Abstract
Despite their predominance in the nervous system, the precise ways in which glial cells develop and contribute to overall neural function remain poorly defined in any organism. Investigations in simple model organisms have identified remarkable morphological, molecular, and functional similarities between invertebrate and vertebrate glial subtypes. Invertebrates like Drosophila and C. elegans offer an abundance of tools for in vivo genetic manipulation of single cells or whole populations of glia, ease of access to neural tissues throughout development, and the opportunity for forward genetic analysis of fundamental aspects of glial cell biology. These features suggest that invertebrate model systems have high potential for vastly improving the understanding of glial biology. This review highlights recent work in Drosophila and other invertebrates that reveal new insights into basic mechanisms involved in glial development.
Introduction
Glia have been historically regarded as simple neuronal support cells; however, increasing evidence emerges demonstrating that glial cells serve crucial roles in a variety of nervous system functions including: providing neurotrophic support [1], guiding neurite outgrowth [2,3], ensheathing axons [4], eliminating cellular debris [5], and facilitating synapse formation, maturation, and plasticity [6]. Despite their remarkable biology, the mechanisms by which glia develop remain largely undefined.
Glia are morphologically and functionally diverse, made up of distinct subtypes. Investigating developmental features of any of these glial cells in mammalian systems can prove challenging due to the difficulty of accessing animals in utero, where critical glial developmental milestones occur. Furthermore, there are limited numbers of molecular-genetic tools to precisely manipulate mammalian glial cells in vivo. Invertebrate model organisms provide simpler systems in which to explore the in vivo mechanisms governing glial cell development and function. The majority of studies probing glial development in invertebrates take advantage of the fruit fly, Drosophila melanogaster (Fig. 1). The Drosophila nervous system is sophisticated, yet relatively simple compared to mammals, and neuronal and glial lineages and subtypes are well defined. Multiple stages of the fly life cycle enable live-imaging throughout most of glial development, and Drosophila glial subtypes are quite similar to their mammalian counterparts. Although Drosophila is the most heavily studied invertebrate glial model, other systems include: C. elegans (worm), medicinal leeches, grasshoppers, moths, and mosquitos. This review highlights recent findings in glial cell development that have capitalized on the use of these invertebrate organisms. However we will not focus on C. elegans glia, as these cells have been the subject of excellent recent reviews [7,8].
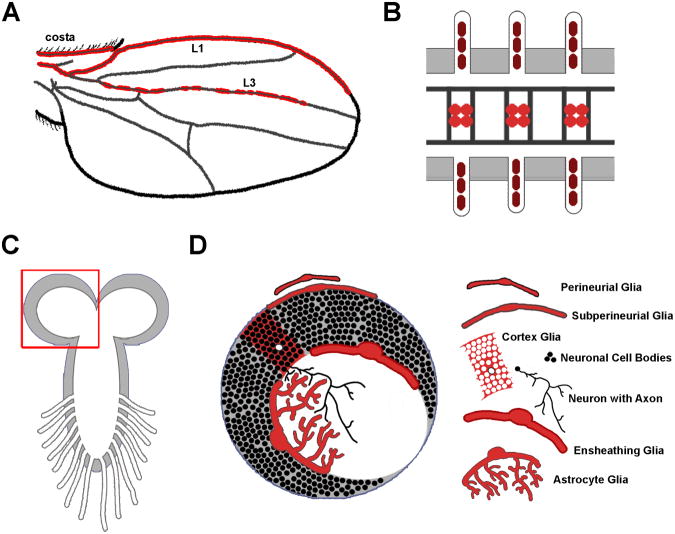
Figure 1. Variation in complexity of Drosophila glia.
A) The Drosophila wing blade provides a simple environment where approximately 130 glia migrate along three sensory nerves (costa, L1, and L3), then mature to wrap and insulate the nerves. B) Three segments of the Drosophila embryonic ventral nerve cord, depicting the ladder-like array of axons that are separated and insulated by midline glia, and peripheral glia that insulate the peripheral nerves. C-D) The Drosophila larval CNS provides even more complexity and glial diversity. (C) depicts the structure of the larval CNS and proximal portions of the nerves that stretch from the VNC to the larval body wall. (D) shows a close-up of the brain lobe outlined in (C), illustrating the diverse morphology of glial cells found within this sophisticated invertebrate model organism.
Glial Development in the Drosophila Embryo
One of the earliest steps in the specification of Drosophila glia involves the activation of the glial-specific gene, glial cells missing (Gcm), which subsequently regulates the expression of a number of Drosophila glial genes [9-11]. One major gene turned on by Gcm expression is reversed polarity (repo), which is considered a definitive marker of glial fate in Drosophila [12,13]. However, repo is not activated in all gcm-positive cells including macrophages [14] and visual system neurons [15]; therefore, additional pro-glial regulatory factors are needed for Gcm to specify a glial fate. Three cis-regulatory elements have been identified in repo: one for dorsolateral epidermis expression, an epidermal repressor, and a glial enhancer [14,16]. The glial enhancer has been further broken down to segments promoting expression only in very specific subsets of glia. The complexity of regulation of even the pan-glial gene repo emphasizes that glial fate in Drosophila is not a simple binary switch, but requires complex regulation whereby Gcm converges with other undefined factors to regulate target glial genes.
Gcm also regulates glial fate through histone aceytyl transferase activity, where gcm enforces low levels of histone 3 lysine 9 acetylation through the downregulation of Drosophila CREB-Binding Protein (dCBP) [17]. Overexpression of dCBP in repo-expressing cells leads to the reduction of glial genes, and eventually results in cell death; however, the dCBP family has been shown to function in more than just glial fate and survival. Nejire, a member of the dCBP family, was shown to regulate migration of embryonic subperineurial glia (SPG) and their distribution along the peripheral nerve [18]. Nejire may work with another HIF-related transcription factor, tango, since tango mutants show a similar defect in SPG migration [18].
Recent studies identified non-autonomous factors in glial migration that highlight the importance of neuron-glia interactions. The adhesion factor Fasciculin 2 (Fas2) on motor axons interacts with a glial Fas2 isoform to regulate glial cell movement. Fizzy-related/Cdh1 (Fzr/Cdh1) is a coactivator of anaphase-promoting complex/cyclosome (APC/C) that functions in peripheral neurons to control glial cell migration. Fzr/Cdh1 downregulates Fas2 in a graded manner along motor neuron axons, and inhibiting endocytosis of Fas2 in motor neurons blocks glial migration [19]; therefore, Fas2 acts as an adhesive break to regulate glial migration along the axonal tract.
Drosophila glia have also been shown to respond to canonical axon guidance cues as they migrate away from the midline. Netrin, a conserved axon guidance molecule, has two isoforms in Drosophila (NetA and NetB) that are expressed in the midline and induce repulsion through the UNC-5 receptor, and attraction through Frazzled (Fra), the Drosophila homolog of the UNC-40/DCC receptor. Two distinct types of embryonic glia rely on Netrin signaling for their migratory patterning [11]. Perineurial glia (PG) express UNC-5 for proper peripheral lateral migration, as a subset of these stall in UNC-5 mutants, and longitudinal glia (LG) located near the CNS midline express Fra to guide their migration medially from an initial lateral position where they are born [20]. Surprisingly, the migration of these glia is not guided by the expected CNS midline, but from a novel source of Netrin release, including neural/glial stem cells called neuroblasts (NBs). Precisely how NBs coordinate this migration is an interesting and open question.
Upon finding their axonal targets, some glia wrap axons. Embryonic CNS midline glia are mesectoderm-derived glial cells that not only guide axons, but also ensheath and maintain the structure of the CNS midline commissures. Through the interaction of glial-specific wrapper with neurexin IV on developing axons, midline glial cells wrap and separate axons that cross the midline, forming a ladder-like array of nerve bundles [4]. Canoe, a PDZ domain-containing protein that binds to wrapper via a DE-Cadherin protein, shotgun, has recently been shown to regulate neuron glia interactions at the midline [21]. Canoe-mutant midline glia show reduced adhesion to axons, resulting in poor migration, and failure to properly separate anterior and posterior axonal commissures. Time-lapse imaging of midline glia revealed that they migrate along three different stereotyped paths and have very different fates: anterior midline glia (AMG) migrate inward posteriorly and ensheath axons; a subset of AMG migrate inward anteriorly and undergo apotosis; and posterior midline glia (PMG) that migrate inward anteriorly and undergo apotosis [22]. The fates of these glial populations are determined through a combination of notch and hedgehog signaling [23]: In response to Notch signaling, AMG express the transcription factor, Runt, whereas PMG express Engrailed, a transcription factor of the hedgehog pathway; the two genes mutually repress each other to split AMG/PMG fates. Interestingly, loss of a notch co-factor, mastermind, can increase the number of midline glia, and consequently decrease midline glial diversity toward AMG fate [24]. However, unlike notch mutants, midline glia form normally in mastermind mutants, and subsequently accumulate increased number of midline glia as development proceeds into early larval development.
Drosophila Glial Development Round 2: Larval Stages
While a good deal of Drosophila nervous system development occurs during embryogenesis, the system is far from mature as it enters larval stages. In embryonic development, neuroblasts divide in a temporally- and molecularly-defined manner to give rise to ganglion mother cells, which divide into neurons [25]. At the end of embryonic development, neuroblasts enter a quiescent state, to be reactivated in larval development. Although it has been known that this reactivation was induced by a fat body derived mitogen (FBDM) [26], two studies recently found that this factor relays this information through neuroblast-associated glial cells (Fig. 2A) [27,28]. Glia cells nearby the quiescent neuroblasts receive FBDM, and in turn secrete Drosophila insulin-like peptide 6 (dILP6) to activate phosphatidylinositol-3 Kinase (PI3K) in neuroblasts, releasing them from quiescence in a nutritionally-dependent manner. Interestingly, glial loss of the Fragile X gene, Fmr1, also increased pAKT—the downstream target of PI3K—in neuroblasts [29]. Therefore, developmental misregulation of Fmr1 in glia could have a profound effect on larval CNS cell populations. Intriguingly, glia can regulate insulin-related growth in other ways. Glia release a secreted decoy of InR (SDR) into the hemolymph, where it mimics the insulin receptor (InR) extracellular domain, and interacts with several dILPs to inhibit signaling [30]. Smaller animals are observed if SDR is overexpressed, whereas SDR mutants exhibit larger bodies. Thus, Drosophila glial appear, surprisingly, to be capable of directly regulating animal body size.
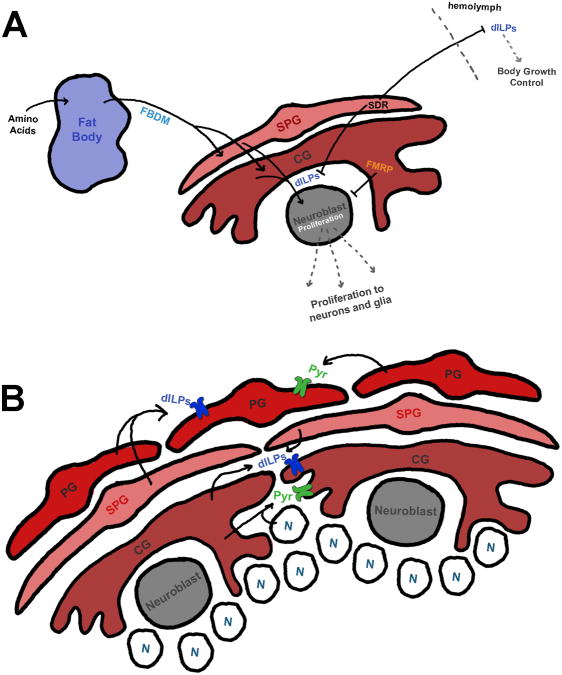
Figure 2. Glial morphogenesis, proliferation, and regulation of neuroblast reactivation through insulin and FGF signaling.
A) Regulation of neuroblast proliferation and animal growth. The fat body is activated by amino acid nutrient signaling, which then releases a fat body derived mitogen (FBDM) to glial cells, which in turn release dILP6, causing neuroblast reactivation and proliferation into neurons and glia. Surface glia also release a secreted decoy of insulin receptor (SDR) to block dILP activity in the brain where it can regulate neuroblast proliferation, or to the hemolymph to regulate animal body growth. FMRP signals non-autonomously in glia to inhibit neuroblast proliferation.
B) Glia respond to drosophila Insulin-Like Peptides (dILPs) through the insulin receptor (InR, purple) to regulate proliferation through PI3K/tor signaling in PGs, and Ras/MAPK signaling in CGs. Likewise, glia respond to pyramus (pyr) through the FGF receptor, Heartless (green) from nearby self-type specific glial cells to regulate morphology and proliferation. CGs, but not PGs, can respond to neuronally-derived pyr as well.
Though it is thought that many glia in the larval nervous system arise from neuroglioblasts (precursors to both neurons and glia) or their daughter GMCs, some glia can proliferate on their own to increase glial numbers. The miRNA, bantam, is required for maintaining the proliferative glial pool in the outer proliferative center (OPC) and glial precursor cell areas of the optic lobe [31]. In the absence of bantam, glial cells exhibit an abnormal distribution and pattern of proliferation, leading to small brains arising from reduced proliferation. At the same time, yorkie, a hippo pathway component, regulates glial proliferation in the nearby optic stalk in part through modulating bantam and myc signaling [32]. Finally, in the central brain lobes PG and cortex glia (CG) can expand their population through self-proliferation during larval stages (Fig. 2B) [33]. PG require parallel fibroblast growth factor (FGF) and Insulin signaling for proliferation and morphogenesis, whereas CG rely more strongly on FGF signaling via neuronally-derived pyramus activation of the FGF receptor, Heartless. InR overexpression in PG caused an increase in PG number via PI3K/TOR signaling; however, this effect was not seen in CG, where it is proposed that InR signals through the Ras/MAPK pathway. Interestingly, expression of dominant negative PI3K or the downstream target, TOR, caused decreased numbers of PG and CG. In line with these studies, a group has recently put forth a striking Drosophila model of glioma, where PI3K/EGFR co-activation in glial cells causes them to overproliferate, grow in size, take over surrounding brain tissue, and can generate tumors that can be transplanted into wild type hosts and then metastasize [34]. Finally, in addition to growth regulation, PI3K/EGFR co-activation can also induce polyploidism. Polyploidism has been found to be developmentally necessary in certain glia, such as SPG, where the level of polyploidy is directly correlated to growth and brain size [35]. For instance, the Drosphila blood-brain barrier (BBB) is comprised of a tight network of SPG that surround the surface of the nervous system, forming septate junctions between them. Inhibition of polyploidism using cdt1 RNAi in SPG cells results in a breakdown of the BBB.
Ensheathment of peripheral nerves is a critical glial function. SPG, PG, and wrapping glia (WG) extend down the peripheral nerves to the synaptic endings at the neuromuscular junction (NMJ), insulating the nerve similarly to mammalian Schwann cells. The developmental and structural maintenance of these insulating glia require two integrin complexes, αPS2βPS and αPS3βPS, that form adhesions with integrin-linked kinase (ILK) and Talin [36]. The loss of the βPS subunit results in a failure of the PG and WG to surround the nerve. However, if development proceeds normally, PG and SPG cells arrive at the NMJ as early as the 2nd instar larval stage. Interestingly, upon reaching the NMJ, glial growth appears to be regulated by NMJ expansion. While the relative size of the NMJ is quite variable, glial size and morphology was adjusted to match synaptic volume in all cases tested [37].
Glia in Drosophila Metamorphosis and Early Adult Development
During pupal development, the fly transforms from a worm-like larva to a fully-formed adult, and the nervous system is significantly rewired to accommodate the structural changes, such as the appearance of the adult wings. Glial cells in the wing migrate along the costal, L1, and L3 sensory nerves from 17-30 hours after pupal formation [38,39]. Four cells at the tip of the glial migratory chain act as pioneers to guide the migrating glia along the axon tracts to their final locations. The pioneer cells cannot move in isolation, and the ability of isolated communities to move directly correlates with cell number; however, smaller groups reestablish connections more easily with other isolated groups. This chain migration appears to be an excellent model for studying glial biology in a simple cellular environment.
Non-Drosophila Invertebrate Models to Study Glial Development
Glia are a poorly understood cell type in any model system, and while Drosophila has been the powerhouse in understanding the molecular basis of invertebrate glial development, a handful of recent studies have started to elucidate glial biology in other invertebrate systems. Like Drosophila, Repo has been shown to mark glial subtypes in Schistocerca gregaria (grasshopper) [40], Camponotus japonicus (carpenter ant) [41], and Aedes aegypti (yellow fever mosquito) [42]. Furthermore, the morphologies of many of these glial cells are highly conserved between these species. For example, while the central complex in the grasshopper develops embryonically, as opposed to adult stages in the fruit fly, both species contain astrocyte-like glia that invade the neuropil after neuronal and tracheal scaffolds have developed [40]. Likewise, an immunohistochemical investigation of the developing nervous system, using cross-reacting antibodies from other species, reveals a number of similarities and differences between the yellow fever mosquito and Drosophila [42]. Although the authors found subtle differences in the developmental timing of the optic lobes between the two species, overall they appear quite similar. The majority of neuronal proliferation occurs during larval stages, with vast neuronal reorganization and new synapse formation during metamorphosis. A pharmacological study in pupating Manduca sexta (moth) revealed that inhibition of the FGF receptor is required to maintain proper proliferation, migration, and survival of glia in the olfactory system [43], arguing that developmental mechanisms are well-conserved among invertebrate species.
The majority of studies in non-Drosophila invertebrate systems use histochemistry or pharmacology, rather than genetic manipulation, to understand invertebrate glial biology and nervous system development; however, genetic and protein manipulation is beginning to be used in non-genetic invertebrate systems. One species of medicinal leech (Hirudo verbana) expresses 15 innexins in different combinations within small subsets of segmental ganglia neurons and glia. Ectopic expression of innexins within individual cells resulted in rewiring and atypical coupling between cells that express the same innexin [44], suggesting a specified program of neuro-glial coordination during development.
Conclusions
Glial biology is still an emerging field, but determining the ways in which glia develop and interact with neurons is paramount in our goal of fully understanding nervous system function. In this brief review we have highlighted novel glial-neuroblast interactions that regulate neural proliferation and animal growth, as well as key factors required for glial cell specification, proliferation, migration, and morphogenesis. Whether these same pathways function in a similar way in mammals remains an open question. Despite the notable conservation observed between invertebrate and vertebrate neurons, and the fundamental insights provided by invertebrate model genetic organisms, the glial field has yet to fully embrace Drosophila, C. elegans, or other invertebrate models. However we anticipate this will change in the very near future. These systems allow for a depth of experimental and genetic scrutiny unmatched in higher organisms, and many glial subtypes (especially in Drosophila) are morphologically or functionally highly analogous to mammalian glia. Moreover, new technologies, such as ZFNs, TALENS and CRISPR [45], are emerging that allow for easier molecular-genetic manipulation even in model organisms that are not currently used for genetic studies. Such approaches should allow researchers to probe glial functions incisively in animals with highly accessible glial cell types—for example, the highly accessible neuropilar astrocytes of the medicinal leech (Hirudo medicinalis) that have cell bodies that are 100 μm across [46] and cellular processes that cover an area of 300-350 μm in diameter [47]. Beyond basic glial biology, a number of recent studies of invertebrate glial have provided novel insights into nervous system dysfunction and human diseases, including glioma [31,32,34,48], Fragile X Syndrome [29], and even Alzheimer's Disease [49]. These findings emphasize the importance of utilizing invertebrate systems to tackle the task of understanding glial biology, as well as their influence on and interaction with the surrounding nervous system.
Highlights.
Invertebrate and mammalian glia are highly homologous
In vivo glial mechanisms can be defined by powerful molecular-genetic approaches
Invertebrate studies identify key intrinsic and extrinsic factors in glial development
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buchanan RL, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10:839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- 2.Hidalgo A, Booth GE. Glia dictate pioneer axon trajectories in the Drosophila embryonic CNS. Development. 2000;127:393–402. doi: 10.1242/dev.127.2.393. [DOI] [PubMed] [Google Scholar]
- 3.Spindler SR, Ortiz I, Fung S, Takashima S, Hartenstein V. Drosophila cortex and neuropile glia influence secondary axon tract growth, pathfinding, and fasciculation in the developing larval brain. Dev Biol. 2009;334:355–368. doi: 10.1016/j.ydbio.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stork T, Thomas S, Rodrigues F, Silies M, Naffin E, Wenderdel S, Klambt C. Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development. 2009;136:1251–1261. doi: 10.1242/dev.032847. [DOI] [PubMed] [Google Scholar]
- 5.Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen NJ, Barres BA. Signaling between glia and neurons: focus on synaptic plasticity. Curr Opin Neurobiol. 2005;15:542–548. doi: 10.1016/j.conb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Oikonomou G, Shaham S. The glia of Caenorhabditis elegans. Glia. 2011;59:1253–1263. doi: 10.1002/glia.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Procko C, Shaham S. Assisted morphogenesis: glial control of dendrite shapes. Curr Opin Cell Biol. 2010;22:560–565. doi: 10.1016/j.ceb.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosoya T, Takizawa K, Nitta K, Hotta Y. glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell. 1995;82:1025–1036. doi: 10.1016/0092-8674(95)90281-3. [DOI] [PubMed] [Google Scholar]
- 10.Jones BW, Fetter RD, Tear G, Goodman CS. glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell. 1995;82:1013–1023. doi: 10.1016/0092-8674(95)90280-5. [DOI] [PubMed] [Google Scholar]
- 11.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–580. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 12.Xiong W, Okano H, Patel N, Blendy J, Montell C. repo encodes a glial-specific homeo domain protein required in the Drosophila nervous system. Genes Dev. 1994;8 doi: 10.1101/gad.8.8.981. [DOI] [PubMed] [Google Scholar]
- 13.Akiyama-Oda Y, Hosoya T, Hotta Y. Alteration of cell fate by ectopic expression of Drosophila glial cells missing in non-neural cells. Dev Genes Evol. 1998;208:578–585. doi: 10.1007/s004270050217. [DOI] [PubMed] [Google Scholar]
- 14.Lee BP, Jones BW. Transcriptional regulation of the Drosophila glial gene repo. Mech Dev. 2005;122:849–862. doi: 10.1016/j.mod.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Chotard C, Leung W, Salecker I. glial cells missing and gcm2 cell autonomously regulate both glial and neuronal development in the visual system of Drosophila. Neuron. 2005;48:237–251. doi: 10.1016/j.neuron.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 16.Johnson RW, Wood JL, Jones BW. Characterization of cis-regulatory elements controlling repo transcription in Drosophila melanogaster. Gene. 2012;492:167–176. doi: 10.1016/j.gene.2011.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Flici H, Erkosar B, Komonyi O, Karatas OF, Laneve P, Giangrande A. Gcm/Glide-dependent conversion into glia depends on neural stem cell age, but not on division, triggering a chromatin signature that is conserved in vertebrate glia. Development. 2011;138:4167–4178. doi: 10.1242/dev.070391. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt I, Franzdottir SR, Edenfeld G, Rodrigues F, Zierau A, Klambt C. Transcriptional regulation of peripheral glial cell differentiation in the embryonic nervous system of Drosophila. Glia. 2011;59:1264–1272. doi: 10.1002/glia.21123. [DOI] [PubMed] [Google Scholar]
- 19.Silies M, Klambt C. APC/C(Fzr/Cdh1)-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nat Neurosci. 2010;13:1357–1364. doi: 10.1038/nn.2656. [DOI] [PubMed] [Google Scholar]
- 20.von Hilchen CM, Hein I, Technau GM, Altenhein B. Netrins guide migration of distinct glial cells in the Drosophila embryo. Development. 2010;137:1251–1262. doi: 10.1242/dev.042853. [DOI] [PubMed] [Google Scholar]
- 21.Slovakova J, Carmena A. Canoe functions at the CNS midline glia in a complex with Shotgun and Wrapper-Nrx-IV during neuron-glia interactions. Development. 2011;138:1563–1571. doi: 10.1242/dev.056192. [DOI] [PubMed] [Google Scholar]
- 22.Wheeler SR, Pearson JC, Crews ST. Time-lapse imaging reveals stereotypical patterns of Drosophila midline glial migration. Dev Biol. 2012;361:232–244. doi: 10.1016/j.ydbio.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson JD, Wheeler SR, Stagg SB, Crews ST. Drosophila hedgehog signaling and engrailed-runt mutual repression direct midline glia to alternative ensheathing and non-ensheathing fates. Development. 2011;138:1285–1295. doi: 10.1242/dev.056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Wheatley R, Fulkerson E, Tapp A, Estes PA. Mastermind mutations generate a unique constellation of midline cells within the Drosophila CNS. PLoS One. 2011;6:e26197. doi: 10.1371/journal.pone.0026197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 26.Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- 27.Chell JM, Brand AH. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **28.Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. This study providesin vivoevidence that neuroblast reactivation after quiescence is regulated by glial insulin secretion stimulated by fat body-derived signal in response to amino acids. This work indicates that both nutritional intake, as well as glial niches, are required to maintain proper neural stem cell activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callan MA, Clements N, Ahrendt N, Zarnescu DC. Fragile X Protein is required for inhibition of insulin signaling and regulates glial-dependent neuroblast reactivation in the developing brain. Brain Res. 2012;1462:151–161. doi: 10.1016/j.brainres.2012.03.042. [DOI] [PubMed] [Google Scholar]
- *30.Okamoto N, Nakamori R, Murai T, Yamauchi Y, Masuda A, Nishimura T. A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev. 2013;27:87–97. doi: 10.1101/gad.204479.112. This work illustrates a new role for glia in long-distance signaling to hemolymph outside of the nervous system, whereby glia secret SDR to inhibit insulin signaling, and ultimately regulate growth of the entire animal in a nutritionally-independent manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Padgett RW. bantam is required for optic lobe development and glial cell proliferation. PLoS One. 2012;7:e32910. doi: 10.1371/journal.pone.0032910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy BV, Irvine KD. Regulation of Drosophila glial cell proliferation by Merlin-Hippo signaling. Development. 2011;138:5201–5212. doi: 10.1242/dev.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Avet-Rochex A, Kaul AK, Gatt AP, McNeill H, Bateman JM. Concerted control of gliogenesis by InR/TOR and FGF signalling in the Drosophila post-embryonic brain. Development. 2012;139:2763–2772. doi: 10.1242/dev.074179. The authors demonstrate that glial subtypes of the drosophila brain are capable of symmetric division and extensive proliferation throughout larval development; furthermore, this proliferation and glial morphogenesis are regulated through InR/PI3K/TOR signaling as well as FGF signaling with subtype-secific mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5:e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *35.Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26:31–36. doi: 10.1101/gad.177436.111. Usingin vivoanalysis ofDrosophilaSPG cells, the authors demonstrated for the first time that polyploidy can directly contribute to organ size and growth. Inhibition of polyploidy in SPG results in poor glial development with smaller cells, disruption of septate junctions, and therefore breakdown of the BBB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Xie X, Auld VJ. Integrins are necessary for the development and maintenance of the glial layers in the Drosophila peripheral nerve. Development. 2011;138:3813–3822. doi: 10.1242/dev.064816. The authors demonstrate the conservation of integrin expression between mammalian andDrosophilaglial cells that wrap peripheral nerves, and further reveal functional conservation as genetic loss of integrins result in the inability to wrap peripheral nerves, akin to myelination defects in mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brink DL, Gilbert M, Xie X, Petley-Ragan L, Auld VJ. Glial processes at the Drosophila larval neuromuscular junction match synaptic growth. PLoS One. 2012;7:e37876. doi: 10.1371/journal.pone.0037876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giangrande A, Murray MA, Palka J. Development and organization of glial cells in the peripheral nervous system of Drosophila melanogaster. Development. 1993;117:895–904. doi: 10.1242/dev.117.3.895. [DOI] [PubMed] [Google Scholar]
- 39.Berzsenyi S, Kumar A, Giangrande A. Homeostatic interactions at the front of migration control the integrity and the efficiency of a migratory glial chain. J Neurosci. 2011;31:13722–13727. doi: 10.1523/JNEUROSCI.2473-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyan G, Loser M, Williams L, Liu Y. Astrocyte-like glia associated with the embryonic development of the central complex in the grasshopper Schistocerca gregaria. Dev Genes Evol. 2011;221:141–155. doi: 10.1007/s00427-011-0366-4. [DOI] [PubMed] [Google Scholar]
- 41.Nasu N, Hara K. Gliogenesis in the mushroom body of the carpenter ant, Camponotus japonicus. Zoolog Sci. 2012;29:800–806. doi: 10.2108/zsj.29.800. [DOI] [PubMed] [Google Scholar]
- 42.Mysore K, Flister S, Muller P, Rodrigues V, Reichert H. Brain development in the yellow fever mosquito Aedes aegypti: a comparative immunocytochemical analysis using cross-reacting antibodies from Drosophila melanogaster. Dev Genes Evol. 2011;221:281–296. doi: 10.1007/s00427-011-0376-2. [DOI] [PubMed] [Google Scholar]
- 43.Gibson NJ, Tolbert LP, Oland LA. Activation of glial FGFRs is essential in glial migration, proliferation, and survival and in glia-neuron signaling during olfactory system development. PLoS One. 2012;7:e33828. doi: 10.1371/journal.pone.0033828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Firme CP, 3rd, Natan RG, Yazdani N, Macagno ER, Baker MW. Ectopic expression of select innexins in individual central neurons couples them to pre-existing neuronal or glial networks that express the same innexin. J Neurosci. 2012;32:14265–14270. doi: 10.1523/JNEUROSCI.2693-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013 doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lohr C, Deitmer JW. Intracellular Ca2+ release mediated by metabotropic glutamate receptor activation in the leech giant glial cell. J Exp Biol. 1997;200:2565–2573. doi: 10.1242/jeb.200.19.2565. [DOI] [PubMed] [Google Scholar]
- 47.Kuffler SW, Potter DD. Glia in the leech central nervous system: physiological properties and neuron-glia relationship. J Neurophysiol. 1964;27:290–320. doi: 10.1152/jn.1964.27.2.290. [DOI] [PubMed] [Google Scholar]
- **48.Read RD, Fenton TR, Gomez GG, Wykosky J, Vandenberg SR, Babic I, Iwanami A, Yang H, Cavenee WK, Mischel PS, et al. A Kinome-Wide RNAi Screen in Drosophila Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma. PLoS Genet. 2013;9:e1003253. doi: 10.1371/journal.pgen.1003253. Using the power of in vivo reverse genetic screens possible in Drosophila, Read et al. utilized an kinome-wide RNAi screen to discover a number of kinases capable of modulating the severity of glioma. The authors demonstrate expression and functional conservation with two of these kinases in both mouse models of glioma as well as human glioma tissue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolkan BJ, Triphan T, Kretzschmar D. beta-secretase cleavage of the fly amyloid precursor protein is required for glial survival. J Neurosci. 2012;32:16181–16192. doi: 10.1523/JNEUROSCI.0228-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]