Abstract
Many cellular processes involve interactions between membrane-associated proteins, and those interactions are enhanced by membrane association. We have used crosslinking reactions to compare the extent and specificity of protein interactions in solution versus on a membrane surface. Cysteine mutants of a soluble cytoplasmic fragment (CF) of the aspartate receptor, a transmembrane receptor involved in bacterial chemotaxis, are used in disulfide bond formation with the thiol-specific oxidant diamide and chemical crosslinking reactions with the trifunctional maleimide TMEA. CF binding to vesicles is mediated by its N-terminal His tag binding to vesicles containing a nickel-chelating lipid, so crosslinking reactions conducted in the presence and absence of vesicles differ only in whether CF is bound to the vesicles or is free in solution. For multiple Cys throughout the CF, membrane association is shown to increase the rate and extent of these reactions. Crosslinking specificity, which is measured as the preference for crosslinking between Cys near each other in the native structure, is also enhanced by membrane association. These results provide an experimental demonstration that membrane binding enhances protein-protein interactions, an important consideration for understanding processes involving membrane-associated proteins. The experiments further demonstrate the importance of crosslinking conditions for these reactions that are often used to probe protein structure and dynamics, and the potential of membrane association to restore native interactions of membrane-associated proteins for crosslinking studies.
Introduction
Numerous processes in the cell take place on the surface of membranes, where the proteins involved often display a reversible interaction with the membrane. Membrane association is expected to significantly enhance protein interactions,1 and is a postulated means to generate quaternary interactions.2 In addition, membrane association is expected to increase the specificity of protein interactions, which could lower the rates of some reactions among the membrane-associated species relative to the rate for proteins in solution.
We used the vesicle-mediated assembly method developed previously in our lab,3-5 to compare the extent and specificity of protein associations in solution versus on a membrane surface. Through the introduction of a unique cysteine, the reactivity of the cysteine in disulfide bond and chemical crosslinking reactions were used to make these comparisons. The approach has been applied to the cytoplasmic domains from the Escherichia coli aspartate and serine receptors, representative members of the large superfamily of chemotaxis receptors.6,7 The structure of the cytoplasmic domain is the defining feature of this receptor superfamily. It is organized as a coiled-coil hairpin (18 heptad repeats in length in the E. coli proteins), which dimerizes to form a four-helix bundle shown in Figure 1.8-10
Figure 1.
X-ray structure of Tsr cytoplasmic domain dimer (1qu7)8 showing the cysteine sites, which are numbered according to the intact Tar sequence. The approximate location of S272 (which is beyond the 1qu7 crystal structure) is also shown. In the top structure, the methylation (red), flexible (green) and signaling (blue) subdomains are emphasized. The color scale in the lower structure depicts B factors of the 1qu7 structure. Note that these B factors are for the 4Q construct; B factors for the 2Q2E construct are higher,26 and are likely to be even higher for the 4E construct used in this study, particularly in the methylation region due to electrostatic repulsion, as previously demonstrated by higher disulfide yields for Cys sites in this region in the 4E vs. the 4Q construct of the intact Asp receptor.27
Cysteine residues were introduced individually at 10 positions throughout the receptor cytoplasmic fragment (CF), at sites with different exposure in the structure (buried at the intramonomer or intermonomer interface, or solvent exposed) and spread throughout the 3 subdomains along the length of the CF. Each mutant CF was subjected to both disulfide formation with the thiol-specific oxidant diamide to produce receptor dimers and to crosslinking with the trifunctional maleimide TMEA to produce receptor dimers and trimers. Reactions were conducted in the presence and absence of extruded vesicles containing the nickel-chelating lipid DGS-NTA, which binds the N-terminal His tag of the CF construct.4 Comparison of CF crosslinking in solution and on vesicles was used to show that (i) vesicle association of the CF increases the rate and extent of crosslinking, and (ii) crosslinking reactivity differences among sites reflect differences in dynamics along the length of the CF. Furthermore, diamide crosslinking reactions between different Cys mutants were used to determine the preference for formation of homodimer crosslinks (between monomers with the same Cys residue) vs. heterodimer crosslinks (between monomers with two different Cys residues), to test whether membrane association of the CF increases the specificity of crosslinking. Suppression of heterodimer formation was observed for CF crosslinking for Cys sites distant from each other in the structure. The results also demonstrated the importance of the choice of crosslinking conditions: stoichiometric crosslinking agent concentrations were important for revealing reactivity differences and excess crosslinking agent concentrations were important for revealing the partitioning between homo and heterodimers.
Materials and Methods
Vesicle Preparation (LUV)
DOPC (1, 2-dioleoyl-sn-glycero-3-phosphocholine) and the nickel-chelating lipid, DGS-NTA-Ni2+ (1, 2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl) were obtained from Avanti Polar Lipids (Alabaster, Alabama). A suspension of multilamellar vesicles (MLVs) was prepared by hydration of a 1:1 DOPC:DGS-NTA-Ni2+ lipid film in kinase buffer (75 mM Tris, 100 mM KCl, pH 7.5) at room temperature, to give a total lipid concentration of 2 mg/mL. When vesicles were prepared for kinase assays, 5mM MgCl2 and 5% DMSO were included in the buffer; for assays under reducing conditions, 2 mM TCEP was also included. Large unilamellar vesicles (LUVs) were prepared by extrusion in a small–volume commercial apparatus (Avanti Mini Extruder). The MLV suspension was passed 15 times through Nucleopore Track-Etch™ polycarbonate membrane with a 100nm pore diameter.
Construction of Cys mutants and truncated mutants of Cytoplamic domain
The histidine-tagged aspartate and serine receptor cytoplasmic domain (TarCF and TsrCF), encoding Glu at the four main sites of methylation (4E), were expressed in the E. coli strain DH5αF’ using plasmids pHTCF11 and pHTCF(tsr).12 Cysteine and stop codon mutations were introduced by site-directed mutagenesis and reagents were purchased from Agilent Technologies, CA (previously Stratagene) and New England Biolabs, MA. Truncation of the C-terminus of the CF removed ~3.7 kDa from Tsr and ~3.3 kDa from Tar; the resulting plasmids expressed Tsr 258-518 and Tar 257-525. Primers were obtained from Integrated DNA Technologies (San Diego, CA). The PCR reaction was carried out in a thermocycler (Mastercycler® personal) and Pfu, dNTPs and DpnI were obtained from Stratagene (Cedar Creek, TX) and New England Bio-Labs for some reactions. The PCR products were treated with DpnI at 37°C for 1hr to digest the template plasmid DNA. Agarose gels were run to verify that the PCR reaction produced a product of the expected length.
The PCR product was transformed into DH5αF’ chemically competent cells, and transformants were selected with LB-ampicillin plates (0.15mg ampicillin/ml). Colonies were randomly selected and grown at 37 °C with and without IPTG induction for expression analysis. For CF truncation, cultures were screened by SDS-PAGE to determine if a truncated CF (faster migrating than wild type CF) was present. For Cys mutagenesis, cultures were subjected to cross-linking conditions to screen for the presence of the reactive cysteine. Two mL cultures were grown at 37°C to an OD600 of 0.4 to 0.5. These cultures were then induced with IPTG (1mM) and grown for an additional 1 to 2hr. The cells were harvested by centrifugation at 13,000 rpm for 15min in a benchtop micro centrifuge (Eppendorf) and the cell pellet was resuspended in 40 μL of water. For each sample, 20μL was treated with 0.15mM Cu(II)phenanthroline diluted from 150 mM stock, and 20 μL was not treated.13-16 After addition of 20 μL of gel loading buffer (without beta-mercaptoethanol) samples were boiled for 3 min. Permanents were prepared and plasmids were isolated for the colonies that showed the expected disulfide dimer or truncation by SDS-PAGE. The presence of the mutation in these plasmids was verified by sequencing (GENEWIZ, www.genewiz.com, South Plainfield, New Jersey). Sequence results were analyzed by alignment in the CLUSTALW program (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
Protein purification
Mutated or truncated CF purification was conducted according to published protocols using nickel affinity chromatography (GE Healthcare).4 CheA, CheW and CheY were purified as previously described.5,17-19 Protein concentration was determined using a modified Lowry assay kit (Thermo Scientific).
Activity assays
To initiate complex assembly, 580 μM lipid (extruded LUVs of 1:1 DOPC:DGS-NTA-Ni2+) were added to a mixture of 30 μM CF, 10 μM CheW, and 1.2 μM CheA in kinase buffer. Samples were incubated at 25°C in a water bath for 4 to 5 h prior to activity measurements. Steady-state kinase activities were measured as described previously,3,5 using a coupled ATPase assay in which the rate of ATP consumption was measured spectrophotometrically as the rate of NADH oxidation. For each assay, the assembled complex was diluted 100-fold into kinase buffer containing 55 μM CheY, 2.2 mM phosphoenolpyruvate, 4 mM ATP, 250 μM NADH and 20 units of PK/LDH enzyme (Sigma –Aldrich, St. Louis, MO) and immediately placed in the spectrophotometer. The background activity of CheY under equivalent conditions without receptor complexes was subtracted prior to the determination of specific activity. Specific activities were calculated from the linear change in absorbance at 340nm (d[ATP]/dt=−6220[dA340/dt]) over a one minute time period and normalized to the total CheA concentration.
The bound fraction of each protein was determined by sedimentation at 60,000 rpm at 25°C for 30 min (Beckman, Optima™TLX Ultracentrifuge) to separate bound (pellet) from unbound (supernatant) protein. The bound CheA, CheW and CF fractions were calculated by analyzing the integrated intensities of Gel-code Blue (Pierce Chem. Co.) stained protein bands from aliquots of total protein (prior to centrifugation) and unbound protein (supernatant) fractions using ImageJ software.
Diamide & TMEA induced cross-linking
Tris(2-maleimidoethyl)amine (TMEA) was obtained from Thermo Scientific; 1,1’-Azobis(N,N-dimethylformamide) (diamide) and N-ethylmaleimide (NEM) were obtained from Sigma. Tris(2-carboxyethyl)phosphine (TCEP) was obtained from Thermo Scientific. Purified CF (300 μM) was treated with 10 mM TCEP at 25 °C for 30 min to reduce disulfide-linked dimers that accumulated during purification and storage. TCEP was removed with 7K, Zeba™ Spin desalting columns (Thermo Scientific), using a Beckman Coulter Allegra™ 6R bench top centrifuge. The spin column was pre-equilibrated with buffer by centrifuging at 2000 rpm for 2min at 4 °C three times. The sample was applied to the pre-equilibrated column and centrifuged 3 min at 2000rpm and 4°C for TCEP removal. In addition, the reduced CF was subjected to 3 buffer exchanges in 10 kDa MWCO filters (Millipore), by centrifugation at 2000 rpm for 20-30min at 4 °C using Beckman Coulter Allegra™ 6R bench top centrifuge. Reduction was confirmed by pre-quenching an aliquot with 10 mM NEM followed by SDS-PAGE analysis on a 10% acrylamide gel. Aliquots of the reduced protein were flash frozen in liquid nitrogen and stored at −80 °C.
For the TMEA crosslinking reactions, aliquots were prepared in kinase buffer from the TCEP-reduced CF stock solutions to give a final concentration of 30 μM CF in the experimental samples. In the vesicle-assembled CF samples, LUVs were added to give 580 μM total lipid concentration (1:1 DOPC:DGS-NTA-Ni2+) and incubated for 5 min at room temperature. An equivalent volume of buffer was added to CF samples to measure cross-linking in the absence of vesicle-mediated assembly. TMEA stock solutions were prepared fresh in 90-100% DMSO. Each reaction was initiated by addition of the appropriate volume of the TMEA stock solution (DMSO was <5% DMSO in final reaction), and samples were incubated at 25 °C for 30 min. The reaction was stopped by quenching the sample with freshly prepared 10 mM NEM. The 100 mM NEM stock solution was prepared by dissolving in 100% DMSO, obtained from Fisher Scientific. SDS-PAGE (10% acrylamide gel concentration prepared from 30% acrylamide/0.8% bisacrylamide stock solution) was performed under reducing conditions to eliminate dimers that form due to air oxidation. The extent of cross-linked product was determined by gel-scanning using a Bio-Rad densitometer and calculating the area of protein bands using ImageJ software.
Sample preparation, reaction, and quenching procedures for diamide crosslinking were similar to those described for the TMEA crosslinking assay. 100 mM diamide (in water) was prepared fresh before each experiment. Each reaction was initiated by addition of the appropriate volume of the diamide stock solution. To measure crosslinking kinetics, the reaction was started by adding diamide to the pre-incubated samples at 25 °C. 10μl aliquots were taken at each time point and immediately quenched with 10 mM NEM. Non-reducing SDS-PAGE (10% gel) was conducted to measure the extent of cross-linking as described above. Each assay included a control sample without diamide to determine the crosslink product produced due to air oxidation during incubation, which was always less than 8%. The kinetic data was fit to an exponential association equation, y = Y0 +A1(1 − exp(−t/ τ1)) + A2(1 − exp(−t/ τ2) in Origin software to obtain time constants (τ1 and τ2) and amplitudes (A1 and A2). TMEA crosslinking kinetics were not characterized because the reaction was much faster than diamide crosslinking, perhaps on a similar timescale to the in vivo crosslinking of Tsr.20
Results
Mutagenesis creates cysteine probe sites throughout the receptor CF
Extensive published Cys mutagenesis studies of the Asp receptor made it possible to choose relatively non-perturbing Cys mutation sites probing buried or solvent-exposed locations throughout the cytoplasmic domain. The 10 chosen sites span the three subdomains as illustrated in Figure 1 (top), which depicts the cytoplasmic domain oriented with the membrane distal tip on the right, the methylation subdomain in red, the flexible subdomain in green, and the signaling subdomain in blue. Previous studies by Falke and coworkers 21-24 have shown for most of these sites that Cys mutant proteins retain at least partial activity and have reactivities indicating burial or solvent exposure. For each subdomain, Cys sites were chosen that were solvent-exposed in the dimer structure (1qu7), or buried either at the dimer interface or within the monomer, as listed in Table 1. Sites are numbered according to the sequence of the intact Asp receptor (Tar) throughout the manuscript except in Figure 7E for the data from a Tsr CF construct. The S364C mutation site was included for comparison with the TMEA trimer crosslinking studies of Parkinson and coworkers.20,25 All mutations were introduced in the 4E construct of CF, containing Glu at all 4 methylation sites, to minimize complications from the oligomerization/aggregation tendencies of the 4Q construct:5 samples of 4E are likely to be the most homogeneous in solution. Results of assays of CheA and CheW binding and kinase activation for this mutant series are reported in Figure S1.
Table 1.
Locations of cysteine mutants in Tar CF constructs.
| Subdomain | |||
|---|---|---|---|
| Accessibility | Methylation | Flexible | Signaling |
| solvent-exposed | S487 | K357 | S364 |
| dimer interface | S272 | A436 | T373 |
| buried | V475 | V426 | A405, A377 |
Figure 7.
Suppression of CF heterodimer formation on vesicles. (A) Mixtures of CFs with Cys at different sites (green and red) are predicted to form less heterodimer crosslinks on vesicles than in solution, due to aligning of CF’s bound to vesicles. The truncated CF construct (trTarS272C or trTsrS274C) is combined with each of the 10 different full length TarCF constructs for crosslinking. (B) Homodimers and heterodimers are resolved on gels, in solution (−) and on vesicles (+). (C) Extent of total dimer formation with trTarS272C, expressed as the percent of CF and trCF in dimeric form, in solution and on vesicles (dark and light columns, respectively). (D) Percent heterodimer formation with trTarS272C in solution and on vesicles (dark and light columns, respectively). CFs were present at 15 μM in each pair (30 μM total), with a diamide concentration of 300 μM for 30 minutes at 25°C. (E) Ratio of heterodimer formation (on vesicles/in solution) for all sites. Intervening number of residues between sites (assuming helical hairpin structure with turn at TarE389=TsrE391) are listed (red) below each site. Averages and standard deviations computed from triplicate samples assayed in separate experiments.
Vesicle template assembly increases the rate and extent of crosslinking
Binding CF to vesicles increases its effective concentration by localizing it on the surface of a membrane and restricting its free rotation. This is expected to increase the rate and extent of crosslinking. Data presented below for CF crosslinking by diamide and by TMEA in the presence and absence of vesicles confirm this prediction.
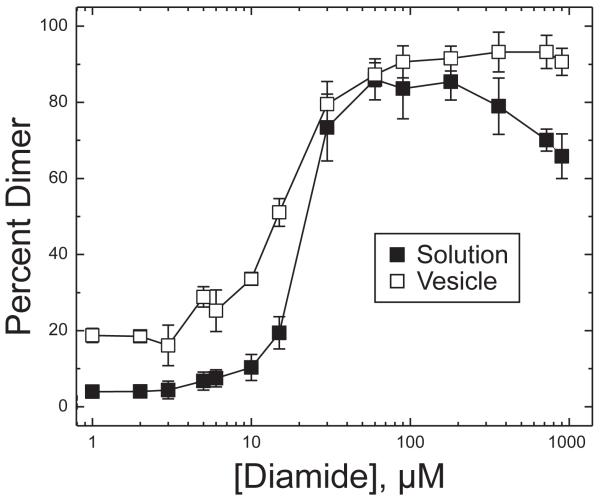
Since the choice of conditions is important in the design of crosslinking experiments with different goals, we first determined the concentration dependence for each crosslinking agent. Crosslinking using a range of concentrations of the oxidant diamide was performed for 30 min at 25 °C and the percent dimer was quantified by SDS-PAGE measurement of the intensity of the dimer band as a percentage of the total (monomer + dimer) CF bands. The results are shown in Figure 2 for 30 μM TarCF(S272C) in solution (black squares) and bound to vesicles (white squares). This plot shows that crosslinking is incomplete under stoichiometric diamide concentration (15 μM), and enables the identification of excess diamide conditions (180 μM) that are sufficient to drive crosslinking to completion (≈90% dimer) both in solution and on vesicles.
Figure 2.
Dimer formation as a function of diamide concentration. Percent dimer formation of 30 μM TarCF(S272C) in reactions at different concentrations of diamide for 30 minutes at 25 °C, in solution (■) or bound to vesicles (□). Vesicles were present at 580 μM total lipid of a 1:1 DOPC:DGS-NTA-Ni2+ composition. Averages and standard deviations computed from triplicate
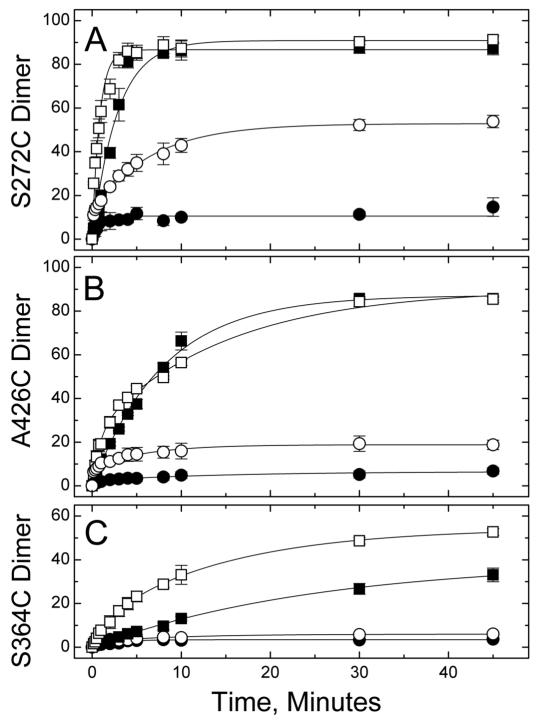
The time course of dimer formation was measured for three representative Cys sites, from each of the three CF subdomains. Each CF was reacted with either stoichiometric or excess diamide, both in solution and on vesicles, and the resulting percent dimer at each time point is plotted in Figure 3. In all cases a higher extent of crosslinking is observed with excess diamide. Differences in the rates and extent of crosslinking with excess diamide reveal reactivity differences between these three sites. The crosslinking reaction with TarCFS272C (a dimer interface site in the methylation subdomain) is the fastest, reaching its final extent by 10 min under all tested conditions. For this most reactive site, excess diamide drives the reaction to near completion, forming ≈ 90% dimer CF in the presence or absence of vesicles. The next most reactive site, V426C (a buried site in the flexible subdomain), requires 30 min to reach its similar final extent of ≈ 90% dimer. The least reactive site measured, S364C (a solvent-exposed site in the signaling subdomain), takes 50 min to reach approximately 50 and 40% crosslinking in the presence and absence of vesicles, respectively. The time course of crosslinked dimer formation was well fit by monophasic (no vesicles) and biphasic (with vesicles) curves, with the amplitude and time constant (τ) values listed in Table 2. The reason for the biphasic kinetics is not clear: perhaps CF bound to vesicles has enhanced dimerization or stability that alters the crosslinking rates of a fraction of the population. The resulting time constants also reflect the reactivity differences described above: maximal values of τ = 392 s (highest reactivity S272C site) < τ = 880 s (intermediate reactivity V426C site) < τ = 1500 s (lowest reactivity S364C site).
Figure 3.
Disulfide formation kinetics with diamide. Disulfide formation of 30 μM TarCFS272C (A), TarCFV426C (B), or TarCFS364C (C) was measured with or without vesicles (open or filled symbols, respectively), using either 15 μM (○,●) or 180 μM (□,■) diamide. Averages and standard deviations computed from triplicate samples assayed in separate experiments. Lines represent best fits, with parameters reported in Table 2.
Table 2.
Kinetics of diamide-induced disulfide formation.
| Site | Diamide (μ M) | +/− Vesicles | Phase | Amplitude (percent) | τ (seconds) | Adj. R-square |
|---|---|---|---|---|---|---|
| 272 | 180 | + | fast | 25.5 ± 3.8 | 8.0 ± 2.6 | 0.995 |
| slow | 64.0 ± 3.6 | 92.7 ± 9.3 | ||||
| 272 | 180 | - | 90.9 ± 3.6 | 168 ± 20 | 0.970 | |
| 272 | 15 | + | fast | 6.4 ± 2.1 | 12.4 ± 0.8 | 0.995 |
| slow | 40.4 ± 1.1 | 392 ± 26 | ||||
| 272 | 15 | - | 10.5 ± 0.6 | 47 ± 12 | 0.821 | |
|
| ||||||
| 426 | 180 | + | fast | 24.3 ± 2.9 | 53.1 ± 9.6 | 0.996 |
| slow | 65.8 ± 2.3 | 880 ± 130 | ||||
| 426 | 180 | - | 87.2 ± 1.5 | 481 ± 20 | 0.996 | |
| 426 | 15 | + | fast | 8.07 ± 0.55 | 8.4 ± 2.2 | 0.984 |
| slow | 10.76 ± 0.65 | 355 ± 52 | ||||
| 426 | 15 | - | 5.46 ± 0.45 | 208 ± 49 | 0.838 | |
|
| ||||||
| 364 | 180 | + | fast | 10.6 ± 2.9 | 119 ± 30 | 0.999 |
| slow | 43.6 ± 2.1 | 860 ± 120 | ||||
| 364 | 180 | - | 39.2 ± 2.5 | 1500 ± 180 | 0.989 | |
| 364 | 15 | + | fast | 2.34 ± 0.19 | 9.1 ± 2.9 | 0.976 |
| slow | 3.65 ± 0.25 | 560 ± 100 | ||||
| 364 | 15 | - | 3.41 ± 0.15 | 143 ± 21 | 0.945 | |
The presence of vesicles increased the extent of crosslinking at all three sites (and at additional sites, as shown in Figure 4). For the least reactive S364C site, small but measurable increases in the extent of crosslinking occurred in the presence of vesicles under both stoichiometric (3 to 6%) and excess (39 to 54%) diamide conditions. For the more reactive sites, vesicles cause no change in the extent of reaction under excess diamide conditions, because the reaction is already driven to completion. Instead, differences in the extent of crosslinking are observed under stoichiometric diamide conditions: vesicles increased crosslinking from 5 to 19% for V426C and 11 to 47% for S272C.
Figure 4.
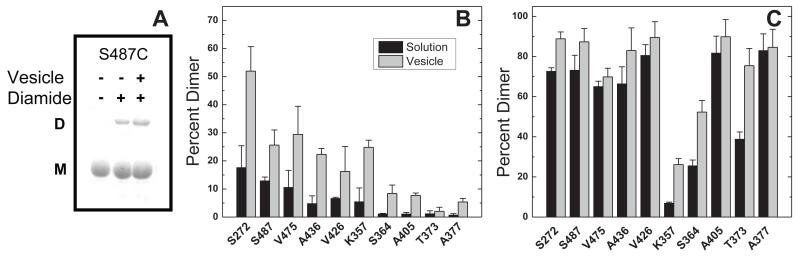
Reactivity of sites with diamide crosslinking. (A) Representative gel depicting crosslinking results for S487C control (pre-quenched with NEM) and for samples treated with diamide without or with vesicles. The extent of dimer formation, under conditions of (B) stoichiometric diamide or (C) excess diamide, after a 30-minute reaction at 25 °C in the absence (black) and presence (gray) of vesicles. Averages and standard deviations were computed from triplicate samples assayed in separate experiments. [CF] = 30 μM, [diamide] = 15 μM (B) or 180 μM (C), [lipid]total = 580 μM (gray columns), 1:1 DOPC:DGS-NTA-Ni2+.
The presence of vesicles also increased the rate of the crosslinking reaction for most reactions. Two cases with little change in extent of crosslinking, S272C and S364C sites under excess diamide conditions, show a clear increase in crosslinking rate with vesicles in Figure 3 (top and bottom plots). This is shown quantitatively by the decrease in time constants (Table 2), from 168 s to 93 and 8 s for the S272C site and from 1500 s to 860 and 119 s for the S364C site. In contrast, the V426C site does not show a significant change in rate or extent of crosslinking under excess diamide conditions: both reactions yield approximately 90% dimer with a time constant of 481 s in the absence of vesicles and an amplitude-weighted average time constant of 656 s in the presence of vesicles. Perhaps the CF-CF encounter rate, which is expected to increase upon binding to vesicles, is not the rate-determining step for V426C under excess diamide conditions.
The reactions with stoichiometric diamide show a low extent of dimer formation in the absence of vesicles (3-11%) and a significant increase (2 to 5-fold) in the presence of vesicles. Under these diamide-limited conditions, the extent of dimer formation may be low because crosslinking at the remaining sites is either too slow to compete with some reaction that is consuming the diamide, or slower than the timescale of the experiment (τ >> 3000 s). Addition of vesicles increases this slower rate to achieve a higher level of dimer formation. The speed of both the fast and slow phases increases in the presence of vesicles: time constants of the fast phase decrease from 47 to 12 s, 208 to 8 s, and 143 to 9 s, and time constants of the slow phase decrease from τ >> 3000 s to 392, 365, and 560 s for the S272C, V426C, and S364C sites, respectively.
These data demonstrate that vesicle association increases the rate and extent of CF crosslinking by diamide. This increase in the extent of crosslinking persists for nearly all of the other Cys sites examined (Figure 4). Results with a trifunctional crosslinker TMEA (Figures 5-6) further demonstrate that vesicle template assembly increases the yield of crosslinked oligomers.
Figure 5.
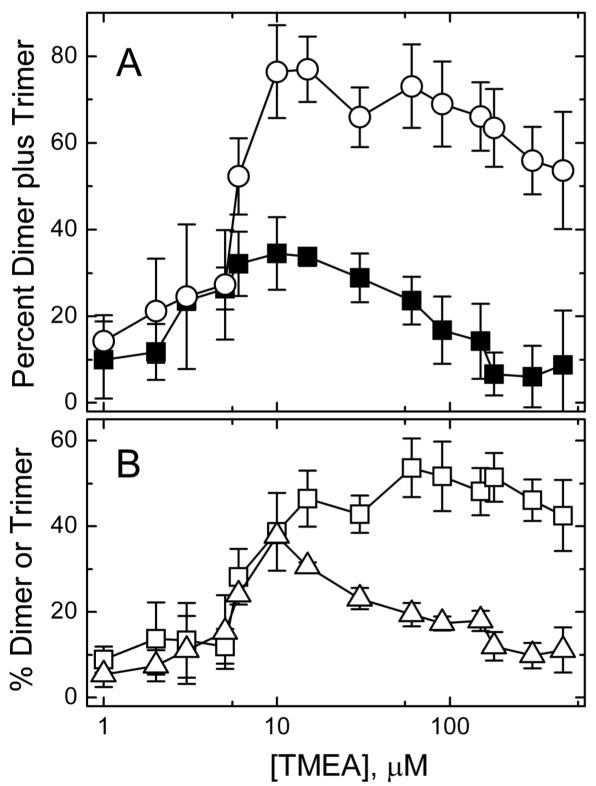
TMEA crosslinking kinetics. Percent crosslinked species formed in reactions of 30 μM TarCF(S364C) as a function of the TMEA concentration (30 min, 25 °C), either in solution or in the presence of vesicles (580 μM total lipid, 1:1 DOPC:DGS-NTA-Ni2+). A) The percent dimer plus trimer formed with vesicles present (◯), and the percent dimer formed in solution (■, only dimers were observed). B) The percent dimer (□) and trimer (△) generated with vesicles. Averages and standard deviations computed from triplicate samples assayed in separate experiments.
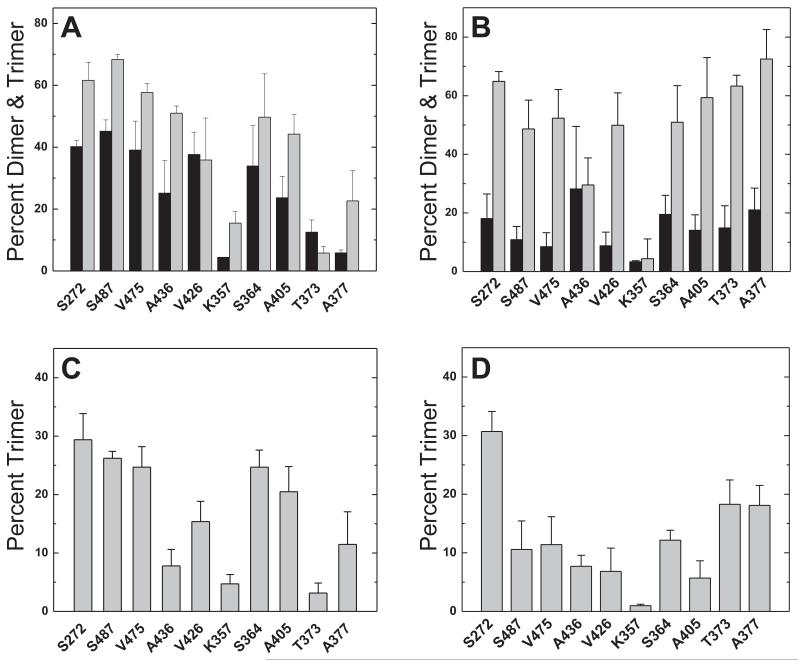
Figure 6.
Reactivity of sites with TMEA crosslinking. Crosslinking reactions conducted with stoichiometric TMEA (10 μM, A and C) or excess TMEA (90μM, B and D). Total percent dimer plus trimer (A and B) produced in reactions, in the presence (light bars) or absence (dark bars) of vesicles. The extent of trimer formation under conditions of (C) stoichiometric or (D) excess TMEA, after a 30-minute reaction at 25 °C with vesicles present. Data are averages and standard deviations from triplicate samples assayed in separate experiments. [CF] = 30 μM, [lipid]total = 580 μM, 1:1 DOPC:DGS-NTA-Ni2+.
Diamide crosslinking reactivity differences probe stability of receptor monomers
Using the set of 10 Cys mutations as probes across the CF, crosslinking reactions can measure reactivity differences among sites. Figure 4 shows the crosslinked dimer yields for 30 minute reactions at 25°C and demonstrates very different reactivity patterns for reaction conditions with stoichiometric (Figure 4B) vs. excess diamide (Figure 4C). Excess diamide conditions drive most of the crosslinking reactions to completion (Figure 4C), which obscures the reactivity differences among the sites. In both cases, a qualitatively similar pattern is observed with or without vesicles (gray and black bars, respectively), suggesting that the reactivity differences are due to intrinsic properties of CF monomers (CF in solution has been shown to be monomeric under similar conditions).17,28
Vesicles enhance crosslinking at nearly all sites under stoichiometric diamide conditions (Figure 4B). This enhancement is not visible under excess diamide conditions (Figure 4C) at sites driven to complete crosslinking. However, sites with low crosslinking under these conditions (357, 364, 373) also show increased crosslinking in the presence of vesicles.
Stoichiometric diamide conditions are best for revealing reactivity differences among the 10 Cys sites. Figure 4B shows that site reactivity decreases progressively from the methylation subdomain to the signaling subdomain (left to right), in both the presence and absence of vesicles. This decrease in reactivity mirrors the decreasing temperature factors observed for the CF crystal structure, as depicted by the rainbow colors on the lower structure of Fig 1 (blue for lowest temperature factors). Thus the progressive decrease in crosslinking reactivity across the structure may be due to a progressive increase in structural stability of the CF toward the signaling domain, which is similar in solution and when bound to vesicles.
Although the experiments were not designed to discriminate between crosslinking within dimers vs. crosslinking between dimers, the reactivity differences shown in Figure 4 suggest that the vesicle conditions may favor crosslinking within dimers. In the methylation subdomain, under stoichiometric diamide conditions (Figure 4B), crosslinking yields with vesicles are higher for the dimer interface site (272) than for the solvent-exposed site (487). Under these stoichiometric conditions, crosslinking yields on vesicles are uniformly intermediate for all three sites within the flexible bundle subdomain and uniformly low for all four sites within the signaling subdomain. However, comparison of yields on vesicles under excess diamide conditions (Figure 4C) reveals that the sites with lowest reactivity are the solvent-exposed sites of the flexible bundle and signaling subdomains (357 and 364), again suggesting that conditions favor diamide crosslinking within rather than between dimers.
TMEA crosslinking reactivity differences confirm the stability gradient
Parkinson, Studdert and coworkers have reported interesting in vivo TMEA crosslinking results that are consistent with the proposed CF organization into trimers of dimers.20,25,29 Using in vitro conditions with vesicle template assembly offers a complementary look at this crosslinking reaction. Initial experiments with this crosslinking agent examined the concentration dependence of dimer and trimer formation to choose optimal concentrations for further experiments. As demonstrated in Figure 5A, the maximal oligomer formation (dimers plus trimers) occurs at stoichiometric TMEA (10 μM) in either the presence (white circles) or absence (black squares) of vesicles. This is due to the covalent attachment of TMEA to CF: excess TMEA increases the yield of TMEA-CF at the expense of dimers (TMEA-CF2) and trimers (TMEA-CF3). Figure 5B shows that trimer formation in particular declines under excess TMEA conditions, which suggests it may be important to use stoichiometric TMEA concentrations for studies probing trimer organization.
As observed for diamide, crosslinking yields with TMEA are enhanced by CF binding to vesicles. The major difference upon addition of vesicles to the crosslinking reactions is the appearance of a trimer band on gels (Figure S2). Crosslinked trimers are produced only in the presence of vesicles, which is typical of all sites under both stoichiometric and excess TMEA conditions. The total crosslinking yield, the sum of the dimer plus trimer fractions, is shown in Figure 6A for stoichiometric (10 μM) TMEA conditions and Figure 6B for excess (90 μM) TMEA conditions. The total crosslinking yield in the absence of vesicles (black bars) is higher under stoichiometric TMEA conditions (Figure 6A), consistent with the data in Figure 5A. Under both conditions, total crosslinking yield is increased by the addition of vesicles (gray bars).
Comparison of crosslinking experiments under stoichiometric vs. excess TMEA reveals interesting differences. As for diamide crosslinking, a reactivity gradient with total crosslinking decreasing from the methylation to the signaling subdomain (left to right) is observed with stoichiometric TMEA, with or without vesicles (Figure 6A), with minor exceptions (K357 and T373 sites). Again this reactivity gradient is not observed under excess TMEA conditions (Figure 6B). Trimer yields for all sites in the presence of vesicles are shown in Figure 6C for reactions with stoichiometric TMEA and Figure 6D for reactions with excess TMEA. Stoichiometric TMEA conditions produce a maximum in trimer formation at sites in the methylation subdomain and at S364C. The latter is consistent with current views of the organization of CF oligomers: Studdert and Parkinson noted that CF with Cys at the analogous site in Tsr have the correct geometry and spacing in the 1qu7 trimer of dimers crystal structure to form TMEA trimers, and experimentally demonstrated trimer formation with in vivo TMEA crosslinking of intact Ser receptors with Cys at this site.20 The high trimer yields we observe at sites in the methylation region may be related to the higher reactivity of these sites as shown for diamide crosslinking in Figure 4B. The trimer formation pattern observed under excess TMEA conditions appears less consistent with current models of the organization of CF oligomers: trimer yields are relatively low at the 364 site and are higher at sites closer to the receptor tip (373 and 377) that have poor geometry for trimer crosslinking (373 is at the dimer interface and 373 is buried). Thus crosslinking under stoichiometric TMEA conditions again reveals a gradient in structural stability (Figure 6A), maximizes trimer formation (Figure 5B), and produces trimers in a pattern more consistent with the current model for organization of CF oligomers (Figure 6C).
Disulfide bond formation is more specific on vesicles
Template assembly is expected to increase the specificity of crosslinking, since binding of CF to vesicles will orient the long axis of the CF and promote parallel, in-register protein-protein interactions, as shown in Figure 7A. This hypothesis was tested by comparing formation of heterodimers between CFs with different cysteine mutations in solution vs. on vesicles. Diamide crosslinking reactions were performed with a truncated CF construct (C-terminal truncation) of one Cys mutant combined with full-length constructs of each of the 10 Cys mutants. Homodimers and heterodimers were resolved on gels, as shown in Figure 7B for the reaction between truncated constructs of the Asp and Ser receptor CF with Cys at equivalent sites, trTarS272C or trTsrA274C, and the full length Asp receptor constructs with Cys at two other sites, TarA377C or TarT373C. We use the term heterodimer to refer to dimers with crosslinks between truncated CF and full length CF, whether the dimer is Tar/Tar or Tar/Tsr. Gels resolving the full set of heterodimer crosslinking reactions for trTarS272C are shown in Figure S3. Crosslinking reaction conditions with excess diamide (300 μM) were chosen to drive crosslinking to completion at all sites, as shown in Figure 7C for the reactions with trTarS272C (results for trTsrA274C are shown in Figure S4). This should reduce the influence of intrinsic reactivity differences between the sites, so that the results instead reflect the partitioning of the same total amount of dimers between homodimers and heterodimers. As shown qualitatively by the gels (Figures 7B and S3) and quantitatively in Figure 7D, the presence of vesicles suppresses heterodimer crosslinking reactions. Formation of heterodimers of the same Cys site, trTarS272C with S272C, occurs with approximately 50% yield, as expected if the truncation does not influence CF association (random association predicts 50% trCF/CF, 25% trCF/trCF, and 25% CF/CF). This reaction is not suppressed by vesicles, as represented by a ratio of heterodimers on vesicles/heterodimers in solution of approximately one (Fig 7E). For all of the other sites, the presence of vesicles significantly suppresses heterodimer formation, with heterodimer ratios ≤ 0.6. This pattern of heterodimer suppression occurs for reactions of the series of Tar CFs with either trTarS272C or trTsrA274C.
This clear heterodimer suppression by vesicle assembly occurs for crosslinking between sites distant from each other in the CF structure. The distance between sites can be estimated based on the residue separation within the structure (in red under Cys sites in Figure 7E). Residue separation between sites on different branches of the CF hairpin structure is determined by using the hairpin alignment7 to find the equidistant site on the same branch, and then computing the residue separation to that site. Heterodimer suppression is excellent for the S272C site because all of the other sites are over 18 residues (× 1.5 Å/residue in helical structures = 28 Å) away from this site. Heterodimer suppression was also tested for two additional sites, S364C and V426C. Results for these sites are shown in Figure S5 and suggest that significant heterodimer suppression by vesicles (ratio ≤ 0.6) occurs for sites that are over 12 residues (18 Å) away. An exception to this is the lack of vesicle suppression of heterodimer formation between trV426C and the distant T373C. Another anomaly is the unexplained absence of heterodimer formation in solution between trS364C and the 272, 487, and 475 sites. Results shown in Figure S5 for both 426 (A, C, E) and 364 (B, D, F) may be complicated by some discrimination against trCF/CF interactions, since heterodimers between equivalent Cys sites, trCF426/CF426 and trCF364/CF364, only form to 28-38% rather than to the expected 50%. In addition, the S364C and V426C sites are intrinsically less reactive, which may indicate that structural fluctuations are required for crosslinking at these sites. Furthermore, these sites are distant from the membrane anchor, which would allow for structural fluctuations. Such fluctuations may reduce or eliminate the preference on vesicles for the crosslink between equivalent Cys sites.
Discussion
We have explored the effects of membrane association on chemical crosslinking reactions that are often used to probe protein structure and dynamics. Crosslinking reactions were performed on a soluble fragment corresponding to the cytoplasmic domain of a membrane protein, the chemoreceptor CF, with an N-terminal His tag that mediates binding to vesicles containing the nickel-chelating lipid DGS-NTA. Using this template directed assembly system,3-5 crosslinking reactions conducted in the presence and absence of vesicles differ only in whether CF is bound to the vesicles or is free in solution. Membrane association should increase the effective concentration of CF by (i) localizing the CF on the surface of the membrane, and (ii) restricting free rotation of the CF. As anticipated, this leads to an increase in the rate and extent of crosslinking by diamide and TMEA. Furthermore, only in the presence of vesicles does the trifunctional crosslinker TMEA yield ‘higher order’ interactions (larger than dimer). This increased crosslinking is an experimental demonstration that membrane binding enhances protein-protein interactions,1 an important consideration for understanding processes involving membrane-associated proteins.
This study also demonstrates some benefits of protein assembly on membranes as a tool for crosslinking studies. More native-like interactions are captured by crosslinking reactions of CF assembled on membranes. Chemotaxis receptors are found in vivo in hexagonal arrays in which the receptors are thought to adopt a trimer of dimers organization. In vivo crosslinking of the intact chemoreceptor Tsr Cys 366 (equivalent to Tar Cys 364) with TMEA has been shown to capture receptor trimers.20 The results reported here show that binding of CF to membranes makes it possible for in vitro TMEA crosslinking to capture trimers. Moreover, membrane binding orients the long axis of the CF, which should increase the specificity of CF-CF interactions. We show that this leads to suppression of crosslink formation between Cys sites distant in the CF structure in the presence of membranes. Binding of the CF to the membrane presumably favors the in-register alignment needed for the homodimer crosslink and disfavors the large displacements needed for crosslinks between sites over 18 Å apart along the length of the CF structure. The proportion of these heterodimers decreases on vesicles, relative to solution, suggesting an increase in more native protein-protein interactions.
The vesicle-bound CF system probably retains dynamics that make crosslinking between sites distant in the native structure possible. Membrane binding does not disfavor out-of register alignment needed for heterodimer crosslinking between nonequivalent sites such as TrTarV426C / TarT373C, requiring a 21 Å displacement, and other sites requiring 7-13 Å displacements. Furthermore, TMEA trimer crosslinking occurs at sites far from the receptor tip, near locations where trimer crosslinking is disfavored in vivo.29 It is possible that formation of membrane-bound complexes4 of CF with CheA and CheW would decrease dynamics and further reduce crosslinking between Cys sites that are distant in the native structure.
The crosslinking results also reflect the challenge of using crosslinking as a probe of protein structure, as many of the observed reactivity differences are not fully consistent with simple patterns predicted by current models for the CF structure. For example, the location of sites within the native dimer structure of the CF predicts greater diamide crosslinking at dimer interface sites (272, 436, 373) than at the buried and solvent-exposed sites. This is only clearly observed in the methylation region, which suggests that the crosslinking differences are influenced by other factors. For example, we have suggested that a gradient of dynamics causes the strong reactivity gradient observed across the length of the CF. Similarly, the TMEA crosslinking reactivities also do not track the expectation for trapping trimers within the trimer of dimers structure, through reactions at the solvent-exposed sites: although the highest trimer formation occurs at two solvent-exposed sites (364 and 487), the other solvent-exposed site (357) has very low trimer yield, and other sites (buried 405, buried 475, and dimer interface 272) have unexpectedly high yields. Performing TMEA crosslinking on a more native-like sample containing CheA and CheW and exhibiting high kinase activity resolved only some of these inconsistencies with the structural model (AKM and RW, unpublished observations). These results emphasize that structural interpretations of crosslinking must be done carefully, since many factors contribute to reactivity differences.
Crosslinking studies of proteins assembled on membrane surfaces are complementary to in vivo crosslinking studies. The template assembly system provides much greater control over variables such as the concentration of the crosslinking agent. We have demonstrated how crosslinking with defined concentrations (stoichiometric vs. excess) can provide key insights into the system. Excess concentrations of TMEA reduce the yield of crosslinked trimers, and obscure the reactivity differences between sites for either TMEA or diamide crosslinking. On the other hand excess concentrations of diamide can be used to drive maximal dimer formation at all sites, regardless of reactivity differences, in order to measure the partitioning between homodimer and heterodimer formation to probe crosslinking specificity. It is more difficult to control the crosslinker concentration for in vivo crosslinking studies, since the cellular concentrations of both the protein and crosslinking agent are not precisely known. If excess TMEA was used in attempts to crosslink receptors in native membranes in vitro, this could account for the lack of trimer formation.20 Template assembly of CF on vesicles restores the ability to form TMEA-linked trimers, albeit the system appears less native than in vivo because TMEA crosslinking occurs at a larger range of Cys sites. Another advantage of using template assembly for crosslinking studies is the ablity to compare crosslinking in solution vs. on membranes. The similarity of the reactivity gradient observed under the two conditions suggests that it arises from a property of the CF monomer, and does not depend on additional structure or organization that occurs on membranes, such as the tethering of one end of the CF molecule to the membrane. The in vivo crosslinking experiment benefits from the presence of native receptor arrays. With further optimization of conditions, crosslinking with template directed assembly has the potential to bring the advantages of in vitro crosslinking (full control of reaction variables) to functional complexes of chemoreceptors and other membrane-bound complexes.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Cedric Bobst and Nguyen Son for assistance with mass spectroscopy to confirm protein modification (TMEA reaction only with Cys), and Sandy Parkinson, Michael Harris, and Libbie Haglin for helpful discussions and comments on the manuscript.
This research was supported by National Institutes of Health grant R01-GM085288.
ABBREVIATIONS
- CF
cytoplasmic fragment of E coli chemotaxis receptor
- DGS-NTA
1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid (Ni-chelating lipid)
- diamide
1,1′-Azobis(N,N-dimethylformamide)
- DMSO
dimethyl sulfoxide
- DOPC
1, 2-dioleoyl-sn-glycero-3-phosphocholine
- LB
Luria-Bertani broth
- LUV
large unilamellar vesicle
- MLV
multilamellar vesicle
- NEM
N-ethylmaleimide
- OD
optical density
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tar
Asp receptor
- TCEP
tris(2-carboxyethyl)phosphine
- TMEA
Tris(2-maleimidoethyl)amine
- tr
truncated
- Tsr
Ser receptor
Footnotes
Activity measurements on the mutant CF series (Figure S1), gels resolving crosslinking products (Figures S2-S3) and additional heterodimer crosslinking results (Figure S4-S5). This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- (1).Grasberger B, Minton AP, DeLisi C, Metzger H. Interaction between proteins localized in membranes. Proc. Natl. Acad. Sci. U.S.A. 1986;83:6258–6262. doi: 10.1073/pnas.83.17.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Kuriyan J, Eisenberg D. The origin of protein interactions and allostery in colocalization. Nature. 2007;450:983–990. doi: 10.1038/nature06524. [DOI] [PubMed] [Google Scholar]
- (3).Montefusco DJ, Asinas AE, Weis RM. Liposome-mediated assembly of receptor signaling complexes. Meth. Enzymol. 2007;423:267–298. doi: 10.1016/S0076-6879(07)23012-5. [DOI] [PubMed] [Google Scholar]
- (4).Shrout AL, Montefusco DJ, Weis RM. Template-Directed Assembly of Receptor Signaling Complexes. Biochemistry. 2003;42:13379–13385. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
- (5).Montefusco DJ, Shrout AL, Besschetnova TY, Weis RM. Formation and Activity of Template-Assembled Receptor Signaling Complexes. Langmuir. 2007;23:3280–3289. doi: 10.1021/la062717r. [DOI] [PubMed] [Google Scholar]
- (6).Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- (9).Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Park S-Y, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- (11).Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- (12).Asinas AE. Protein interactions and kinase regulation in template-assembled complexes of chemotaxis receptor fragments. In: Weis RM, editor. ProQuest Dissertations and Theses. University of Massachusetts Amherst; United States -- Massachusetts: 2007. [Google Scholar]
- (13).Careaga CL, Falke JJ. Structure and dynamics of Escherichia coli chemosensory receptors. Engineered sulfhydryl studies. Biophysical Journal. 1992;62:209–219. doi: 10.1016/S0006-3495(92)81806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Careaga CL, Falke JJ. Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. Journal of Molecular Biology. 1992;226:1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Careaga CL, Sutherland J, Sabeti J, Falke JJ. Large amplitude twisting motions of an interdomain hinge: a disulfide trapping study of the galactose-glucose binding protein. Biochemistry. 1995;34:3048–3055. doi: 10.1021/bi00009a036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Popa MO, Lerche H. Cu2+ (1,10 phenanthroline)3 is an open-channel blocker of the human skeletal muscle sodium channel. Br. J. Pharmacol. 2006;147:808–814. doi: 10.1038/sj.bjp.0706667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wu J, Long DG, Weis RM. Reversible dissociation and unfolding of the Escherichia coli aspartate receptor cytoplasmic fragment. Biochemistry. 1995;34:3056–3065. doi: 10.1021/bi00009a037. [DOI] [PubMed] [Google Scholar]
- (18).Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- (19).Kott L, Braswell EH, Shrout AL, Weis RM. Distributed subunit interactions in CheA contribute to dimer stability: a sedimentation equilibrium study. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2004;1696:131–140. doi: 10.1016/j.bbapap.2003.10.001. [DOI] [PubMed] [Google Scholar]
- (20).Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Danielson MA, Bass RB, Falke JJ. Cysteine and disulfide scanning reveals a regulatory alpha-helix in the cytoplasmic domain of the aspartate receptor. Journal of Biological Chemistry. 1997;272:32878–32888. doi: 10.1074/jbc.272.52.32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Bass RB, Falke JJ. Detection of a conserved alpha-helix in the kinase-docking region of the aspartate receptor by cysteine and disulfide scanning. J. Biol. Chem. 1998;273:25006–25014. doi: 10.1074/jbc.273.39.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bass RB, Coleman MD, Falke JJ. Signaling Domain of the Aspartate Receptor Is a Helical Hairpin with a Localized Kinase Docking Surface: Cysteine and Disulfide Scanning Studies. Biochemistry. 1999;38:9317–9327. doi: 10.1021/bi9908179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Winston SE, Mehan R, Falke JJ. Evidence that the Adaptation Region of the Aspartate Receptor Is a Dynamic Four-Helix Bundle: Cysteine and Disulfide Scanning Studies. Biochemistry. 2005;44:12655–12666. doi: 10.1021/bi0507884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Kim S-H, Wang W, Kim KK. Dynamic and clustering model of bacterial chemotaxis receptors: structural basis for signaling and high sensitivity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11611–11615. doi: 10.1073/pnas.132376499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Starrett DJ, Falke JJ. Adaptation Mechanism of the Aspartate Receptor: Electrostatics of the Adaptation Subdomain Play a Key Role in Modulating Kinase Activity †. Biochemistry. 2005;44:1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Long DG, Weis RM. Escherichia coli aspartate receptor. Oligomerization of the cytoplasmic fragment. Biophysical Journal. 1992;62:69–71. doi: 10.1016/S0006-3495(92)81782-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Massazza DA, Parkinson JS, Studdert CA. Cross-Linking Evidence for Motional Constraints within Chemoreceptor Trimers of Dimers. Biochemistry. 2011;50:820–827. doi: 10.1021/bi101483r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.