Abstract
The proper function of the nervous system depends on precise production and connection of distinct neurons and glia. Cell fate determination of neurons and glia is tightly controlled by complex gene expression regulation in the developing and adult nervous system. Emerging evidence has demonstrated the importance of noncoding microRNAs (miRNAs) in neural development and function. This review highlights current discoveries of miRNA functions in specifying neuronal and glial cell fate. We summarize the roles of miRNAs in expansion and differentiation of neural stem cells, specification of neuronal subtypes and glial cells, reprogramming of functional neurons from embryonic stem cells and fibroblasts, and left-right asymmetric organization of neuronal subtypes. Investigating the network of interactions between miRNAs and target genes will reveal new gene regulation machinery involved in tuning the cell fate decisions of neurons and glia.
Introduction
One fascinating phenomenon in the nervous system of invertebrates and vertebrates is the precise regulation of cell fate determination. Distinct neurons and glia are derived from neural stem cells (NSCs) or specific neural progenitors (NPs) and glial progenitors, respectively, in different regions of the developing central nervous system (CNS) by complex temporospatial gene regulation [1–5]. Even in adult brains, tightly controlled and diverse neurogenesis is critical for proper brain functions [6–8]. Resulting from extensive investigations of molecular mechanisms of cell fate determination, NSCs, specific progenitors, embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs) and even fibroblasts have been programmed or reprogrammed into specific neuronal and glial types for treatments of neurological disorders [9–12].
Emerging studies have shown that like protein coding genes, microRNAs (miRNAs) play essential roles in cell fate determination. miRNAs, found in almost all eukaryotic cells, are a group of 18–22 nucleotide (nt) highly conserved small noncoding RNAs, which normally negatively regulate target gene expression by binding to messenger RNAs (mRNAs), typically in the 3’ untranslated region (3’UTR) [13,14]. Exciting studies have demonstrated important roles of miRNAs in neural development and neurological diseases [15–18]. In this review, we will highlight miRNA-mediated neuronal and glial specification from NSCs, specific progenitors, ESCs, iPSCs and fibroblasts, and left-right organization of specific neuronal subtypes in the nervous system.
miRNAs regulate expansion and differentiation of NSCs and NPs
A feature of NSCs is their ability to self-renew to expand the NSC pool. Some miRNAs have been identified that promote self-renewal and proliferation of NSCs and NPs, and inhibit differentiation in both the developing and adult nervous system (Figure 1 and Table 1). In the embryonic mouse cerebral cortex, miR-19 in the miR-17-92 cluster has been found to promote NSC proliferation and radial glial cell (RGC) expansion by targeting Pten [19●]. Interestingly, miR-92, another miRNA in the miR-17-92 cluster, has been shown to inhibit transition of intermediate progenitors (IPs) from RGCs by targeting Tbr2 [19,20]. Dual regulation by members of the miR-17-92 cluster on numbers of RGCs and IPs is critical for controlling the proper progenitor pool and brain sizes [19]. miR-134 has been shown to be essential for the maintenance of cortical NPs by targeting doublecortin (Dcx) and/or Chordin-like 1 (Chrdl-1) [21].
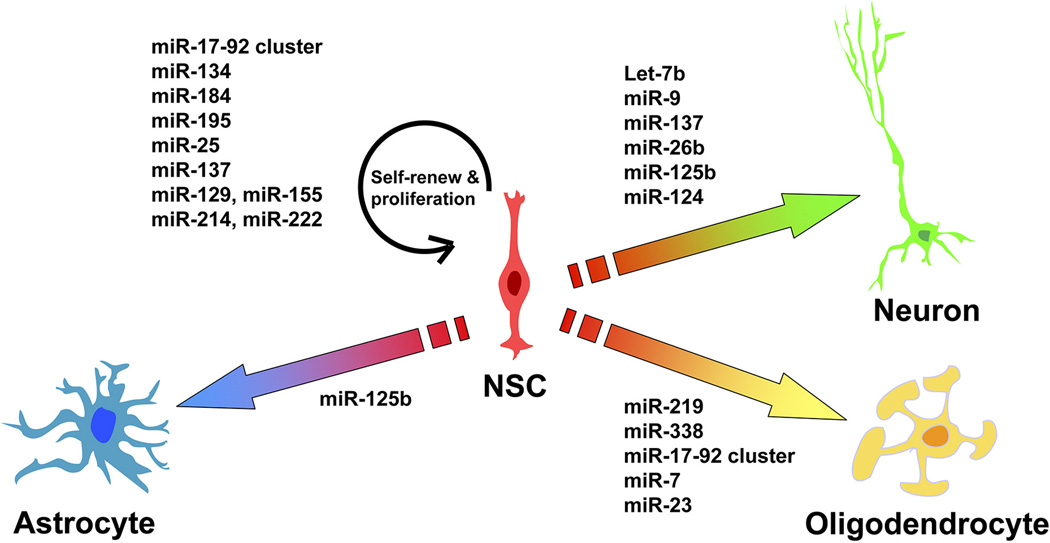
Figure 1.
A scheme of the roles of miRNAs in cell fate determination. miRNAs that regulate neural stem cell (NSC) self-renewal and proliferation, neuronal differentiation, astrogliogensis, and oligodendrocyte differentiation are listed.
Table 1.
A list of miRNAs that regulate neuronal and glial cell fate determination.
| Cell fate determination | miRNAs | Species and tissues | Targets | Reference | |
|---|---|---|---|---|---|
| NSC proliferation | miR-17-92 cluster | Mouse embryonic cortex | Pten, Tbr2 | [19, 20] | |
| miR-134 | Cultured embryonic mouse cortical NSCs | Chrdl-1, Dcx | [21] | ||
| miR-184 | Cultured adult mouse forebrain NSCs | Numbl | [22] | ||
| miR-195 | Cultured adult mouse forebrain NSCs | MBD1 | [23] | ||
| miR-25 | Cultured adult mouse forebrain NSCs | ? | [24] | ||
| miR-137 | Cultured adult mouse forebrain NSCs | Ezh2 | [25] | ||
| miR-129, miR-155, miR-214, miR-222 | Xenopus retina progenitor | Oxt2, Vsx1 | [26] | ||
| NSC differentiation | Let-7b | Adult and embryonic mouse forebrain NSCs | TLX, Cyclin D1, lin-28 | [28, 29] | |
| miR-9 | Adult and embryonic mouse forebrain NSCs NPs in Zebrafish NPs in Xenopus |
TLX Fgf8-1, FgfR1 Hairy1 |
[30] [32] [33] |
||
| miR-137 | Embryonic mouse forebrain NSCs | LSD1 | [31] | ||
| miR-26b | Zebrafish neural tube | Ctdsp2 | [34,35] | ||
| miR-125b | Cultured mouse forebrain NSCs | Nestin | [36] | ||
| miR-124 | Mouse neuroblastoma cell linesAdult SVZ NSCs | PTBP1 Sox9 |
[37] [38,39] |
||
| Neuronal subtype specification | Dopaminergic neurons (DN) | miR-133b | Embryonic mouse midbrain | Pitx3 | [41] |
| miR-132 | Embryonic stem cells | Nurr1 | [42] | ||
| miR-7a | Suppressing DN differentiation | Pax6 | [43] | ||
| Projection neurons | miR-9-2/3 | Embryonic mouse cortex | Foxg1, Pax6, Gsh2 | [44] | |
| Olfactory neurons | miR-200 family | Mouse and zebrafish | Foxg1, Zfhx1, Lfng | [45] | |
| Spinal motor neurons | miR-17-3p | Mouse spinal motor neuron progenitors | Olig2 | [46] | |
| miR-9 | Chick spinal motor neurons | FoxP1 | [47,48] | ||
| Gliogenesis | Astrogliogenesis | miR-125b | Neural cell line | ? | [49] |
| Oligodendrocyte differentiation | miR-219 and miR-338 | Postnatal rat and mouse CNS | PDGFRα, Sox6, FoxJ3, ZFP238, Hes5 | [50,51] | |
| miR-17-92 cluster | Neonatal mouse brain | ? | [52] | ||
| miR-7 | Embryonic mouse brain | Pax6, NeuroD4 | [53] | ||
| miR-23 | Mouse fibroblast and neural cell lines | Lamin B1 | [54] | ||
NSCs: Neural stem cells; NPs: neural progenitors; SVZ: subventricular zone; CNS: central nervous system.
In adult NSCs, Liu et al. have shown that miR-184 promotes adult NSC proliferation by repressing Numb-like (Numbl) [22]. Meanwhile, miR-184 is suppressed by methyl-CpG binding protein 1 (MBD1), suggesting that a regulatory network of miRNAs controls adult NSC expansion [22]. A similar regulatory loop has been identified between MBD1 and miR-195, which also positively regulates adult NSC proliferation [23●]. Moreover, miR-25, a member of the miR-106-25 cluster, has been shown to promote adult NSC proliferation, potentially through regulation of genes in the insulin/insulin-like growth factor-1 pathway [24]. miR-137, which is regulated by DNA methyl-CpG-binding protein (MeCP2) and transcription factor (TF) Sox2, promotes adult NSC proliferation and inhibits differentiation by targeting Ezh2, a histone methyltransferase and Polycomb group protein [25]. In the Xenopus retina, miR-129, miR-155, miR-214 and miR-222 have been found to promote progenitor proliferation by targeting Oxt2 and Vsx1 [26].
An interesting observation of miRNA regulation is that it often forms a feedback loop with its target genes in the process of controlling cell fate. Schwamborn et al. have shown that Let-7 is a target of TRIM32 and suppresses NSC proliferation [27]. Let-7b enhances differentiation by targeting the nuclear receptor TLX and the cell cycle regulator cyclin D1 [28]. Interestingly, further investigation has shown that let-7 normally suppresses lin-28 protein expression, and lin-28 also blocks let-7 expression by binding to the let-7 precursor and inhibiting its biogenesis [29]. In addition, miR-9, a CNS-enriched miRNA, has been shown to suppress mouse NSC expansion and induce differentiation through a feedback regulation of TLX [30]. TLX further recruits histone lysine-specific demethylase 1 (LSD1), which is a target of miR-137, and modulates proper expression of miR-137, which normally suppresses NSC proliferation [31]. These studies suggest that miRNAs play a critical role in ensuring proper numbers of NSCs and NPs by either directly silencing target genes, or forming a regulatory loop with targets.
miRNAs that inhibit NSC self-renewal and enhance differentiation have also been identified. In addition to targeting TLX, miR-9 has been shown to inhibit NP proliferation and elevate differentiation by suppressing several genes in the fibroblast growth factor signaling pathway such as Fgf8-1 and FgfR1 in zebrafish, and by targeting hairy1 in Xenopus [32,33]. miR-26b has been reported to induce neuronal differentiation by suppressing its host gene ctdsp2 in the zebrafish neural tube [34●,35]. In NSC cultures, miR-125b has been found to inhibit NSC proliferation by repressing the neural precursor marker Nestin [36]. miR-124 is another well-studied CNS-enriched miRNA that has been shown to induce differentiation of embryonic NSCs and NPs by targeting the global splicing repressor PTBP1 and to promote nervous system-specific alternative splicing [37]. In the adult brain, miR-124 plays a positive role in regulating neuronal differentiation of adult NSCs in the subventricular zone (SVZ) by suppressing Sox9 expression [38,39].
As studies that examine the function of miRNAs in NSC/NP proliferation and differentiation are accumulating, it is becoming clear that most miRNAs can be characterized into two groups based on their general roles: they promote either proliferation such as the miR-17-92 cluster, or differentiation such as miR-9 and miR-124 (Figure 1). However, the diversity and complexity of individual miRNAs in cell fate determination appear to rely on different species, specific regions in the nervous system, distinct cell context, and mostly the availability and direct physical interaction of their target genes.
Cell fate determination of neuronal subtypes by miRNAs
The complex functions of the nervous system depend on circuit formation built upon specification and connection of distinct neuronal subtypes. miRNAs have also been shown to play important roles in specifying neuronal subtypes. A profiling study has shown specific expression of miRNAs in glutamatergic and GABAergic neurons, and subtypes of GABAergic neurons [40●]. Due to their role in dopamine production, dopaminergic neurons (DNs) are essential for normal cognitive functions and voluntary movement. A negative feedback loop between paired-like homeodomain TF Pitx3 and miR-133b has been identified in the midbrain during DN differentiation and maturation: Pitx3 induces the expression of miR-133b, which also represses Pitx3 expression and suppresses DN maturation and function [41]. miR-132 has been shown to be highly expressed in tyrosine hydroxylase (TH)-positive DNs and to inhibit DN differentiation from ESCs by targeting Nurr1, one of the key TFs in DN differentiation [42]. miR-7a has been found to suppress DN differentiation by targeting Pax6 in the SVZ of postnatal mouse brains [43].
Genetic deletion of miR-9-2 and miR-9-3 results in malformation of the cerebral cortex in mice, suggesting that miR-9 plays a role in regulating projection neuron development [44●]. Members of the miR-200 family have been shown to be critical for neurogenesis of olfactory neurons by targeting Foxg1, Zfhx1 and Lfng [45]. In the spinal cord, miR-17-3p is required for patterning of motor neuron progenitors by targeting Olig2 [46]. miR-9 has been shown to modify spinal motor neuron subtype specification by balancing FoxP1 expression levels [47,48]. Because a miRNA can have multiple targets, neuronal subtype specification is likely achieved through cell type-specific miRNA expression and a balanced outcome of overall target gene expression, which eventually favors generation of a specific cell type.
miRNAs and gliogenesis
Many miRNAs are also involved in gliogenesis, including astrogliogenesis and oligodendrocyte differentiation. An in vitro study has shown that miR-125b positively regulates astrogliogenesis and promotes astrocyte proliferation [49]. Deletion of Dicer in the oligodendrocyte linage causes impaired oligodendrocyte differentiation, which can be partially rescued by ectopic expression of oligodendrocyte lineage-specific miR-219 [50]. miR-219 and miR-338 promote oligodendrocyte differentiation by suppressing PDGFRα, Sox6, FoxJ3, ZFP238 and Hes5 [50,51]. Moreover, studies have shown that miR-19b in the miR-17-92 cluster promotes oligodendrocyte precursor proliferation by regulating Akt signaling, and miR-7 enhances the generation of oligodendrocyte lineage cells by targeting proneuronal differentiation factors such as Pax6 and NeuroD4 [52,53]. Furthermore, miR-23 has been shown to suppress Lamin B1 expression and promote oligodendrocyte differentiation [54]. Compared to studies of miRNAs in neuronal specification, reports of miRNAs in glial development are still sparse (Table 1). Whether miRNAs are directly involved in the cell fate switch between neurons and glia is unclear. Identifying more glial-specific miRNAs will further advance our understanding of glial cell fate determination [55].
miRNAs promote the neuronal fate from ESCs, iPSCs and fibroblasts
Because of the therapeutic potential, induction of different types of neuronal cells from ESCs, iPSCs and fibroblasts has become a hot topic. Excitingly, miRNAs have been found to play roles in the reprogramming process. A miRNA profiling study has shown that miR-9 and miR-124a are highly expressed in ESC-derived culture during neuronal differentiation, which is consistent with their roles in promoting differentiation of NSCs and NPs [56], although one study has shown that miR-9 elevates proliferation of NPs that are derived from human ESCs [57]. Let-7 and miR-125 have been found to be strongly induced during neuronal differentiation from ESCs [29,58]. Further investigation has shown that miR-125 promotes neural conversion of human ESCs into SOX1-positive NPs by repressing SMAD4, suggesting that miR-125 is involved in the Bone Morphogenetic Protein (BMP)-mediated classic signaling transduction of neural lineage commitment from ESCs [59]. A study that analyzes 13 human ESC lines and 26 human iPSC lines has found that an increased miR-371-3 expression level favors the neurogenic differentiation propensity of human ESC and iPSC lines [60●].
Together with other TFs, some neural-specific miRNAs have been used to directly induce reprogramming of fibroblasts into neuronal lineages. miR-9/9* and miR-124, along with three neurogenic TFs NeuroD2, Ascl1 and Mytl1, have been shown to efficiently convert human fibroblasts into functional neurons [61●]. Another investigation has shown that miR-124, together with two TFs MYT1L and BRN2, is able to reprogram postnatal and adult human fibroblasts into functional neurons [62●]. These studies suggest that although miRNAs alone are not sufficient for neuronal reprogramming of ESCs, iPSCs and fibroblasts, neural-enriched miRNAs elevate the efficiency of reprogramming.
miRNAs specify neuronal left-right asymmetry
The nervous system is mostly bilaterally symmetric at the anatomical level, but also displays morphological and functional left-right asymmetry to some extent. miRNAs have been shown to play a role in neuronal left-right asymmetry in the nervous system of C. elegans. lsy-6 was the first identified miRNA that controls the left-right asymmetry of ASE left (ASEL) and ASE right (ASER) gustatory neurons by targeting cog-1, a negative regulator of ASEL neuronal cell fate [63]. Further investigation has shown that a C2H2 zinc finger TF lsy-2 regulates ASEL/R asymmetry by modulating transcription of lsy-6 in ASEL neurons [64]. Another zinc-finger TF die-1 has been shown to activate the expression of lsy-6 only in ASEL, but not in ASER. miR-273, an ASER-specific miRNA, has been found to repress die-1 expression to determine ASEL/R asymmetry [65]. Furthermore, miR-71 has been observed to play a role in regulating left/right identification of Amphid Wing Cell C (AWC) olfactory neurons by repressing TIR-1/SARM1 adaptor proteins in the calcium signaling pathway [66]. A most recent study has further identified a mechanism by which the lsy-6 locus is primed in the precursor for the left neuron by chromatin decompaction [67●]. However, the role of miRNAs in asymmetric neuronal organization in the vertebrate nervous system is still unknown.
Conclusions
It is becoming evident that cell fate determination of neurons and glia is tightly controlled by both protein coding genes and noncoding miRNAs. Because one miRNA has multiple target genes, cell fate specification by miRNAs is likely an overall outcome of balanced protein outputs, even though one or a few targets are probably major players. miRNA-mediated gene expression regulation shares similarities with that of TFs, which normally have binding motifs on promoters of multiple genes, including miRNAs, even though TFs act as both activators and repressors, while miRNAs largely negatively control target gene expression. For example, genome-wide quantification has revealed a range of genes that are directly or indirectly regulated by TFs such as Pax6 and Tbr1 in the mouse cortex [68–70]. Similarly, perturbing miRNA expression also affects many target genes, including TFs, in neurons [71–73].
Therefore, miRNAs, in parallel with TFs, form networks with target genes in gene expression regulation. Using genome-wide approaches in combination with functional analyses should advance mechanistic knowledge of miRNA actions in cell fate determination. Moreover, identifying more tissue- and cell type-specific miRNAs and uncovering how their expression is regulated by TFs will further accelerate research in miRNA-mediated cell fate determination.
Highlights.
miRNAs are small noncoding RNAs that normally silence target gene expression.
Many miRNAs have enriched expression in the central nervous system.
miRNAs are required for proliferation and differentiation of neural stem cells.
Neuronal subtypes are specified by miRNAs by suppressing specific target genes.
Networks of miRNA-target are critical in cell fate determination of neurons and glia.
Acknowledgements
We thank Jennifer Knauss for critical reading of the manuscript. This work was supported by the Hirschl/Weill-Caulier Trust (T. S.), an NPRP grant from the Qatar National Research Fund (T. S.) and an R01-MH083680 grant from the NIH/NIMH (T. S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
● of special interest
- 1.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008 doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 4.Kessaris N, Pringle N, Richardson WD. Ventral neurogenesis and the neuron-glial switch. Neuron. 2001;31:677–680. doi: 10.1016/s0896-6273(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 5.Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 6.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Lledo PM, Merkle FT, Alvarez-Buylla A. Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 2008;31:392–400. doi: 10.1016/j.tins.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdullah AI, Pollock A, Sun T. The path from skin to brain: generation of functional neurons from fibroblasts. Mol Neurobiol. 2012;45:586–595. doi: 10.1007/s12035-012-8277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aboody K, Capela A, Niazi N, Stern JH, Temple S. Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a Rosetta stone. Neuron. 2011;70:597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 11.Yang N, Ng YH, Pang ZP, Sudhof TC, Wernig M. Induced neuronal cells: how to make and define a neuron. Cell Stem Cell. 2011;9:517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman SA, Nedergaard M, Windrem MS. Glial progenitor cell-based treatment and modeling of neurological disease. Science. 2012;338:491–495. doi: 10.1126/science.1218071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 14.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang MF, Shi Y. Dynamic Roles of microRNAs in Neurogenesis. Front Neurosci. 2012;6:71. doi: 10.3389/fnins.2012.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Mol Neurobiol. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bian S, Hong J, Li Q, Schebelle L, Pollock A, Knauss JL, Garg V, Sun T. MicroRNA Cluster miR-17-92 Regulates Neural Stem Cell Expansion and Transition to Intermediate Progenitors in the Developing Mouse Neocortex. Cell Rep. 2013;3:1398–1406. doi: 10.1016/j.celrep.2013.03.037. ● This paper reported a fine-tuning regulation of miRNAs in specifying radial glial cells and intermediate progenitors in the developing mouse cortex. The authors generated miR-17-92 cortical specific conditional knockout mice and demonstrated a positive control of miR-17-92 in NSC expansion.
- 20.Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci U S A. 2013;110:7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- 22.Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C, Teng ZQ, McQuate AL, Jobe EM, Christ CC, von Hoyningen-Huene SJ, Reyes MD, Polich ED, Xing Y, Li Y, et al. An epigenetic feedback regulatory loop involving microRNA-195 and MBD1 governs neural stem cell differentiation. PLoS One. 2013;8:e51436. doi: 10.1371/journal.pone.0051436. ● Together with [22] and [25], these three papers showed an epigenetic regulation of DNA binding proteins on expression of different miRNAs, which play a positive role in NSC proliferation.
- 24.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3:108–124. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci U S A. 2009;106:21179–21184. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 30.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, Li W, Fu C, Yin J, Wang A, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leucht C, Stigloher C, Wizenmann A, Klafke R, Folchert A, Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11:641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 33.Bonev B, Pisco A, Papalopulu N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev Cell. 2011;20:19–32. doi: 10.1016/j.devcel.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dill H, Linder B, Fehr A, Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26:25–30. doi: 10.1101/gad.177774.111. ● This paper presented the regulation of an intronic miRNA on its host gene expression in modulating neurogenesis. The repression of neurogenesis of the RNA polymerase II C-terminal domain small phosphatase 2 (ctdsp2) can be inhibited by its intronic miR-26b through a targeting effect.
- 35.Han J, Denli AM, Gage FH. The enemy within: intronic miR-26b represses its host gene, ctdsp2, to regulate neurogenesis. Genes Dev. 2012;26:6–10. doi: 10.1101/gad.184416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui Y, Xiao Z, Han J, Sun J, Ding W, Zhao Y, Chen B, Li X, Dai J. MiR-125b orchestrates cell proliferation, differentiation and migration in neural stem/progenitor cells by targeting Nestin. BMC Neurosci. 2012;13:116. doi: 10.1186/1471-2202-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. ● This study used a miRNA tagging and affinity-purification technique to investigate distinct miRNA profiling in glutamatergic and GABAergic neurons and subtypes of GABAergic neurons. This work provided useful information of miRNA expression in specific neuronal subtypes.
- 41.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang D, Li T, Wang Y, Tang Y, Cui H, Zhang X, Chen D, Shen N, Le W. miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J Cell Sci. 2012;125:1673–1682. doi: 10.1242/jcs.086421. [DOI] [PubMed] [Google Scholar]
- 43.de Chevigny A, Core N, Follert P, Gaudin M, Barbry P, Beclin C, Cremer H. miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons. Nat Neurosci. 2012;15:1120–1126. doi: 10.1038/nn.3142. [DOI] [PubMed] [Google Scholar]
- 44. Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. ● The authors generated miR-9-2/3 double knockout mice, which show severe cortical defects, and demonstrated the essential role of miR-9 in cortical development.
- 45.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JA, Huang YP, Mazzoni EO, Tan GC, Zavadil J, Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69:721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otaegi G, Pollock A, Hong J, Sun T. MicroRNA miR-9 Modifies Motor Neuron Columns by a Tuning Regulation of FoxP1 Levels in Developing Spinal Cords. J Neurosci. 2011;31:809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otaegi G, Pollock A, Sun T. An Optimized Sponge for microRNA miR-9 Affects Spinal Motor Neuron Development in vivo. Front Neurosci. 2011;5:146. doi: 10.3389/fnins.2011.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pogue AI, Cui JG, Li YY, Zhao Y, Culicchia F, Lukiw WJ. Micro RNA-125b (miRNA-125b) function in astrogliosis and glial cell proliferation. Neurosci Lett. 2010;476:18–22. doi: 10.1016/j.neulet.2010.03.054. [DOI] [PubMed] [Google Scholar]
- 50.Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137:2127–2132. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X, Wu J, Zheng M, Gao F, Ju G. Specification and maintenance of oligodendrocyte precursor cells from neural progenitor cells: involvement of microRNA-7a. Mol Biol Cell. 2012;23:2867–2878. doi: 10.1091/mbc.E12-04-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin ST, Fu YH. miR-23 regulation of lamin B1 is crucial for oligodendrocyte development and myelination. Dis Model Mech. 2009;2:178–188. doi: 10.1242/dmm.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi-Carter R. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33:5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB. MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell. 2010;6:323–335. doi: 10.1016/j.stem.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cimadamore F, Amador-Arjona A, Chen C, Huang CT, Terskikh AV. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139:1247–1257. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 60. Kim H, Lee G, Ganat Y, Papapetrou EP, Lipchina I, Socci ND, Sadelain M, Studer L. miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell. 2011;8:695–706. doi: 10.1016/j.stem.2011.04.002. ● The authors analyzed 13 human embryonic stem cell lines and 26 human induced pluripotent stem cell lines and detected miR-371-3 that can be used to predict neural differentiation propensity by its expression level.
- 61. Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. ● This paper, along with the following, used a combination of brain-enriched miRNAs and neurogenic transcription factors to directly reprogram human fibroblasts into functional neurons, which provides a tool for stem cell-based therapy of neurological diseases using miRNAs.
- 62. Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. ● This paper, along with the previous, used a combination of brain-enriched miRNAs and neurogenic transcription factors to directly reprogram human fibroblasts into functional neurons, which provides a tool for stem cell-based therapy of neurological diseases using miRNAs.
- 63.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 64.Johnston RJ, Jr, Hobert O. A novel C. elegans zinc finger transcription factor, lsy-2, required for the cell type-specific expression of the lsy-6 microRNA. Development. 2005;132:5451–5460. doi: 10.1242/dev.02163. [DOI] [PubMed] [Google Scholar]
- 65.Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 66.Hsieh YW, Chang C, Chuang CF. The microRNA mir-71 inhibits calcium signaling by targeting the TIR-1/Sarm1 adaptor protein to control stochastic L/R neuronal asymmetry in C. elegans. PLoS Genet. 2012;8:e1002864. doi: 10.1371/journal.pgen.1002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cochella L, Hobert O. Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell. 2012;151:1229–1242. doi: 10.1016/j.cell.2012.10.049. ● This paper revealed a mechanism of the two step activation of miRNA lsy-6 in specifying neuronal left-right asymmetry.
- 68.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, Smith J, van Heyningen V, Rubenstein JL, Livesey FJ. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holm PC, Mader MT, Haubst N, Wizenmann A, Sigvardsson M, Gotz M. Loss- and gain-of-function analyses reveal targets of Pax6 in the developing mouse telencephalon. Mol Cell Neurosci. 2007;34:99–119. doi: 10.1016/j.mcn.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 70.Elsen GE, Hodge RD, Bedogni F, Daza RA, Nelson BR, Shiba N, Reiner SL, Hevner RF. The protomap is propagated to cortical plate neurons through an Eomes-dependent intermediate map. Proc Natl Acad Sci U S A. 2013;110:4081–4086. doi: 10.1073/pnas.1209076110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dorval V, Smith PY, Delay C, Calvo E, Planel E, Zommer N, Buee L, Hebert SS. Gene network and pathway analysis of mice with conditional ablation of Dicer in post-mitotic neurons. PLoS One. 2012;7:e44060. doi: 10.1371/journal.pone.0044060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawase-Koga Y, Low R, Otaegi G, Pollock A, Deng H, Eisenhaber F, Maurer-Stroh S, Sun T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J Cell Sci. 2010;123:586–594. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]