Abstract
Highly mineralized tooth enamel develops from an extracellular matrix chiefly comprised of amelogenins formed by splicing of 7 (human) or 9 (rodent) exons secreted from specialized epithelia cells known as ameloblasts. Here we examined the role of the 59 amino acid alternatively spliced amelogenin known as Leucine Rich Amelogenin Peptide (LRAP) on enamel formation, using transgenic murine models in which LRAP overexpression is driven by an amelogenin promotor (TgLRAP). Beginning in the secretory stage of mouse amelogenesis, we found a reduced thickness of enamel matrix and a loss of Tomes’ processes, followed by upregulated amelogenin mRNA expression, inhibited amelogenin secretion and loss of cell polarity. In the prescretory stage (P0) amelogenin m180 mRNA expression was increased 58 fold along with a 203 fold increase in MMP-20 expression and 3.5 and 3.2 fold increased in respectively enamelin and ameloblastin. When LRAP was overexpressed on an amelogenin knockout mouse model, the ameloblasts were not affected. Further, expression of the global chromatin organizer and transcription factor SATB1 was reduced in secretory stage TgLRAP ameloblasts. These findings identify a cellular role for LRAP in enamel formation that is not directly related to directing enamel crystal formation as is reported to be the primary function of full length amelogenins. The effect of LRAP overexpression in upregulating amelogenins, MMP-20 and SATB1, suggests a role in protein regulation critical to ameloblast secretion and matrix processing, to form a mineralized enamel matrix.
Keywords: Ameloblasts, Amelogenin, Leucine Rich Amelogenin Peptide (LRAP), Enamel, transgenic mice, MMP20, SATB1
1. Introduction
Dental enamel is formed by mineralization of a protein matrix, which is subsequently removed to allow final mineralization of intercrystaline spaces, resulting in the hardest mineralized tissue in the body. This initially secreted protein matrix is made up primarily of amelogenins, which are alternatively spliced proline rich hydrophobic proteins and which comprise approximately 90% of all proteins secreted by the enamel forming ameloblast cells (Simmer, 1995; Termine et al., 1980). As these proteins are cleaved and degraded, mineral deposition in the form of hydroxyapatite crystals occurs in a well-ordered pattern (Wen et al., 2001). Therefore, amelogenins appear to have a major role in enamel biomineralization as they function to regulate the orientation, shape and length of enamel hyroxyapatite crystals (Fincham and Moradian-Oldak, 1993; Fincham et al., 1994). In fact, mice lacking the amelogenin gene form only thin mineralized enamel like matrix, lacking the prismatic structure of normal enamel (Gibson et al., 2001).
The primary RNA transcript of amelogenin can be alternatively spliced to form at least 16 mRNAs, which translate into proteins varying in abundance with proportions that change during stage of tooth formation (Hu et al., 1997; Li et al., 1995; Li et al., 1998; Simmer et al., 1994). The major expressed amelogenin isoform in the mouse enamel is M180, which is encoded by exons 2–7, skipping exon 4 with exon 1 functioning as a non-coding region (Gruenbaum-Cohen et al., 2009). Despite the quite well known variance of amelogenin proteins, the exact function of each one of them in enamel formation is not fully understood yet.
One of the alternatively spliced amelogenins known as leucine rich amelogenin peptide, LRAP, was first purified from secretory enamel matrix by Fincham and co-workers (Fincham et al., 1981) and was later identified by Vies and colleagues as the factor responsible for dentin matrix protein induced osteogenesis (Nebgen et al., 1999). The function of LRAP in dental enamel formation remains elusive. LRAP and full length amelogenin (M180) share many common characteristics in their sequence. The 59 amino acid murine LRAP (M59) contains of the first 33 N-terminus and the last 26 C-terminus amino acids from the full length amelogenin (Gibson et al., 1991). Previous studies of an LRAP overexpressing transgenic mouse generated by Gibson and co-workers showed enamel rods characteristic of normal enamel structure. Furthermore, they found that the LRAP transgene over expressed in amelogenin null mice did not rescue the lack of enamel matrix formation observed in amelogenin null mice, suggesting that LRAP does not have a direct structural role in enamel mineralization (Chen et al., 2003; Gibson et al., 2009; Habelitz et al., 2006).
Cell signaling activities of LRAP in enamel organ epithelial cells was shown by Le et al. (Le et al., 2007) who used human ameloblast lineage cells to show that exogenous LRAP increased amelogenin protein synthesis and reduced notch protein. Addition of exogenous LRAP to mouse enamel organ derived LS8 cells showed co-localization with LRAP (referred to as A-4) and lysosomal-associated membrane protein 1 (LAMP1), as well as an activation of the nitric oxide signaling pathway (Iacob and Veis, 2008; Le et al., 2007)
In this study, we used the TgLRAP mouse model generated by Gibson and co-worker (Chen et al., 2003), which overexpresses LRAP using a bovine amelogenin promotor, to address questions of the role of LRAP in tooth enamel formation. We analyzed developing molars of mice at postnatal day 0, 2, 5, 8 and 10, spanning pre-secretory (day 0) to maturation stage ameloblasts (day 10), to determine stage specific effects of LRAP on ameloblast function and enamel formation.
2. Results
2.1 TgLRAP mice produced a distinct phenotype not found in mice overexpressing full length amelogenin (M180), amelogenin null, or LRAP transgene expressed on amelogenin null mice (TgLRAP/amelogenin null)
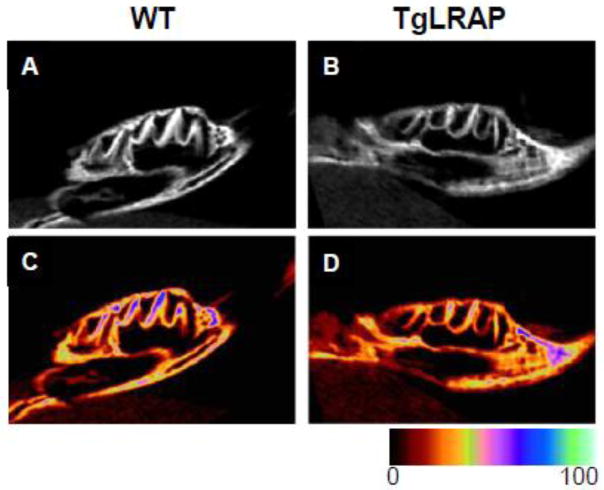
MicroCT analysis showed a lack of enamel matrix in molars from TgLRAP mice as compared to WT mice with a possible reduction in mineralization of enamel and overlying alveolar bone (Fig 1). Postnatal day 5 (P5) WT mice showed the presence of a secreted enamel matrix, and ameloblasts demonstrated the elongated polarized appearance typical of secretory ameloblasts with Tomes’ processes present (Fig 2A&B). M180 overexpression (TgM180) did not cause a morphological change in ameloblasts and enamel matrix compared to WT (Fig 2C&D). In contrast, P5 TgLRAP mice had a dramatic reduction in secretion of enamel matrix with a loss of Tomes’s processes. The alignment of TgLRAP ameloblasts was disorganized, and increased in the late secretory cells found over the developing cusp slopes (Fig 2E&F). Enamel matrix was minimally present in TgLRAP, amelogenin null (Fig 2G&H) and TgLRAP/amelogenin null (Fig 2I&J) mouse molars. However, in contrast to the disrupted ameloblast phenotype in the TgLRAP mouse, ameloblasts in both the amelogenin null and TgLRAP/amelogenin null mice lacked Tomes’ processes, but otherwise appeared unaffected (Fig 2G–J).
Figure 1.
Micro-CT images of representative first molars of WT and TgLRAP P5 mice. Sagittal views of mandibular first molar from P5 WT (A and C) and TgLRAP (B and D) show similar morphologies. WT (A) molars had a thicker enamel layer as compared to TgLRAP (B) (seen as bright white layer) on the original images. (C and D) Images converted from the original data to color range further confirmed reduced enamel thickness and a decrease in mineralization of enamel in the TgLRAP molar.
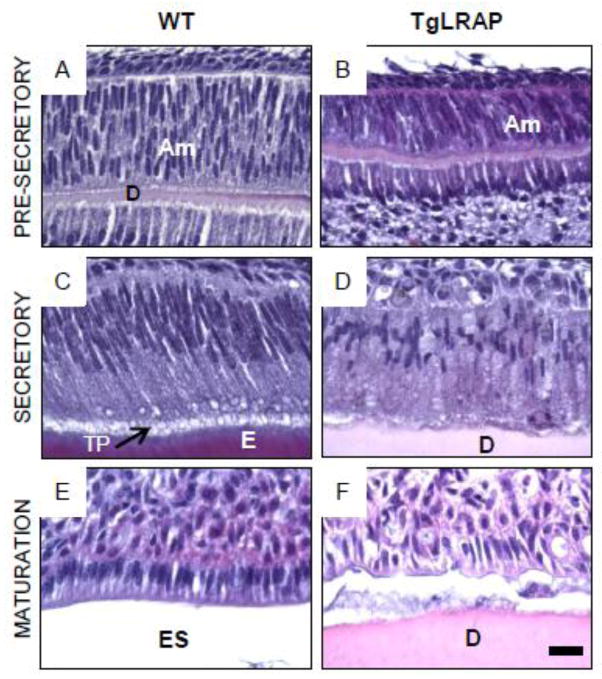
Figure 2.
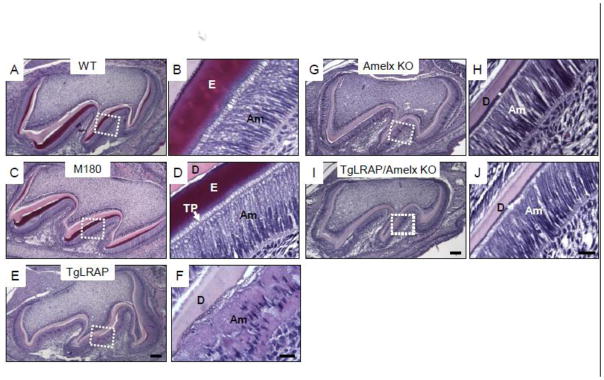
Hematoxylin and eosin (H&E) staining of 5 day post natal (P5) molars from various amelogenin transgenic mouse models. (A and B) Secretory ameloblasts on distal cusp slope in WT molar (a boxed area in A) showed the typical polarized appearance of the long cylindrical ameloblast cells with Tomes’ process extensions which have laid down enamel matrix. (C and D) M180 secretory ameloblasts appeared morphologically similar to those of WT. Tomes’ process extensions were noted throughout the secretory stage. (E and F) TgLRAP secretory stage ameloblasts showed a disorganized pattern of the ameloblasts with multiple layers of cells and less elongated nuclei. Only a small amount of enamel matrix formed, and surface of dentin had an irregular pattern. (G and H) Amelogenin null molar had very thin enamel like matrix (*) and normal appearing elongated ameloblasts, but lacked Tomes’ process extensions. (I and J) Ameloblasts of LRAP transgene overexpression in amelogenin null background did not show morphological differences similar to those in amelogenin null mice. Am, ameloblast; D, dentin; E, enamel; TP, Tomes’ process. Scale bar 100 μm for A,C,E; 20 μm for B,D,F; 100 μm for G,I; 20μm for H,J.
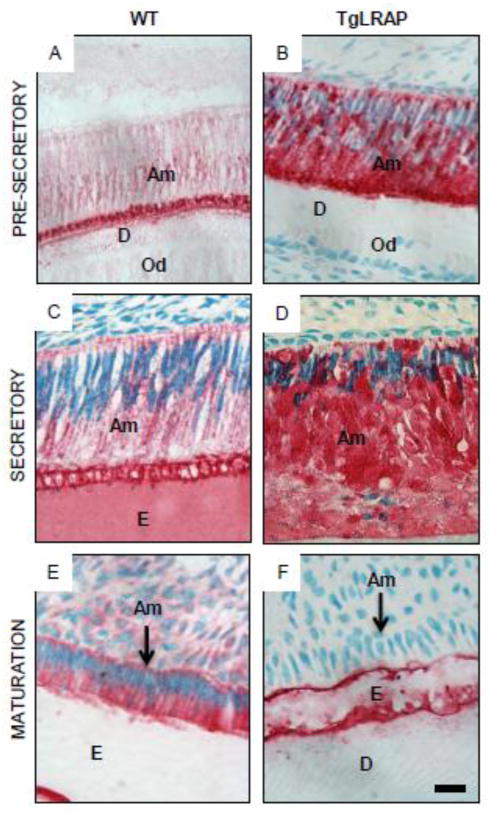
In the earlier pre-secretory stage of the molar, ameloblast morphology was not obviously altered in TgLRAP as compared to WT mice (Fig 3A&B). At this earlier developmental stage enamel matrix has yet to be deposited and only a thin layer of dentin matrix was present in both the WT and TgLRAP tissues. In the secretory stage the WT ameloblasts (Fig 3C) had elongated compared to pre-secretory cells and were well polarized with Tomes’ processes facing the enamel matrix at their apical end. In contrast, the TgLRAP ameloblasts layer (Fig 3D) was less organized and only a minimal amount of enamel matrix was secreted. Tomes’ processes were not detectable in the TgLRAP cells. In the maturation stage of development WT ameloblasts (Fig 3E) had transitioned to a shortened cell that was still well polarized with a widened enamel space left by the demineralized enamel matrix. The TgLRAP ameloblasts (Fig 3F) had also transitioned to a shortened cell phenotype in the maturation stage, but remained disorganized in appearance. The lack of an enamel space suggests that only a thin layer of enamel was formed.
Figure 3. Stage specific phenotype of WT and TgLRAP ameloblasts.
(A and B) At the pre-secretory stage, both WT (A) and TgLRAP (B) ameloblasts were not polarized and appeared similar. At this stage enamel matrix had yet to be deposited and only a thin layer of dentin matrix was present. (C and D) At the secretory stage the WT ameloblasts (C) had elongated and were well polarized with Tomes’ process facing the enamel matrix at their apical end. In contrast, the TgLRAP ameloblasts layer (D) was less organized and only a minimal amount of enamel matrix was secreted. Tomes’ processes were not detectable in the TgLRAP cells. (E and F) In WT mice maturation stage ameloblasts (E) had transitioned to a shortened cell that was still well polarized with a widened enamel space left by the demineralized enamel matrix. The TgLRAP ameloblasts (F) had also transitioned to a shortened cell phenotype, but remained disorganized appearance. The lack of an enamel space suggests that only a thin layer of enamel was formed. Am, ameloblast; E, enamel; ES, enamel space; D, dentin; TP, Tomes’ process. Scale bar 20 μm.
2.2 Multiple spliced variants of amelogenin and other enamel matrix proteins are upregulated in P0 TgLRAP molars, but not in P5 TgLRAP molars, as compared to WT molars
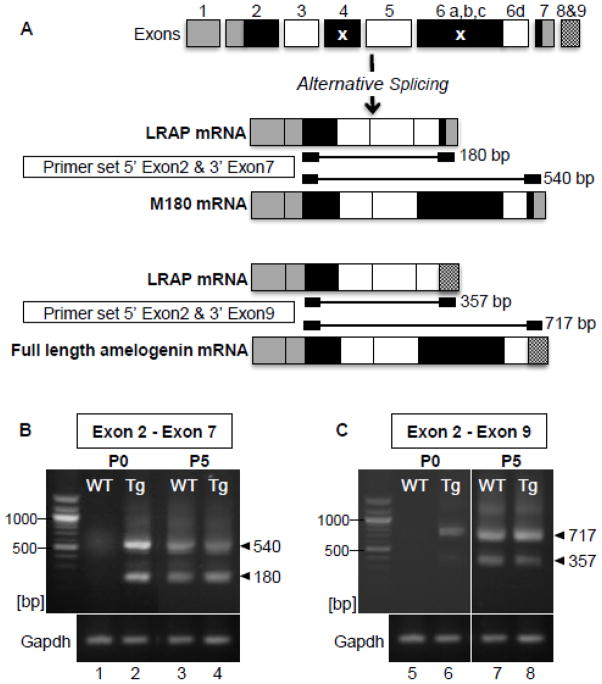
As enamel matrix formation was altered in molars of TgLRAP mice, two primer sets based on specific spliced variants of amelogenin mRNA were designed for RT-PCR, to compare the expression profile of amelogenin mRNAs (Fig 4A). LRAP lacks exon 4 and the majority of exon 6 (6 a, b, and c), and inclusion of exons 8 and 9, occurs when exon 7 is sliced out, ending with the alternative stop codons in exon 9 (Li et al., 1998). RT-PCR showed the TgLRAP mice had an upregulation of both M180 (as 540bp) and LRAP (as 180 bp) mRNA in P0 pup molars as compared to WT mouse molars (Fig 4B). The larger variant ending in exon 7 (Fig 4B lane 2) was then sequenced to confirm that indeed amelogenin mRNA corresponding to the M180 variant including the full coding region from exon 2 through 7, but excluding exon 4, was upregulated. At P5, which contains primarily secretory stage ameloblasts, there was no obvious difference in expression of LRAP (180 bp) and M180 (540 bp) mRNA between WT and TgLRAP molars. (Fig 4B lane 3&4).
Figure 4. Expression of multiple spliced variants of amelogenin was upregulated in molars from TgLRAP mice at P0.
(A) Spliced variants of amelogenin mRNA and PCR products size using two different primer sets are shown. LRAP lacks exon 4 and the majority of exon 6 (6 a, b, and c) as indicated by x. Inclusion of exons 8 and 9, occurs when exon 7 is sliced out, ending with the alternative stop codons in exon 9 (LI et al). (B) In P0 WT mice, M180 mRNA expression was barely detected (lane 1), whereas in P0 TgLRAP mice both LRAP (180 bp) and M180 (540 bp) variants were clearly upregulated compared to WT (lane 2). At P5, both splice variants were observed in WT and TgLRAP with a similar relative intensity (lane 3 and 4). Sequencing results confirmed the dominant upper band from lane 2 matched the M180 amelogenin. (C) A primer set (5′ exon 2 and 3′ exon9) allowed detection of endogenous amelogenin mRNA but not the LRAP transgene. At P0, RT-PCR products for full length amelogenin (717 bp) and LRAP8,9 (357 bp) were observed only in TgLRAP mice(lane 6) but not WT (lane 5). At P5, both WT and TgLRAP mice showed two spliced variants at similar level (lane 7 and 8).
To confirm the upregulation of amelogenin mRNA expression, quantitative PCR (qPCR) was further performed and it showed a 58 fold upregulation of M180 in P0 TgLRAP tooth buds compared to WT (Table 1). At P5 qPCR did not show a significant difference in M180 expression (data not shown). Interestingly MMP-20, a primary amelogenin degradation enzyme (Lu et al., 2008), was dramatically upregulated (over 200 fold). Amongst the major enamel matrix proteins besides amelogenin (Iwasaki et al., 2005; Lee et al., 2003; Uchida et al., 1991), both enamelin and ameloblastin were upregulated approximately 3 fold, based on quantitative PCR whereas amelotin expression was relatively unchanged (Table 1).
Table 1.
| Real Time PCR fold change P0 TgLRAP vs. P0 WT | ||
|---|---|---|
| Gene | Fold Change | |
| M180 | ⇑ | 58** |
| ENAMELIN | ⇑ | 3.5* |
| AMELOBLASTIN | ⇑ | 3.2* |
| MMP20 | ⇑ | 203** |
| AMELOTIN | — | 1.3 |
p-value<0.05,
p-value<0.001
To assess if the TgLRAP mouse had a true upregulation of both the (endogenous/murine) LRAP and full length isoforms, rather than just an increase in the transgene, primers were then designed to include exons 8–9 at the 3′ end which were not included in the transgenic LRAP sequence. At P0 in TgLRAP molars, both full length (717 bp) and (endogenous/murine) LRAP 8,9 (357 bp) variants were upregulated as compared to WT molars when exon 8–9 was included in the primer sequences (Fig 4C lane 4&5). Similar to amplification of exon 2–7 (Fig 4A), both full length and LRAP 8,9 variants were expressed at similar level in P5 WT and TgLRAP molars (Fig 4C lane 6&7).
2.3 In situ hybridization showed that LRAP was overexpressed in both ameloblasts and odontoblasts at P0
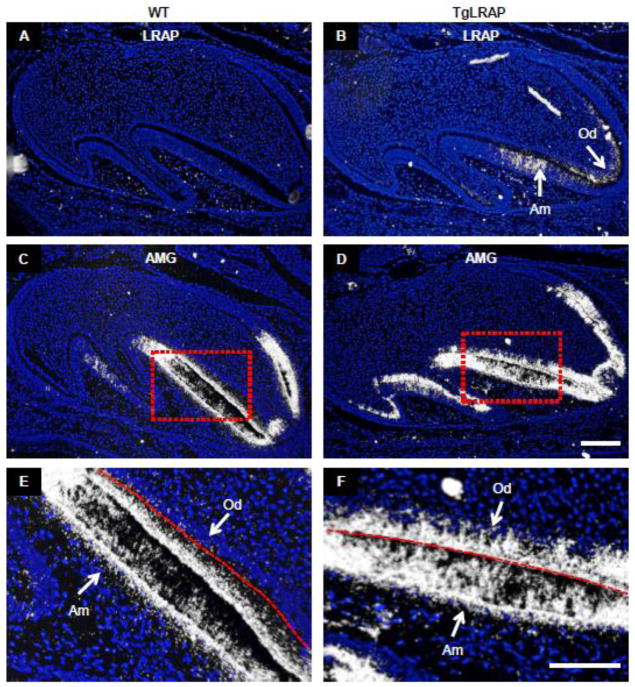
The mRNA analyzed in the TgLRAP was extracted from the whole tooth bud. To further analyze the spatial expression pattern of amelogenin mRNA in ameloblasts and odontoblasts in P0 molars, we completed in situ hybridization against both the bovine LRAP transgene and mouse amelogenin. In P0 molars, the LRAP transgene was first evident in the pre-odontoblasts adjacent to the pre-ameloblasts (Fig 5B). Transgene expression in the ameloblasts was not detected in the pre-ameloblasts but detected only in the more differentiated pre-secretory ameloblasts located along the tooth cusps at P0. Probes specific to full length amelogenin demonstrated a markedly increased expression of amelogenin mRNA in pre-odontoblasts of the TgLRAP tooth organ in addition to a greater expression of those in pre-ameloblasts and pre-secretory ameloblasts comparing to WT (Fig 5C–F). The appearance of LRAP transgene mRNA in the mesenchyme before the ameloblasts, suggests that LRAP in the mesenchyme may have a role in driving the differentiation of ameloblasts with a corresponding increase in amelogenin expression in TgLRAP molar.
Figure 5. Spatial expression pattern of amelogenin mRNA by in situ hybridization (ISH) in P0 molars.
(A) LRAP mRNA was not detected in WT tissue. (B) In the TgLRAP mice the LRAP transgene probably as well as endogenous LRAP mRNA was localized in the odontoblasts adjacent to pre-ameloblasts, and in the pre-secretory ameloblasts. (C and E) ISH with a full-length mouse amelogenin (AMG) probe showed abundant expression in pre-ameloblasts and pre-secretory ameloblasts in WT molar. Higher magnification of boxed area (E) showed amelogenin mRNA was detected only in the pre-secretory ameloblasts with the red dashed line indicating the future enamel-dentin junction between the ameloblasts and odontoblasts which have yet to form matrix. (D and F) In the TgLRAP mice AMG mRNA was abundantly expressed in pre-ameloblasts and pre-secretory ameloblasts, as well as in adjacent odontoblasts. Am, ameloblast; Od, odontoblast. Scale bars 100 μm.
2.4 TgLRAP mice showed increased amelogenin protein in pre-secretory and secretory ameloblasts with a loss of regulation of amelogenin secretion
To further examine if amelogenin protein synthesis or secretion was altered, amelogenin was immunolocalized in P2 (pre-secretory), P5 (late secretory) and P10 (early maturation) molars. Amelogenin (AMG) immunolocalization showed that prescretory ameloblasts found in P2 molars of TgLRAP mice had increased amounts of amelogenin protein as compared to WT mice, but no obvious alteration of ameloblast morphology (Fig 6A&B). Secretory ameloblasts found in P5 WT molars showed elongated shape and began to secrete amelogenin proteins into the extracellular enamel matrix (Fig 6C). However P5 TgLRAP mouse molars showed less elongated and disorganized nuclei and an apparent lack of continued secretion of amelogenin protein into the extracellular enamel space (Fig 6D). At P10, WT ameloblasts were maturation stage with shorter the height, but still polarized location of nucleus, while in contrast the TgLRAP mouse ameloblasts layer was less organized and showed a loss of polarization as compared to WT (Fig 6E&F). Amelogenin protein could be detected in the WT maturation stage ameloblasts, possibly due to re-uptake of hydrolyzed amelogenin fragments from the matrix as reported previously (Reith and Cotty, 1967; Smith, 1979), while the P10 TgLRAP ameloblasts showed no amelogenin positive staining.
Figure 6. Immunohistochemical localization of amelogenin protein on molar ameloblasts.
(A) Presecretory ameloblasts in P2 WT mice showed amelogenin protein mainly localized on the apical region of cytoplasm. (B) Besides the strong immunoreaction on the apical region, entire cytoplasm of presecretory stage ameloblasts in P2 TgLRAP mice showed highly intense amelogenin protein positive reaction. (C) Amelogenin protein was detected on entire cytoplasm and Tomes’ process of secretory stage ameloblast of P5 WT mice. (D) Secretory stage ameloblasts of P5 TgLRAP mice formed a disorganized layer and demonstrated amelogenin protein highly retained in the cytoplasm. (E) At maturation stage, ameloblasts of P10 WT mice were amelogenin immunopositive. (F) However ameloblasts in P10 TgLRAP mouse molars lacked polarity and were amelogenin immunonegative. Am, ameloblast; E, enamel; Od, odontoblast; D, dentin. Scale bar, 20μm.
2.5 Apoptosis was increased in the late secretory stage ameloblasts of TgLRAP mouse molars
As apoptosis is found in transition and maturation stage ameloblasts a TUNEL assay was performed to determine if stage specific patterns of apoptosis differed between WT and TgLRAP tissues. The late secretory ameloblasts found on the distal slope of the mesial cusp of the P5 mouse molar showed TUNEL positive staining, which was not present in the WT mouse ameloblasts (Fig 7A&B). This area is the location where cells dramatically lose organized layer and showed greatest accumulation of amelogenin protein in the cytoplasm. The transitional stage ameloblasts present at P8 in the WT mouse molar showed positive TUNEL stain, but no TUNEL stain was present in P8 TgLRAP molars (Fig 7C&D). The evidence of earlier apoptosis in ameloblasts from TgLRAP molars as compared WT mice, suggests earlier timing of ameloblast differentiation in TgLRAP mouse molars.
Figure 7. TUNEL staining on P5 and P8 molar ameloblasts.
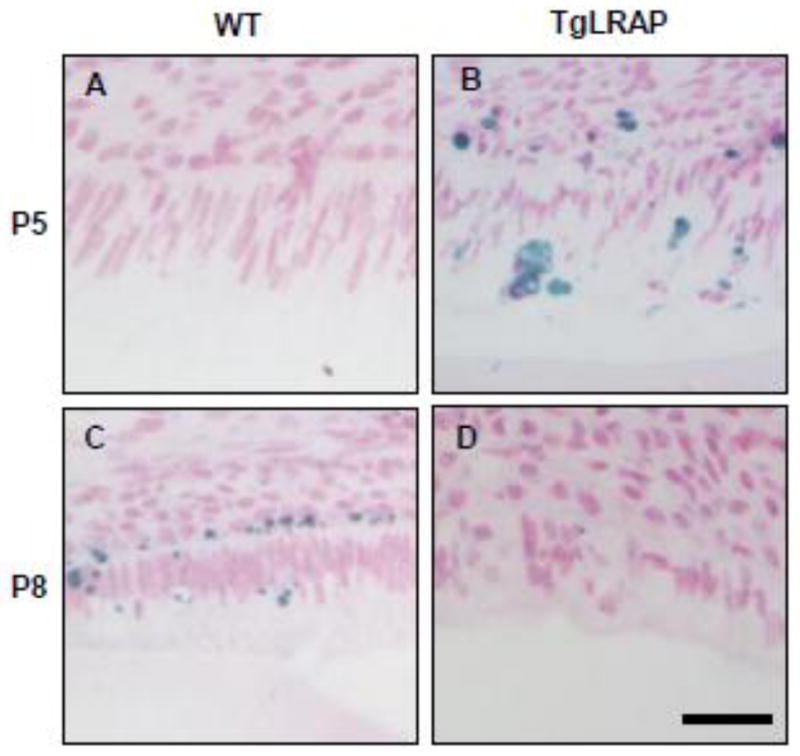
(A) P5 WT molars had no TUNEL positive ameloblasts. (B) P5 TgLRAP molar showed TUNEL positive staining (in blue) in the region corresponding to the most differentiated secretory stage ameloblasts (C) AT P8 WT molars had post secretory stage ameloblasts where TUNEL positive staining was observed. (D) Ameloblasts of P8 TgLRAP mice showed post secretory morphology and slight TUNEL positive staining was observed. Scale bar 30μm.
2.6 Expression of the tissue specific gene expression regulator, special adenine and thymine-rich sequence binding protein 1 (SATB1), was decreased in TgLRAP mice ameloblasts
SATB1, a global chromatin organizer and transcription factor in progenitor cells (Cai et al., 2006; Pavan Kumar et al., 2006), was observed in the nucleus of pre-secretory and secretory stage ameloblasts. Pre-secretory WT and TgLRAP ameloblasts expressed the highest levels of SATB1 (Fig 8A&B). Early secretory and pre-secretory WT ameloblasts showed similar levels of SATB1 nuclear staining, and SATB1 was dramatically reduced at the late secretory stage (Fig 8A,C,E). In contrast, TgLRAP mouse molars showed a dramatic reduction in SATB1 immunostaining in early secretory stage ameloblasts to levels similar to those found in late stage WT molars (Fig 8D&E), and was further reduced in the late secretory TgLRAP ameloblasts below the levels found in WT molar ameloblasts (Fig 8E&F).
Figure 8. Expression of the gene regulation protein SATB1 was decreased in TgLRAP ameloblasts.
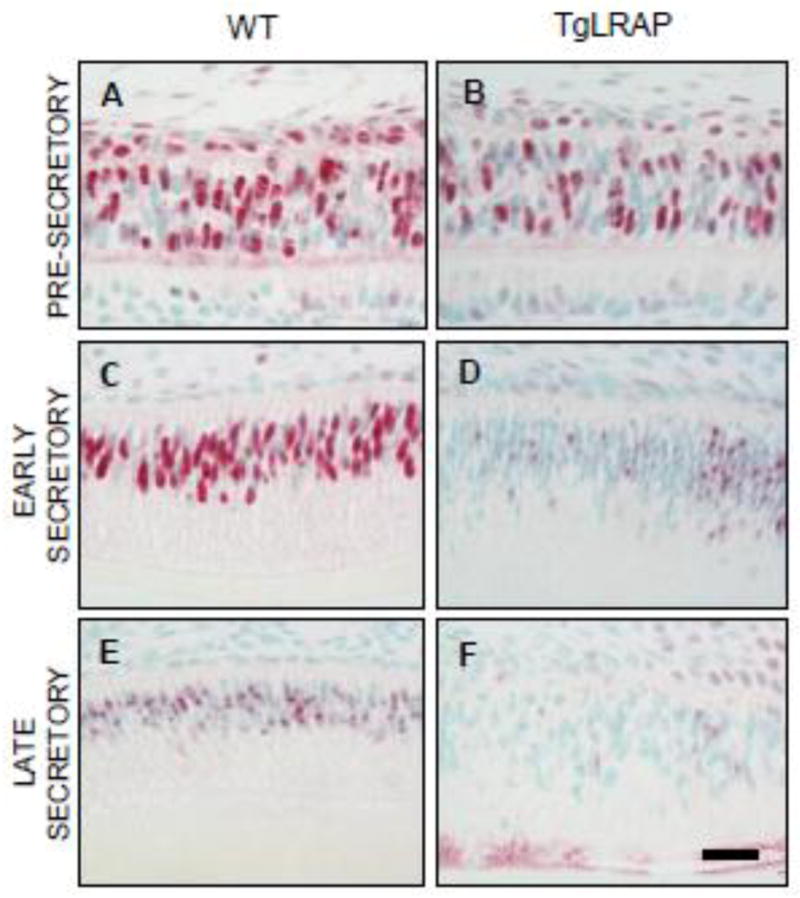
(A and B) At the pre-secretory stage, ameloblasts as well as stratum intermedium cells of both WT (A) and TgLRAP (B) mice strongly expressed SATB1 protein in the nucleus. (C and D) In the early secretory stage, SATB1 expression in the WT ameloblasts (C) remained strong in the nucleus similar to pre-secretory stage, but there was a marked decrease in the TgLRAP ameloblasts (D). (E and F) In the differentiated late secretory ameloblasts, SATB1 expression was reduced in the WT ameloblasts (E) compared to its earlier stage, and the corresponding stage of TgLRAP ameloblasts (F) demonstrated very weak expression. Scale bar, 20um.
3. Discussion
TgLRAP mice were generated by Gibson and co-workers (Chen et al., 2003) as a part of a larger question to determine whether LRAP could rescue the amelogenin null phenotype where enamel matrix formation is inhibited. In studies using this mouse model, they reported that the molars of TgLRAP mice had a normal enamel structure, but with a slightly irregular enamel surface characterized by pitting (Chen et al., 2003). Our histological analyses and microXCT showed that in addition to an irregular enamel surface, the entire enamel layer of molar was thin, suggesting a defect in matrix protein secretion. We then compared the enamel of TgM180 mice to that of the TgLRAP, and found that unlike TgLRAP, the TgM180 mouse molars formed a matrix with a thickness similar to WT mice, no obvious changes in ameloblast phenotype.
The difference between LRAP and M180 amelogenin is that the M180 transcript contains exon 6 a,b,c (540 base pair), which codes for a large central hydrophobic amino acid sequence in the synthesized protein. (Gibson et al., 1991). Neither LRAP or M180 contain exon 4 or exons 8&9 The disruption in enamel formation when LRAP is overexpressed, suggests a specific effect of LRAP on ameloblast mediated amelogenesis.
When we compared the ameloblasts of TgLRAP mouse molars to WT mouse molars, these changes appeared to be stage specific. The pre-secretory ameloblasts found in P0 and P2 mouse molars appeared similar in the TgLRAP and WT mice, even though in the TgLRAP molars both full length amelogenin mRNA and protein expression was upregulated. Early secretory stage ameloblasts located on the outer cusp surfaces also appeared relatively normal, and the first obvious signs of an abnormal ameloblast phenotype occurred in late secretory ameloblasts on the distal slope of the mesial cusp of P5 molars. These ameloblasts appeared to show less elongated nuclei, form disorganized layer, and have increased apoptosis. Further differentiation of ameloblasts at the maturations stage (P10 molars) was completely disrupted and the cells overlying the thin enamel matrix appeared as disorganized epithelial cells. This is the first report of such a significant effect of LRAP overexpression on ameloblast function and differentiation.
This phenotype, showing loss of organized cell layers and protein retention was even more striking in that it only occurred in the TgLRAP mice, in which the LRAP transgene was overexpressed on a wild type background. When LRAP was overexpressed in the amelogenin null background, consistent with reports of the amelogenin null mice, ameloblasts did not have Tomes’ processes, but appeared otherwise largely unaffected. Also consistent with previous reports, we found that the amelogenin null mice secreted a poorly mineralized thin layer of enamel (Gibson et al., 2001), and we found that this enamel matrix morphology was not altered by the LRAP overexpression.
This observation, led us to further investigate the effect of LRAP overexpression on amelogenin synthesis, as well mRNA expression of other enamel matrix proteins and proteinases. Immunohistochemical staining of TgLRAP mice showed that amelogenin protein synthesis was increased in pre-secretory ameloblasts as compared to WT mice. This was consistent with in situ hybridization, which also showed both LRAP and amelogenin upregulated in both ameloblasts as well as odontoboasts. These results suggest a specific effect on LRAP in upregulating amelogenin protein synthesis that is not cell specific.
To further investigate the effects of stage specific upregulation of LRAP in ameloblasts, we completed PCR analysis of amelogenin mRNA synthesis in molars containing primarily pre-secretory ameloblasts (P0) and molars containing primarily secretory stage ameloblasts (P5). PCR showed an upregulation of both LRAP and M180 amelogenin splice variants in P0 mouse molars, with no further upregulation of amelogenin mRNA in P5 TgLRAP as compared to WT mouse molars, suggesting a specific role of transgene LRAP in early stage of ameloblast differentiation.
To determine whether native LRAP was upregulated as opposed to increased expression of the transgene, we then amplified PCR products ending with the alternate C terminus coded by exons 8 and 9 (Li et al., 1998), as these exons were not included in the transgene LRAP. This PCR analysis showed that the larger amelogenin splice variant containing all exons with the exception of exon 4, was specifically upregulated in TgLRAP pre-secretory ameloblasts. These PCR results showing an upregulation of full length (M180) amelogenin in response to increased LRAP are consistent with reports by Iacob and Veis, who found that when exogenous LRAP protein was added to the LS8 ameloblast lineage cell line M180 mRNA was upreguated 13.6 fold upregulation after 24hrs (Iacob and Veis, 2008). Xu and co-workers reported that amelogenin full length protein, but not LRAP can stabilize amelogenin mRNA. (Xu et al., 2006a). Therefore, we suggest that the upregulation of amelogenin expression by LRAP is a true effect on transcription, rather than simply due to mRNA stabilization.
In secretory stage P5 ameloblasts, it is interesting that while enamelin and ameloblastin are upregulated 3 fold, MMP-20 is upregulated more that 200 fold. This specific upregulation of MMP-20 in the TgLRAP mouse model, may relate to enhanced amelogenin expression and synthesis. In previous unpublished studies in our laboratory, we found that a retention of amelogenin, as occurs in an amelogenin P70T mutation, also results in an upregulation of MMP-20 mRNA. Therefore, it seems that MMP-20 expression may be co-regulated regulated with amelogenin at the secretory stage of amelogenesis.
We found that increased amelogenin expression during the pre-secretory stage (P0) did not result in an affect on ameloblast morphology. In TgLRAP mouse molars, amelogenin secretion was limited to the early secretory stage. At the late secretory stage, ameloblasts did not appear to secrete amelogenin, and their morphology was dramatically affected with a less elongated nuclei and disorganized cell layer. Previous in vitro studies by Tompkins and co-workers (Tompkins et al., 2005) of tooth organs grown in culture, found that addition of exogenous LRAP (A-4) to the culture medium, inhibited pre-ameloblast polarization. Our in vivo studies suggest that when LRAP is present intracellularly in pre-secretory ameloblasts, polarization is not affected; possibly indicating that an effect of LRAP on ameloblasts is related to an overexpression of LRAP secreted protein. In support of this possibility is the finding that when LRAP was over expressed on an amelogenin null background mice, no phenotypic changes were observed.
When only M180 was overexpressed we found no ameloblast phenotype, further indicating the unique function of secreted LRAP in altering ameloblast morphology. Ravindranath and co-workers showed that addition of exogenous LRAP protein to E16 molar explants decreased ameloblast height, suggesting LRAP may directly enhance differentiation and promote early ameloblast maturation (Ravindranath et al., 2007). We found that the non-amelogenin matrix proteins, enamelin and ameloblastin, also exhibited increased early gene expression in the TgLRAP mice, further supporting the possibility that LRAP promotes earlier ameloblast differentiation.
As apoptosis is only present in transitional and maturation stage ameloblasts (Smith and Warshawsky, 1977) our finding of a positive TUNEL stain at P5 suggests that the TgLRAP ameloblasts may have differentiated toward transitional stage ameloblasts. Our findings demonstrate an increase in cell death specifically in the late secretory stage ameloblasts at P5. At this age most ameloblasts in the rodent molar would be expected to be secretory stage cells, however, the potential increase in apoptosis suggests that transition stage related apoptosis occurred at an earlier time point. Indeed at P8 TUNEL positive reactions were observed transitional stage ameloblasts in WT mouse molars, while the TgLRAP P8 ameloblasts did not show any TUNEL positive reactions.
If LRAP modulates ameloblast differentiation, it seems that LRAP overexpression may trigger a more overarching mechanism controlling ameloblast differentiation. Recent studies in our laboratory have pointed to the role of special AT sequence binding protein (SATB1) in ameloblast differentiation. SATB1 is known to regulate large-scale chromatin remodeling in certain cell types and progenitor cells (Agrelo et al., 2009; Cai et al., 2003; Cai et al., 2006; Han et al., 2008; Kumar et al., 2007). In epidermal tissue, SATB1 functions as one of the direct downstream components for the global regulatory network of p63 and contributes in epidermal tissue morphogenesis by establishing tissue-specific chromatin organization and gene expression in progenitor cells (Fessing et al., 2011; Han et al., 2008). Epidermis of SATB1 null mice show impaired morphology and downregulation of cell proliferation (Fessing et al., 2011). We found that ameloblasts of the SATB1 null mouse lack Tomes’ processes and have an earlier transformation into maturation stage ameloblasts (unpublished data, Zhang et al, submitted). Downregulation of SATB1 in the LRAP overexpressor mice, suggests the possibility that interactions between LRAP protein and SATB1 protein may modulate ameloblast differentiation.
Along with the altered epithelial cell signaling reported here, LRAP has also been reported to alter cellular pathways in mesenchymal cells, including MAPK signaling in cememtoblasts (Boabaid et al., 2004) and activation of the WNT canonical pathway in bone marrow stem cells (Warotayanont et al., 2009; Wen et al., 2011). These findings and our report of LRAP effects on ameloblast differentiation, clearly show that this alternatively spliced amelogenin does alter cell signaling. Additional studies are needed to further understand how this small alternatively spliced amelogenin protein regulates gene expression and ameloblast differentiation in tooth formation.
4. Methods
4.1 Animal Models
All animal procedures were performed after approval by the University of California San Francisco and University of Pennsylvania IACUC. Amelogenin null, TgLRAP, and TgM180 mice used for these studies were provided by Dr. Carolyn Gibson and as previously described (Chen et al., 2003; Gibson et al., 2001; Gibson et al., 2007) both TgLRAP, and TgM180 mouse models were made by incorporating a plasmid containing the amelogenin promotor with either cDNA encoding for LRAP (TgLRAP) or cDNA containing amelogenin exons 1,2,3,5,6 and 7 (TgM180). TgLRAP mice in an amelogenin null background were made by mating TgLRAP+ males with amelogenin null females. As the murine amelogenin gene is sex linked on the X chromosome this mating resulted in all males being amelogenin null with ~50% expressing the LRAP transgene. Wild type (WT) mice were generated as littermates.
4.2 MicroComputed Tomography (micro-CT)
Level of mineralization of first molar was accessed on non-decalcified hemi-mandibles of 3 samples from P5 WT and TgLRAP respectively by micro-computed tomography (SkyScan1076; Bruker-microCT, Kontich, Belgium) with x-ray source operating settings at 100 kV and 0.1 mA. After reconstitution of images by NRecon software (Bruker-microCT), appropriate sagittal imaging planes were selected from three orthogonal sections centered at a level containing three buccal cusps inside the reconstructed space using Data Viewer software (Bruker-microCT).
4.3 Immunohistochemistry and H&E staining
Maxillae at P0, 2, 5, 8, and 10 were prepared for immunohistochemistry, by immediate immersion in 4% paraformaldehyde (PFA)/0.06M Cacodylate buffer (pH 7.3) for 1 day at 4 °C followed by decalcification in 8% EDTA (pH 7.3) at 4 °C for 10 days, The samples were then dehydrated through a graded series of ethanol followed by routine embedding in paraffin and sectioning.
After deparaffinization, the sections of maxillae were incubated with 10% swine and 5% goat sera followed by incubation with rabbit anti- human full length recombinant amelogenin (1:500; Le et al., 2007) dilution or rabbit anti-SATB1 antibody (1:200; Abcam Inc., Cambridge, MA) overnight at room temperature. A biotinylated swine anti-rabbit IgG F(ab’)2 fraction (Dako Cytomation Inc., Carpinteria, CA) was used as the secondary antibody for 1 h at room temperature incubation. Following incubation with alkaline phosphatase conjugated streptavidin (Vector Laboratories Inc., Burlingame, CA) for 30 min, immunoreactivity was visualized using a Vector ® Red kit (Vector Laboratories Inc.) resulting in pink/red color for positive staining. Counter-staining was performed with methyl green. Negative control was done with normal rabbit sera. Additional sections were utilized for standard hematoxilyn and eosin (H&E) staining after deparaffinization.
4.4 PCR analysis
Total mRNA for each sample, maxillary first molars were dissected from a postnatal day 0 and day 5 WT and transgenic mice. Molars from four pups (n=4) were included for each timepoint. Total RNA was isolated using RNeasy Mini kit (Qiagen, Germantown, MD). An aliquot containing 1μg of total RNA was reverse transcribed to cDNA using SuperScript® III Reverse Transcriptase (Invitrogen, Carlsbad, CA) (see Table 2 for list of primers). Polymerase chain reaction amplification for RT-PCR was performed with the Hot Start Taq kit (Qiagen) by first incubating the reaction mixture at 95° for 5 min, followed by 94°C, 57°C, and 72°C for 1min each for 30 cycles and then 72°C for 10 min with the same condition used for all primers. The products were visualized on a 2% agarose gel with ethidium bromide staining. Real-time PCR gene expression was characterized by quantitative PCR using the ABI 7500 system (Applied Biosystems, Carlsbad, CA). cDNA was amplified with the Fast Start SYBR Green master mix (Roche. Indianapolis, IN). Relative expression levels of target genes were analyzed by the ΔΔCt method as published previously (Thomsen et al., 2010). All data were analyzed by student t-test by using Prism software (GraphPad Software Inc, San Diego, CA, USA).
Table 2.
| Gene | Primer Pair |
|---|---|
| Amelogenin Exon 2–7 | F: 5′GCTATGCCCCTACCACCTC 3′ R: 5′ GTCCACTTCTTCCCGCTTG 3′ |
| Amelogenin Exon 2–9 | F: 5′AACCATCAAGAAATGGGGACC 3′ R: 5′ ACTACATGCCATTGTGTTCTG 3′ |
| Full length Amelogenin (for qPCR) | F: 5′CAGCAACCAATGATGCCAGTTCCT 3′ R: 5′ ACTTCTTCCCGCTTGGTCTTGTCT 3′ |
| Ameloblastin | F: 5′ CTGTTACCAAAGGCCCTGAA 3′ R: 5′ GCCATTTGTGAAAGGAGAGC 3′ |
| Amelotin | F: 5′ ATCAGCCCAGTCATTACCAAAG 3′ R: 5′ AGGTCTGACCCCAGAGTGAG 3′ |
| Enamelin | F: 5′ GCTTTGGCTCCAATTCAAAA3′ R: 5′ AGGACTTTCAGTGGGTGT 3′ |
| MMP20 | F: 5′ CTCGTCCTTTGATGCAGTGA 3′ R: 5′ TGGACATTAGCTGGGGAAAG 3′ |
| Gapdh | F: 5′ GGA CGC ATT GGT CGT CTG G 3′ R: 5′ TTT GCA CTG GTA CGT GTT GAT 3 |
4.5 In situ hybridization
Maxillae from P0 mice were fixed with 4% PFA/0.06M cacodylate buffer (pH 7.3) for 1 day at 4 °C then dehydrated through a graded series of ethanol followed by routine embedding in paraffin and sectioning. Complementary DNAs corresponding to LRAP and full length amelogenin were used to generate antisense and sense riboprobes as described (Albrecht et al., 1997). The transgenic LRAP probe was a 42 bp probe that crossed the boundary between exon 5 and 6d excluding exon 6 domains a,b,c. The amelogenin probe was a 512 bp probe corresponding to the region from exon 2 to 6d of full length amelogenin. The sequences of the probes were confirmed by automated DNA sequencing. The riboprobes were labeled with 35S and in situ hybridization was done as previously described (Albrecht et al., 1997; Ferguson et al., 1999; Sundin et al., 1990). Following hybridization and washing emulsion-dipped slides were exposed to beta emissions from the 35SUTP-labeled riboprobes for 4–6 days. The sections were counterstained with a nuclear stain (Hoechst Stain; Sigma Aldrich, St. Louis, MO). Hybridization signals were detected by darkfield optics, and the nuclear stain was visualized by epifluorescence. The images of the signals and counter-stain were superimposed using Adobe Photoshop software (Adobe, San Francisco, CA) to facilitate identification of cells expressing a particular gene
4.6 TUNEL Assay
To label and detect DNA strand breaks, in vitro terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) staining was performed on the P5 paraffin embedded molar sections using TACS·XL®(Trevigen Inc., Gaithersberg, MD) according to kit instructions.
4.7 Imaging
Histological images were taken with a Nikon Eclipse E3800 microscope (Melville, NY) using a digital camera (QImaging Inc., Surrey, Canada) and SimplePCI imaging software version 5.3.1.
Highlights.
An LRAP transgenic mouse was generated leading to disorganized ameloblasts.
LRAP overexpression increased full length amelogenin gene expression at P0.
Ameloblasts lacked Tomes’ Processes and had increased amelogenin protein.
Pre-ameloblasts had increased expression of non-amelogenin matrix genes.
LRAP is a modulator of early stage ameloblast differentiation.
Acknowledgments
This study was supported by NIH/NIDCR grants R01 DE013508 (P Den Besten) and R03 DE019682 (T Le). We thank the Schneider Laboratory at University of California San Francisco for assistance with the in situ hybridization.
Abbreviations
- LRAP
Leucine Rich Amelogenin Peptide
- MMP20
Matrix Metalloproteinase 20
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, Wutz A. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Developmental cell. 2009;16:507–516. doi: 10.1016/j.devcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht BE, Breitenbach U, Stuhmer T, Harvey RJ, Darlison MG. In situ hybridization and reverse transcription--polymerase chain reaction studies on the expression of the GABA(C) receptor rho1- and rho2-subunit genes in avian and rat brain. Eur J Neurosci. 1997;9:2414–2422. doi: 10.1111/j.1460-9568.1997.tb01658.x. [DOI] [PubMed] [Google Scholar]
- Boabaid F, Gibson CW, Kuehl MA, Berry JE, Snead ML, Nociti FH, Jr, Katchburian E, Somerman MJ. Leucine-rich amelogenin peptide: a candidate signaling molecule during cementogenesis. J Periodontol. 2004;75:1126–1136. doi: 10.1902/jop.2004.75.8.1126. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Chen E, Yuan ZA, Wright JT, Hong SP, Li Y, Collier PM, Hall B, D’Angelo M, Decker S, Piddington R, Abrams WR, Kulkarni AB, Gibson CW. The small bovine amelogenin LRAP fails to rescue the amelogenin null phenotype. Calcif Tissue Int. 2003;73:487–495. doi: 10.1007/s00223-002-0036-7. [DOI] [PubMed] [Google Scholar]
- Ferguson C, Alpern E, Miclau T, Helms JA. Does adult fracture repair recapitulate embryonic skeletal formation? Mechanisms of development. 1999;87:57–66. doi: 10.1016/s0925-4773(99)00142-2. [DOI] [PubMed] [Google Scholar]
- Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, Gordon KB, Smorodchenko AD, Poterlowicz K, Ferone G, Kohwi Y, Missero C, Kohwi-Shigematsu T, Botchkarev VA. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol. 2011;194:825–839. doi: 10.1083/jcb.201101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham AG, Belcourt AB, Termine JD, Butler WT, Cothran WC. Dental enamel matrix: sequences of two amelogenin polypeptides. Biosci Rep. 1981;1:771–778. doi: 10.1007/BF01114799. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J. Amelogenin post-translational modifications: carboxy-terminal processing and the phosphorylation of bovine and porcine “TRAP” and “LRAP” amelogenins. Biochem Biophys Res Commun. 1993;197:248–255. doi: 10.1006/bbrc.1993.2468. [DOI] [PubMed] [Google Scholar]
- Fincham AG, Moradian-Oldak J, Simmer JP, Sarte P, Lau EC, Diekwisch T, Slavkin HC. Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J Struct Biol. 1994;112:103–109. doi: 10.1006/jsbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Golub E, Ding WD, Shimokawa H, Young M, Termine J, Rosenbloom J. Identification of the leucine-rich amelogenin peptide (LRAP) as the translation product of an alternatively spliced transcript. Biochem Biophys Res Commun. 1991;174:1306–1312. doi: 10.1016/0006-291x(91)91564-s. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Li Y, Daly B, Suggs C, Yuan ZA, Fong H, Simmons D, Aragon M, Kulkarni AB, Wright JT. The leucine-rich amelogenin peptide alters the amelogenin null enamel phenotype. Cells Tissues Organs. 2009;189:169–174. doi: 10.1159/000151384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Li Y, Daly B, Suggs C, Aragon MA, Alawi F, Kulkarni AB, Wright JT. Transgenic mice that express normal and mutated amelogenins. J Dent Res. 2007;86:331–335. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- Gruenbaum-Cohen Y, Tucker AS, Haze A, Shilo D, Taylor AL, Shay B, Sharpe PT, Mitsiadis TA, Ornoy A, Blumenfeld A, Deutsch D. Amelogenin in cranio-facial development: the tooth as a model to study the role of amelogenin during embryogenesis. J Exp Zool B Mol Dev Evol. 2009;312B:445–457. doi: 10.1002/jez.b.21255. [DOI] [PubMed] [Google Scholar]
- Habelitz S, DenBesten PK, Marshall SJ, Marshall GW, Li W. Self-assembly and effect on crystal growth of the leucine-rich amelogenin peptide. Eur J Oral Sci. 2006;114(Suppl 1):315–319. doi: 10.1111/j.1600-0722.2006.00312.x. discussion 327–319, 382. [DOI] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Hu CC, Ryu OH, Qian Q, Zhang CH, Simmer JP. Cloning, characterization, and heterologous expression of exon-4-containing amelogenin mRNAs. J Dent Res. 1997;76:641–647. doi: 10.1177/00220345970760020401. [DOI] [PubMed] [Google Scholar]
- Iacob S, Veis A. Identification of the functional activity of the [A-4] amelogenin gene splice product in newborn mouse ameloblasts. Bone. 2008;42:1072–1079. doi: 10.1016/j.bone.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K, Bajenova E, Somogyi-Ganss E, Miller M, Nguyen V, Nourkeyhani H, Gao Y, Wendel M, Ganss B. Amelotin--a Novel Secreted, Ameloblast-specific Protein. J Dent Res. 2005;84:1127–1132. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- Kumar PP, Bischof O, Purbey PK, Notani D, Urlaub H, Dejean A, Galande S. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nature cell biology. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- Le TQ, Zhang Y, Li W, Denbesten PK. The effect of LRAP on enamel organ epithelial cell differentiation. J Dent Res. 2007;86:1095–1099. doi: 10.1177/154405910708601114. [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim SM, Lee YJ, Yamada KM, Yamada Y, Chi JG. The structure of the rat ameloblastin gene and its expression in amelogenesis. Molecules and cells. 2003;15:216–225. [PubMed] [Google Scholar]
- Li R, Li W, DenBesten PK. Alternative splicing of amelogenin mRNA from rat incisor ameloblasts. J Dent Res. 1995;74:1880–1885. doi: 10.1177/00220345950740121101. [DOI] [PubMed] [Google Scholar]
- Li W, Mathews C, Gao C, DenBesten PK. Identification of two additional exons at the 3′ end of the amelogenin gene. Arch Oral Biol. 1998;43:497–504. doi: 10.1016/s0003-9969(98)00013-2. [DOI] [PubMed] [Google Scholar]
- Lu Y, Papagerakis P, Yamakoshi Y, Hu JC, Bartlett JD, Simmer JP. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 2008;389:695–700. doi: 10.1515/BC.2008.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebgen DR, Inoue H, Sabsay B, Wei K, Ho CS, Veis A. Identification of the chondrogenic-inducing activity from bovine dentin (bCIA) as a low-molecular-mass amelogenin polypeptide. J Dent Res. 1999;78:1484–1494. doi: 10.1177/00220345990780090201. [DOI] [PubMed] [Google Scholar]
- Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Molecular cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Ravindranath RM, Devarajan A, Bringas P., Jr Enamel formation in vitro in mouse molar explants exposed to amelogenin polypeptides ATMP and LRAP on enamel development. Arch Oral Biol. 2007;52:1161–1171. doi: 10.1016/j.archoralbio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Reith EJ, Cotty VF. The absorptive activity of ameloblasts during the maturation of enamel. Anat Rec. 1967;157:577–587. doi: 10.1002/ar.1091570404. [DOI] [PubMed] [Google Scholar]
- Simmer JP. Alternative splicing of amelogenins. Connect Tissue Res. 1995;32:131–136. doi: 10.3109/03008209509013715. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Hu CC, Lau EC, Sarte P, Slavkin HC, Fincham AG. Alternative splicing of the mouse amelogenin primary RNA transcript. Calcif Tissue Int. 1994;55:302–310. doi: 10.1007/BF00310410. [DOI] [PubMed] [Google Scholar]
- Smith CE. Ameloblasts: secretory and resorptive functions. J Dent Res. 1979;58:695–707. doi: 10.1177/002203457905800221011. [DOI] [PubMed] [Google Scholar]
- Smith CE, Warshawsky H. Quantitative analysis of cell turnover in the enamel organ of the rat incisor. Evidence for ameloblast death immediately after enamel matrix secretion. Anat Rec. 1977;187:63–98. doi: 10.1002/ar.1091870106. [DOI] [PubMed] [Google Scholar]
- Sundin OH, Busse HG, Rogers MB, Gudas LJ, Eichele G. Region-specific expression in early chick and mouse embryos of Ghox-lab and Hox 1.6, vertebrate homeobox-containing genes related to Drosophila labial. Development. 1990;108:47–58. doi: 10.1242/dev.108.Supplement.47. [DOI] [PubMed] [Google Scholar]
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- Thomsen R, Solvsten CA, Linnet TE, Blechingberg J, Nielsen AL. Analysis of qPCR data by converting exponentially related Ct values into linearly related X0 values. J Bioinform Comput Biol. 2010;8:885–900. doi: 10.1142/s0219720010004963. [DOI] [PubMed] [Google Scholar]
- Tompkins K, Alvares K, George A, Veis A. Two related low molecular mass polypeptide isoforms of amelogenin have distinct activities in mouse tooth germ differentiation in vitro. J Bone Miner Res. 2005;20:341–349. doi: 10.1359/JBMR.041107. [DOI] [PubMed] [Google Scholar]
- Uchida T, Tanabe T, Fukae M, Shimizu M. Immunocytochemical and immunochemical detection of a 32 kDa nonamelogenin and related proteins in porcine tooth germs. Archives of histology and cytology. 1991;54:527–538. doi: 10.1679/aohc.54.527. [DOI] [PubMed] [Google Scholar]
- Warotayanont R, Frenkel B, Snead ML, Zhou Y. Leucine-rich amelogenin peptide induces osteogenesis by activation of the Wnt pathway. Biochem Biophys Res Commun. 2009;387:558–563. doi: 10.1016/j.bbrc.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen HB, Fincham AG, Moradian-Oldak J. Progressive accretion of amelogenin molecules during nanospheres assembly revealed by atomic force microscopy. Matrix Biol. 2001;20:387–395. doi: 10.1016/s0945-053x(01)00144-5. [DOI] [PubMed] [Google Scholar]
- Wen X, Cawthorn WP, MacDougald OA, Stupp SI, Snead ML, Zhou Y. The influence of Leucine-rich amelogenin peptide on MSC fate by inducing Wnt10b expression. Biomaterials. 2011;32:6478–6486. doi: 10.1016/j.biomaterials.2011.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Harada H, Taniguchi A. The exon 6ABC region of amelogenin mRNA contribute to increased levels of amelogenin mRNA through amelogenin protein-enhanced mRNA stabilization. J Biol Chem. 2006a;281:32439–32444. doi: 10.1074/jbc.M605406200. [DOI] [PubMed] [Google Scholar]
- Xu L, Harada H, Yokohama-Tamaki T, Matsumoto S, Tanaka J, Taniguchi A. Reuptake of extracellular amelogenin by dental epithelial cells results in increased levels of amelogenin mRNA through enhanced mRNA stabilization. J Biol Chem. 2006b;281:2257–2262. doi: 10.1074/jbc.M507695200. [DOI] [PubMed] [Google Scholar]