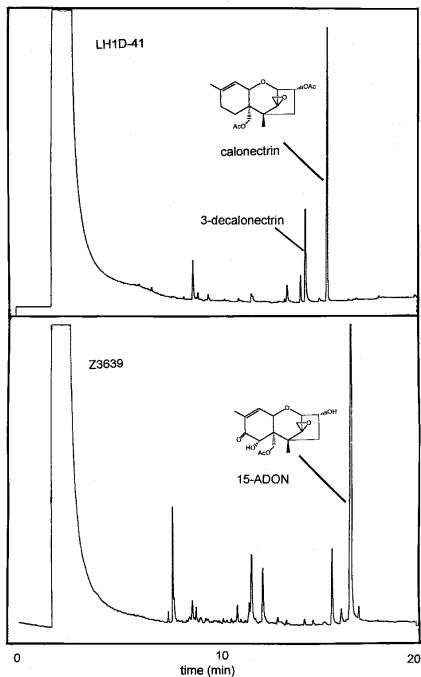
Seven of 72 hygromycin-resistant colonies resulting from transformation of Z-3639 with pLH1D-1 had a trichothecene phenotype that differed from that of Z-3639 and accumulated two compounds in larger relative amounts than did wild-type strain Z-3639: calonectrin and 3-decalonectrin. (Fig. 3). In addition, these seven transformant strains produced no detectable 15-ADON or DON. The remaining 65 hygromycin-resistant strains produced 15-ADON and a small amount of 3-decalonectrin and were indistinguishable from wild-type Z-3639 (Fig. 3).
FIG. 3.
GC chromatograms of extracts from 7-day-old liquid cultures of F. graminearum mutant strain LH1D-41 and wild-type strain Z-3639. OAc or AcO, acetate.
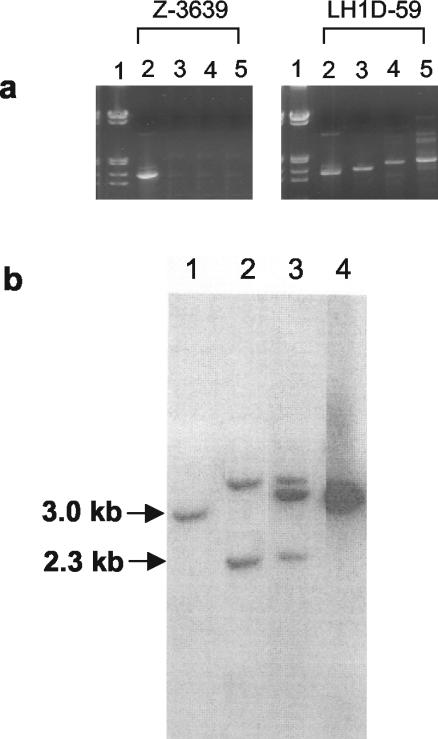
The seven transformants with the altered trichothecene phenotype were confirmed to be LH1 disruptants by PCR analysis (Fig. 4a). Primers 1457 and 1464 (Fig. 1) made a band of approximately 1.5 kb in both wild-type strain Z-3639 and transformant LH1D-59 (Fig. 4a, lanes 2). When primer 1457 (located outside of the region cloned into the disruption vector) was paired with primer 247 (located in the hyg sequence), no band was seen in Z-3639 while a band just slightly larger than 1.5 kb was seen in LD1-59 (Fig. 4a, lanes 3). When primer 1466, located downstream of the 3′ end of the gene (Fig. 1), was paired with primer 1328 (located in the vector sequence), no band was seen in Z-3639 while a band of approximately 1.7 kb was seen in the transformant (Fig. 4a, lanes 4). A control reaction for the presence of the hygB gene (primers 1211 and 1212; Fig. 4a, lanes 5) showed a band in LH1D-59 but not in Z-3639. The sizes of the PCR products are consistent with disruption of LH1 via a single homologous recombinational event between the plasmid and chromosomal LH1 sequences. Both hygromycin and plasmid sequences were detected within the coding region of LH1, showing that the gene had been disrupted.
FIG. 4.
Analysis of Z-3639 and LH1D transformants. (a) Ethidium bromide-stained gel of PCR products. Lanes: 1, molecular weight standards of lambda DNA cut with EcoRI and HindIII; 2, primers 1457 and 1464; 3, primers 1457 and 247; 4, primers 1466 and 1328; 5, primers 1211 and 1212. Locations of primers are indicated in Fig. 1. (b) Southern blot analysis of Z-3639, transformants LH1D-59 and LH1D-3, and vector pLH1D-1. Genomic and plasmid DNAs were digested with XbaI/HindIII and probed with an LH1 fragment prepared by using primers 1459 and 1462. Lanes: 1, Z-3639; 2, LH1D-59; 3, LH1D-3; 4, pLH1D-1.
The PCR results were confirmed by Southern analysis (Fig. 4b). The wild-type F. graminearum strain gave a band of about 3 kb (lane 1). Transformant FgLH1D-59 (lane 2) gave bands of about 4.4 and 2.3 kb. As shown in Fig. 1c, there is a HindIII site located in the plasmid just after the 3′ end of the hygB gene so that an XbaI/HindIII fragment identified by the probe would be a total of 2.5 kb (hygB) plus 1.9 kb (LH1). The smaller 2.3-kb band seen is the 5′ end of the insertion or gene (Fig. 1c). The 3-kb band seen in the wild type is not present in the transformant. Ectopic insertions of the plasmid did occur in some transformants, as can be seen in Fig. 4b (lane 3). In this transformant, there are the expected 4.4- and 2.3-kb bands, as well as a third band of 3.8 kb. This extra band results from an ectopic insertion of the plasmid as digestion of the plasmid alone gives a 3.8-kb band (Fig. 4b, lane 4). Transformants selected for further study carried the single insertion that caused disruption of LH1.
LH1D-41 and LH1D-59 were selected for feeding studies (Table 2). These cultures accumulate calonectrin and could convert 7,8-dihydroxycalonectrin to 15-ADON but could not convert 8-hydroxy-3-decalonectrin to 15-ADON (Table 2). The GzTri3D and GzTri11D mutant strains, which accumulate 15-decalonectrin and isotrichodermin, respectively (see Fig. 6), could convert calonectrin, 8-hydroxy-3-decalonectrin, and 7,8-dihydroxycalonectrin to 15-ADON (Table 2).
TABLE 2.
Summary of feeding experiments with mutant strains blocked in trichothecene biosynthesis
| Strain | Gene blocked | Acetone | Isotrichodermin | Calonectrin | 8-Hydroxy-3-decalonectrin | 7,8-Dihydroxycalonectrin |
|---|---|---|---|---|---|---|
| GzTri3D-11 | Tri3 | 15-Decal | 15-Decal | 15-ADON | 15-ADON | 15-ADON |
| GzTri11D-12 | Tri11 | 3-OAca | 3-OAc | 15-ADON | 15-ADON | 15-ADON |
| LH1D-41 | LH1 | Calonectrin | Calonectrin | Calonectrin | Calonectrin | 15-ADON |
| LH1D-59 | LH1 | Calonectrin | Calonectrin | Calonectrin | Calonectrin | 15-ADON |
OAc, acetate.
FIG. 6.
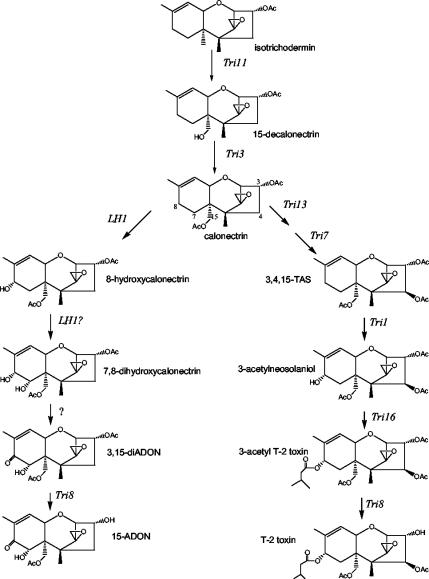
Proposed pathway from calonectrin to 15-ADON used by F. graminearum (left side) or to T-2 toxin used by F. sporotrichioides (right side). OAc or AcO, acetate. 3,15-diADON, 3,15-diacetyl-DON; 3,4,15-TAS, 3,4,15-diacetoxyscirpenol.
Sequence characteristics of LH1.
The sequence including and surrounding LH1 was examined. LH1 is found on 79.5-kb contig 1.4 (scaffold 1) of the F. graminearum genome sequence (Fusarium graminearum Sequencing Project, Whitehead Institute/MIT Center for Genome Research [http://www.genome.wi.mit.edu]). LH1 contains four introns located at bp 340 (54 bp long), bp 500 (62 bp long), bp 630 (48 bp long), and bp 1254 (52 bp long). One of the LH1 clones, Fg05_04m07, was generated by the cDNA library construction method that selects for the isolation of capped messages. If the 5′ end of clone Fg05_04m07 represents the transcription start site, there is a 48-bp 5′ leader sequence in front of the predicted translation start site and a TATA box sequence (general eukaryotic transcription binding site) at position −41. There are three TRI6 binding sites (5′YNAGGCC at −157, −213, and −307) (8), all in the inverse orientation, and one PacC binding site (5′GCCARG at −439) within 500 bp of the transcription start site of LH1. TRI6 is a zinc finger transcriptional activator of the trichothecene biosynthetic pathway (8), and PacC is a general fungal zinc finger transcription factor that regulates gene expression by ambient pH (27).
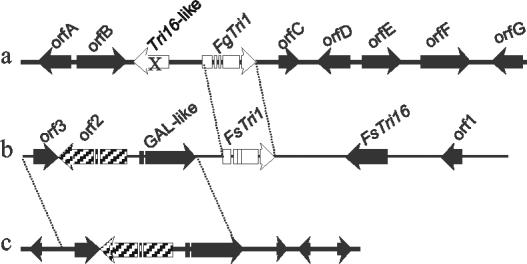
The flanking sequences contain numerous open reading frames (Fig. 5). However, there are no public EST sequences representing any of the predicted genes within 20 kb of LH1. Open reading frames immediately upstream of the region illustrated in Fig. 5 exhibit similarity to sugar utilization genes. An examination of the published F. graminearum genome sequence suggests that a TRI16-like sequence is present next to LH1, but as a pseudogene (Fig. 5).
FIG. 5.
Genomic maps of the FsTri1-Tri16 gene cluster and homologous regions in F. graminearum. Open reading frames and direction of transcription are indicated by arrows. Dotted lines indicate the sequence conserved between F. sporotrichioides and F. graminearum, although the sequences conserved between FsTri16 and the FgTri16-like region are not indicated to simplify the figure. (a) F. graminearum genomic contig 1.4 sequence, nucleotides 26000 to 44000. Translated and predicted proteins: orfA, GAL4-like transcription factor; orfB, sugar transporter; orfC, unknown; orfD, WW domain-containing oxidoreductase; orfF, flavodoxin reductase; orfG, amino acid transporter. (b) F. sporotrichioides Tri1-Tri16 gene cluster (17,529 bp; GenBank accession no. AY217783). (c) F. graminearum genomic contig 1.196, nucleotides 273000 to 284000.
DISCUSSION
Fusarium sporotrichioides Tri1 (FsTri1) encodes a P450 oxygenase that converts triacetoxyscirpenol to 3-acetylneosolaniol in T-2 toxin biosynthesis (Fig. 6). The isolation of other 8-hydroxytrichothecenes (e.g., 8-hydroxyisotrichodermin, 8-hydroxydecalonectrin, 2,3,8,11-isotrichotetraol) from Fusarium suggested that TRI1 may have a wide substrate specificity (18). FsTri1 is located in a small cluster adjacent to an acyltransferase gene responsible for adding the isovalerate group to the C-8 position. Although there is great similarity between the major trichothecene gene clusters of F. sporotrichioides and F. graminearum, DON-producing strains of F. graminearum lack functional Tri7 and Tri13 genes that are involved in hydroxylation and esterification at C-4 (2, 3, 12). The structures of T-2 toxin and 15-acetyldeoxynivalneol (Fig. 6) also significantly differ at the C-7 and C-8 positions. 15-ADON and DON have a keto group at C-8 and a hydroxyl at C-7, while T-2 toxin has no substituent at C-7 and an isovaleryl ester at C-8. The location of the genes involved with the C-8 position in F. sporotrichioides in a small, unique cluster suggests that other Fusarium species may have significant differences or changes in these enzymatic capabilities.
LH1 was represented by six cDNA clones derived from a DON-producing F. graminearum liquid culture and one clone isolated from F. graminearum grown on cornmeal. Northern analysis revealed detectable expression in trichothecene-producing liquid culture and infected wheat heads but not in cultures grown on rich medium. Expression of LH1 was not detected by Northern analysis in mycelia grown on cornmeal even though the specific culture yielding this RNA contained 210 μg of DON per ml (M. Savard, personal communication). However, these Northern results mirror the relative abundance of Tri genes in the Fg03 and Fg05 cDNA libraries. Other trichothecene biosynthetic genes are highly represented in the trichothecene-producing Fg03 library (e.g., there are 13 FgEST clones representing the Tri4 gene), but the Fg05_04m07 clone is the only putative trichothecene biosynthetic gene observed in the sequenced clones of the Fg05 library. It is possible that the fungal culture grown on cornmeal has progressed past the stage of LH1 gene expression.
The 17.5-kb genomic sequence containing the Tri1 and Tri16 gene cluster from F. sporotrichioides (AY217783) also contains three other open reading frames, as well as a potential coding region for a GAL4-like protein, in the following order: orf3, orf2, GAL4-like, Tri1, Tri16, orf1. Compared to F. graminearum, there is only the Tri16-like sequence near LH1. The orf3, orf2 (accession no. AA064249), and GAL4-like sequences in F. graminearum are conserved in the same order as in F. sporotrichioides; however, this group is located on contig 1.196 (scaffold 3) and is therefore separated from LH1 and the Tri16-like sequence. The predicted orf1 sequence from F. sporotrichioides has not been conserved in F. graminearum.
When aligned with FsTRI16, the F. graminearum Tri16-like genomic sequence exhibits greater than 20 insertions and deletions ranging from 1 to more than 400 bp. As a result, the F. graminearum Tri16-like sequence could not code for a gene product with any significant homology with FsTRI16 or other acetyltransferases. When the LH1 protein sequence is aligned with the FsTRI1 protein sequence, the predicted first residue of the LH1 protein aligns with residue 23 of the FsTRI1 protein and there is a single three-amino-acid gap in the LH1 protein after residue 266. Although the sequence identity between LH1 and FsTri1 is only 59%, the fact that all four introns line up exactly suggests that the two sequences do have homology.
A comparison of the heme-binding motifs of several different P450s involved in the trichothecene biosynthetic pathway from F. sporotrichioides and F. graminearum shows the conservation of residues in this 20-amino-acid region. TRI4, a P450 that oxygenates C-2 early in the pathway, is the same in both species, while TRI11, which oxygenates C-15, differs at three amino acids and FsTRI1 and LH1 differ at five. Although it is not known how many amino acid changes it is possible to make within this 20-amino-acid region and still have a functional protein, a comparison of the sequences of F. sporotrichioides and F. graminearum TRI4, TRI11, and TRI1 indicates that 15 of the 20 amino acids may be changed and heme binding will still occur. The consensus sequence among the six proteins is FxxGxxxCxGxxxAxxxxxx. However, there are more significant differences when the total protein or nucleotide sequences are compared with those of the other P450s identified in trichothecene biosynthesis, as sequence conservation between the two species is usually greater than 75% (2). FsTRI11 (492 amino acids) and FgTRI11 (488 amino acids) exhibit a predicted protein sequence identity of 90%. Similarly, TRI4 proteins from both species have a protein identity of 87%. In contrast, the protein identity of FsTRI1 and LH1, at 59%, is much lower than expected, but this may explain why other researchers (18) were unable to detect a Tri1 homologue in F. graminearum.
F. graminearum Z-3639 normally produces 15-ADON (Fig. 3) in liquid culture, but disruption of LH1 results in the accumulation of calonectrin (Fig. 3) and 3-decalonectrin. 3-Decalonectrin is the predicted product of the Tri8 esterase, which removes the C-3 acetyl group from a variety of trichothecenes (14), but also may result from acid hydrolysis of the C-3 acetyl group in the medium. The final sequence of the oxygenation and oxidation steps in 15-ADON-producing strains of F. graminearum is not known. Both 7,8-dihydroxycalonectrin (Fig. 6) and 15-deacetyl-7,8-dihydroxycalonectrin were precursors of 3-ADON in F. culmorum (7) cultures that also accumulated calonectrin and 7-hydroxycalonectrin. On the basis of these data, the authors proposed the following pathway: calonectrin → 7-hydroxycalonectrin → 7,8-dihydroxycalonectrin → 3,15-diacetyl-DON → 3-ADON. However, we have not found 7-hydroxycalonectrin in our mutant cultures derived from Z-3639. F. graminearum Tri8 mutants accumulate several 3-acetylated trichothecenes: 3,15-diacetyl-DON, calonectrin, 8-hydroxycalonectrin, and 7,8-dihydroxycalonectrin (14). This pattern suggests that in F. graminearum Z-3639, oxygenation of calonectrin occurs first at C-8 and is followed by C-7 hydroxylation and oxidation of the C-8 hydroxyl group (Fig. 6).
The sequence differences between FsTRI1 and LH1 could result in LH1 having functions in addition to C-8 hydroxylation. The feeding experiments with 8-hydroxy-3-decalonectrin were consistent with this hypothesis since mutants blocked at Tri3 and Tri11 could convert 8-hydroxy-3-decalonectrin and 7,8-dihydroxycalonectrin to 15-ADON, but LH1 mutants could convert only 7,8-dihydroxycalonectrin (Fig. 6; Table 2). These results indicate that conversion to the carbonyl at C-8 is not blocked in the LH1 mutant strains and that an additional P450 not encoded by LH1 catalyzes this final oxidation step. If LH1 were limited to C-8 hydroxylation, the LH1 mutants would be expected to convert 8-hydroxy-3-decalonectrin to 15-ADON. For example, multifunctional oxygenases are used in successive oxygenations at the same carbon, as well as at different carbons, in G. fujikuroi gibberellin biosynthesis (6, 23, 29). Heterologous expression of LH1 in Fusarium verticillioides or yeast and feeding experiments with additional substrates will allow us to definitively determine if LH1 encodes a multifunctional oxygenase for both C-8 and C-7 hydroxylations or if an additional P450 gene is required for the second hydroxylation in 15-ADON biosynthesis. However, on the basis of the sequence similarity between LH1 and FsTri1 and the similarity in function of C-8 hydroxylation, we propose that LH1 is the homologue of FsTri1.
Acknowledgments
We thank Laurian Robert, Dave Sprott, Julie Chapados, Anick DeMoors, and Catherine Lacroix for sequencing support; Nick Tinker, Jiro Hattori, and Philippe Couroux for bioinformatics support; and Kimberly MacDonald, Stephanie Folmar, Troy Larson, Hélène Rocheleau, and Bob Watson for technical assistance.
Names are necessary to report factually on available data; however, the U.S. Department of Agriculture neither guarantees nor warrants the standard of the products, and the use of the name by the U.S. Department of Agriculture implies no approval of the product to the exclusion of others that may also be suitable.
Footnotes
Contribution 03-290 from Ottawa/Agriculture & Agri-Food Canada.
REFERENCES
- 1.Bowden, R. L., and J. F. Leslie. 1992. Nitrate-nonutilizing mutants of Gibberella zeae (Fusarium graminearum) and their use in determining vegetative compatibility. Exp. Mycol. 16:308-315. [Google Scholar]
- 2.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 36:224-233. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2001. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and F. graminearum. Fungal Genet. Biol. 32:121-133. [DOI] [PubMed] [Google Scholar]
- 4.Desjardins, A. E., R. H. Proctor, G. Bai, S. P. McCormick, G. Shaner, G. Beuchley, and T. M. Hohn. 1996. Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant-Microbe Interact. 9:775-781. [Google Scholar]
- 5.Diatchenko, L., Y.-F. C. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helliwell, C. A., P. M. Chandler, A. Poole, E. S. Dennis, and W. J. Peacock. 2001. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 98:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hesketh, A. R., L. Gledhill, B. W. Bycroft, P. M. Dewick, and J. Gilbert. 1993. Potential inhibitors of trichothecene biosynthesis in Fusarium culmorum epoxidation of a trichodiene derivative. Phytochemistry 32:93-104. [Google Scholar]
- 8.Hohn, T. M., R. Krishna, and R. H. Proctor. 1999. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26:224-235. [DOI] [PubMed] [Google Scholar]
- 9.Jurgenson, J. E., R. L. Bowden, K. A. Zeller, J. F. Leslie, N. J. Alexander, and R. D. Plattner. 2002. A genetic map of Gibberella zeae (Fusarium graminearum). Genetics 160:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura, M., I. Kaneko, M. Komiyama, A. Takasuki, H. Koshino, K. Yoneyama, and I. Yamaguchi. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. J. Biol. Chem. 273:1654-1661. [DOI] [PubMed] [Google Scholar]
- 11.Kimura, M., T. Tokai, K. O'Donnell, T. J. Ward, M. Fujimura, H. Hamamoto, T. Shibata, and I. Yamaguchi. 2003. The trichothecene biosynthesis gene cluster of Fusarium graminearum F15 contains a limited number of essential pathway genes and expressed non-essential genes. FEBS Lett. 539:105-110. [DOI] [PubMed] [Google Scholar]
- 12.Lee, T., D. W. Oh, H. S. Kim, J. Lee, Y. H. Kim, S. H. Yun, and Y. W. Lee. 2001. Identification of deoxynivalenol- and nivalenol producing chemotypes of Gibberella zeae by using PCR. Appl. Environ. Microbiol. 67:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, T., Y.-K. Han, K.-H Kim, S.-H Yun, and Y.-W. Lee. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 68:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCormick, S. P., and N. J. Alexander. 2002. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 68:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick, S. P., and T. M. Hohn. 1997. Accumulation of trichothecenes in liquid cultures of a Fusarium sporotrichioides mutant lacking a functional trichothecene C-15 hydroxylase. Appl. Environ. Microbiol. 63:1685-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormick, S. P., T. M. Hohn, and A. E. Desjardins. 1996. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 62:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormick, S. P., N. J. Alexander, S. E. Trapp, and T. M. Hohn. 1999. Disruption of TRI101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 65:5252-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meek, I. B., A. W. Peplow, C. Ake, Jr., T. D. Phillips, and M. N. Beremand. 2003. Tri1 encodes the cytochrome P450 monooxygenase for C-8 hydroxylation during trichothecene biosynthesis in Fusarium sporotrichioides and resides upstream of another new Tri gene. Appl. Environ. Microbiol. 69:1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. D., and B. A. Blackwell. 1986. Biosynthesis of 3-acetyldeoxynivalenol and other metabolites by Fusarium culmorum HLK 1503 in a stirred jar fermentor. Can. J. Bot. 64:1-5. [Google Scholar]
- 20.Nirenberg, H. I., and K. O'Donnell. 1998. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 90:434-458. [Google Scholar]
- 21.Peplow, A. W., I. B. Meek, M. C. Wiles, T. D. Phillips, and M. N. Beremand. 2003. Tri16 is required for esterification of position C-8 during trichothecene mycotoxin production by Fusarium sporotrichioides. Appl. Environ. Microbiol. 69:5935-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593-601. [DOI] [PubMed] [Google Scholar]
- 23.Rojas, M. C., P. Hedden, P. Gaskin, and B. Tudzynski. 2001. The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc. Natl. Acad. Sci. USA 98:5838-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Sinha, R. C., and M. E. Savard. 1996. Comparison of immunoassay and gas chromatography methods for the detection of the mycotoxin deoxynivalenol in grain samples. Can. J. Plant Pathol. 18:233-236. [Google Scholar]
- 26.Stevens, R. R. 1974. Mycology guidebook, p. 703. University of Washington Press, Seattle.
- 27.Tilburn, J., S. Sarker, D. A. Widdick, E. A. Espeso, M. Orejas, J. Mungroo, M. A. Penalva, and H. N. Arst, Jr. 1995. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 14:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trail, F., J.-R. Xu, P. San Miguel, R. G. Halgren, and H. C. Kistler. 2003. Analysis of expressed sequence tags from Gibberella zeae (anamorph Fusarium graminearum). Fungal Genet. Biol. 38:187-197. [DOI] [PubMed] [Google Scholar]
- 29.Tudzynski, B., M. C. Rojas, P. Gaskin, P., and P. Hedden. 2002. The gibberellin 20-oxidase of Gibberella fujikuroi is a multifunctional monooxygenase. J. Biol. Chem. 277:21246-21253. [DOI] [PubMed] [Google Scholar]
- 30.Turgeon, B. G., R. C. Garber, and O. C. Yoder. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 17:3297-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]