Abstract
DNA exposures to electrophilic methylating agents that are commonly used during chemotherapeutic treatments cause diverse chemical modifications of nucleobases, with reaction at N7-dG being the most abundant. Although this base modification frequently results in destabilization of the glycosyl bond and spontaneous depurination, the adduct can react with hydroxide ion to yield a stable, ring-opened MeFapy-dG and this lesion has been reported to persist in animal tissues. Results from prior in vitro replication bypass investigations of the MeFapy-dG adduct had revealed complex spectra of replication errors that differed depending on the identity of DNA polymerase and the local sequence context. In this study, a series of nine site-specifically modified MeFapy-dG-containing oligodeoxynucleotides were engineered into a shuttle vector and subjected to replication in primate cells. In all nine sequence contexts examined, MeFapy-dG was shown to be associated with a strong mutator phenotype, predominantly causing base substitutions, with G to T transversions being most common. Single and dinucleotide deletions were also found in a subset of the sequence contexts. Interestingly, single-nucleotide deletions occurred not only at the adducted site, but also one nucleotide downstream of the adduct. Standard models for primer-template misalignment could account for some, but not all mutations observed. These data demonstrate that in addition to mutagenesis predicted from replication of DNAs containing O6-Me-dG and O4-Me-dT, the MeFapy-dG adduct likely contributes to mutagenic events following chemotherapeutic treatments.
Introduction
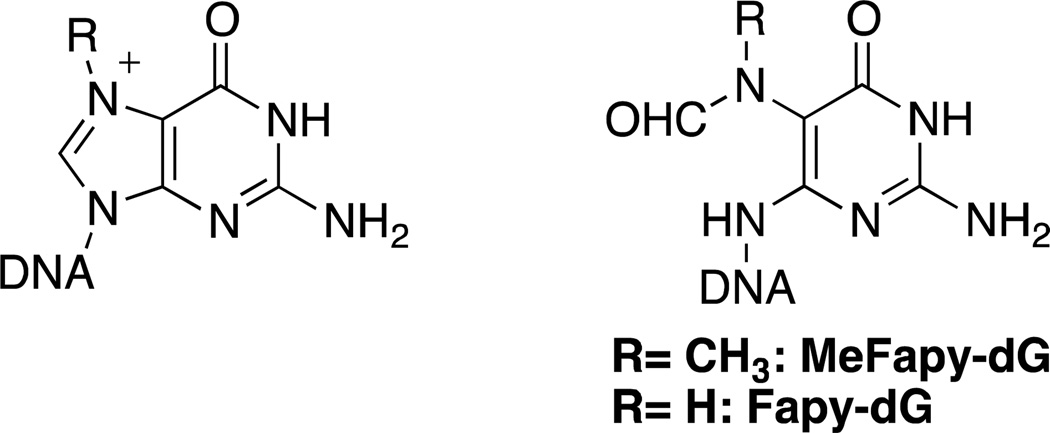
Reaction of electrophilic methylating agents with DNA results in modification of various O-and N-atoms of the bases as well as phosphate O-atoms of the DNA backbone [reviewed1,2], with the most abundant lesion being at N7-dG (Figure 1). Human exposures to methylating agents arise not only from treatments with chemotherapeutic drugs such as temozolomide, dacarbazine and procarbazine, but also environmental toxicants, and food sources. Even though the major cytotoxic and mutagenic adducts are presumed to be O6-Me-dG and O4-Me-dT, the cationic N7-Me-dG lesion can react with hydroxide ion, yielding a stable ring-opened N6-(deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine (MeFapy-dG) (Figure 1).3,4 This lesion has been identified as a stable adduct in rats treated with N,N-dimethylnitrosamine, N-methyl-N-nitrosourea or 1,2-dimethylhydrazine.5,6 The persistence of MeFapy-dG suggests that if the fidelity of translesion DNA synthesis (TLS) past this adduct is low, it could contribute to mutagenesis and potentially carcinogenesis.
Figure 1.
Structures of N7-dG and Fapy-dG adducts.
We have developed chemistry for site-specific incorporation of MeFapy-dG in synthetic oligodeoxynucleotides and in order to evaluate the miscoding potential of this lesion, performed in vitro replication bypass assays using the Escherichia coli exonuclease-deficient Klenow fragment of polymerase (pol) I and Sulfolobus solfataricus P2 polymerase, Dpo4.7,8 Since these data demonstrated that MeFapy-dG was strongly blocking and when bypassed was miscoding, the ability of a panel of human and yeast polymerases to catalyze TLS past this adduct was examined.9 The efficiencies of nucleotide incorporation opposite the MeFapy-dG adduct were measured in two sequence contexts, 5'-TXT-3' and 5'-TXG-3'. Pols α, β, δ, ζ, ι, and ν demonstrated very poor insertion abilities opposite the lesion for all nucleotides and except for pol ζ that was proficient in extension from dC, further polymerization was either not observed or very inefficient. In contrast, Rev 1 was both efficient and accurate at the nucleotide insertion step, while human pols η and κ were moderately efficient in the insertion and extension steps, but error-prone. Relative to control dG, pol η and κ inserted dC opposite MeFapy-dG with ~ 7-fold reduced efficiency and demonstrated ~10-fold lower fidelity of the nucleotide insertion. Mass spectrometric analyses of extension products revealed that in the 5'-TXT-3' sequence context, pols η and κ yielded 71 and 78% error-free DNAs, respectively. In the 5'-TXG-3' context, error-free rates were higher (81 and 92%, respectively). Pols η and κ misincorporated dA opposite the MeFapy-dG in the 5'-TXT-3' sequence context, predicting a G to T transversion at the lesion site if not corrected by one of the DNA proof-reading systems. Single-nucleotide deletions were also observed that occurred one nucleotide downstream of the adduct following correct incorporation of dC opposite the lesion. Furthermore, pol η also misincorporated dT opposite the MeFapy-dG in this sequence context. No deletions were detected for either polymerase in the 5'-TXG-3' context; however, misinsertions of dT and dG were observed for pol η and misinsertions of dT for pol κ.
Based on the data of the in vitro analyses, we hypothesized that although intracellular bypass of MeFapy-dG could be relatively accurate, both base substitutions and deletions were likely to be induced, and that mutagenic outcome of TLS past this lesion could be influenced by the local sequence context. In order to establish the mutation frequencies and spectra for MeFapy-dG in different sequence contexts in mammalian cells, site-specifically modified oligodeoxynucleotides were synthesized, ligated in a single-stranded shuttle vector and mutagenesis was evaluated following replication through African green monkey kidney cells.
Materials and Methods
1. Oligodeoxynucleotide synthesis
Protocols for the synthesis of oligodeoxynucleotides containing a site-specific MeFapy-dG adduct have been reported previously.8 The stereochemistry of the adduct was a mixture of the α and β anomers and cis and trans isomers with respect to the formamide group. Based on prior analyses,10 it is predicted that the anomeric α:β ratios ranged from ~36:64 to 21:79. The sequences of the adducted 12-mer oligodeoxynucleotides were as follows: 5'-GCTAGCXAGTCC-3', 5'-GCTAGNXGGTCC-3', 5'-GCTAATXGGTCC-3', and 5'-GCTAGTXNGTCC-3', where X = dG or MeFapy-dG and N = dT, dA, dC, or dG.
2. Replication of site-specifically MeFapy-dG-modified DNAs in AGM kidney cells, and evaluation of the mutation spectra and frequencies
Experimental protocols to assess the mutagenic spectra and frequencies arising from replication of single-stranded pMS2 DNAs containing either control or MeFapy-dG-containing oligodeoxynucleotides were carried out as previously described.11 The shuttle vector, pMS2, was the gift of Dr. M. Moriya, SUNY, Stony Brook, NY. Briefly, a hairpin loop in a single-stranded circular pMS2 DNA was cut with EcoRV, followed by ligation of control or adducted 12-mer DNAs using a 56-mer scaffolding oligodeoxynucleotide. The flanking regions of scaffolding oligodeoxynucleotides hybridized to the ends of the linearized pMS2 molecules, while the internal 12-nucleotide sequence was complementary to the sequences under investigation. Following transfection of control or lesion-containing vectors into African green monkey kidney cells (COS-7), the progeny DNAs were isolated after 48 h, and E. coli DH5α cells were transformed and selected for ampicillin resistance. Individual colonies were grown in 96-well plates containing LB-ampicillin for 16 h at 37°C, and aliquots were transferred to Hybond N membrane (Amersham) and processed for DNA hybridization. 32P-labeled oligodeoxynucleotide probes were used to identify clones that contained the inserted 12-mer sequences and those that had no mutations. DNAs that hybridized to the insert probe but not to the error-free sequence probes were isolated and sequenced. The data were generated with one set of vectors. A minimum of three independent transfections of COS-7 were performed for each particular sequence.
Results
MeFapy-dG-induced mutagenesis in the 5'-CXA-3' sequence context
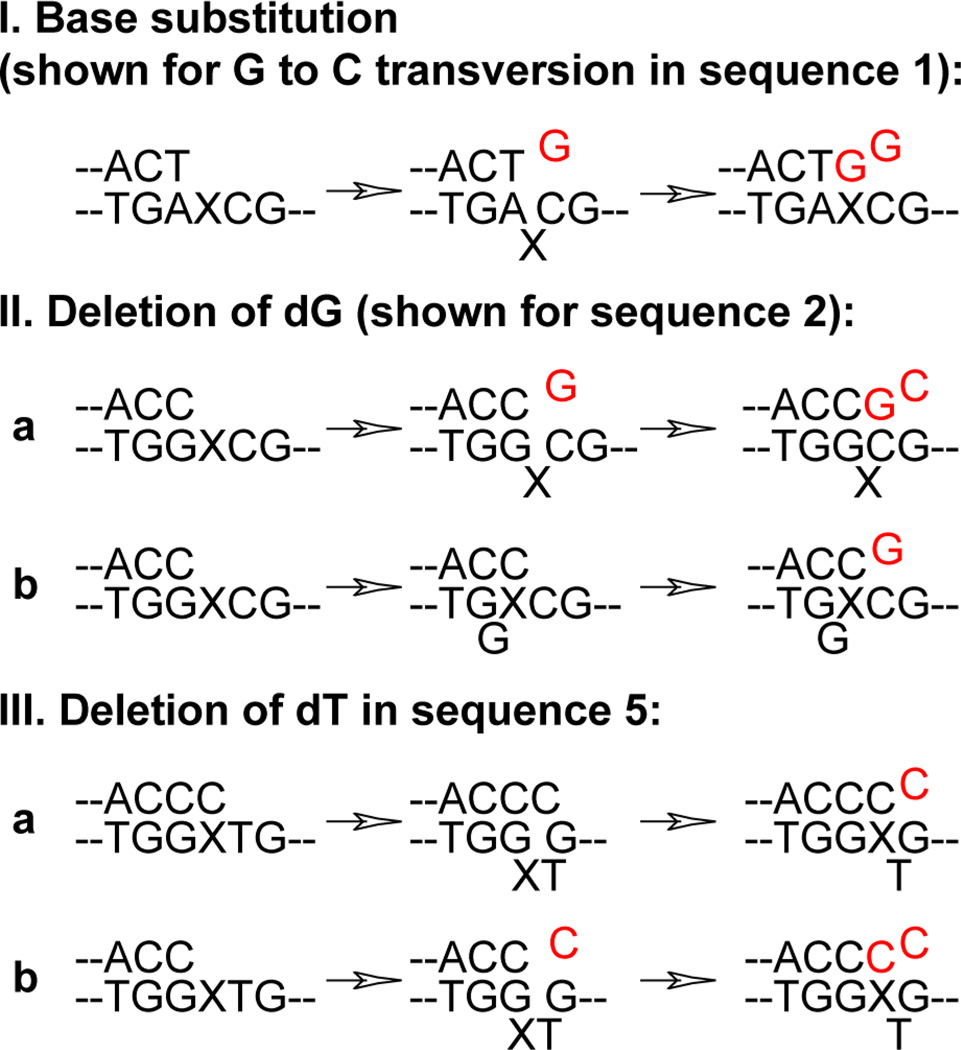
The initial set of experiments to assess the mutagenic properties of MeFapy-dG was conducted using the 5'-GCTAGCXAGTCC-3' sequence (sequence 1), where X was either a control dG or MeFapy-dG. Previously, this sequence was used to assess the mutagenic potential of the N2-dG adducts induced by various enal compounds, such as acrolein,11,12 crotonaldehyde,13 and 4-hydroxynonenal.14 Oligodeoxynucleotides were incorporated into single-stranded pMS2 vectors and following vector replication in COS-7 cells, the progeny DNAs were analyzed for mutations in the insert sequences. No mutations were found in 217 clones that originated from the non-damaged control vectors. In contrast, replication of lesion-containing DNAs resulted in base substitutions that were found at ~9% frequency with G to T (6.3%) and G to C (2.0%) transversions being most common (Table 1). This frequency of mutations was comparable or even higher than that observed for the enal-derived N2-dG adducts, although the spectra were similar. The occurrence of G to C mutations suggested that dC, the 5'-neighbor of MeFapy-dG, could have served as a template during both steps of the bypass reaction. In such a model, MeFapy-dG would be bypassed by a mechanism that involved the dNTP-stabilized misalignment15,16 followed by primer-template realignment (Figure 2, model I).
Table 1.
Base substitutions, deletions, and insertions at the sites of MeFapy-dG or the neighboring nucleotides.
| Sequence | Plasmid tested, number |
G to T, number (%) |
G to C, number (%) |
G to A, number (%) |
Deletion, number (%) |
Insertion, number (%) |
Total , % |
|
|---|---|---|---|---|---|---|---|---|
| 1 | GCTAGCXAGTCC | 205 | 13 (6.3) | 4 (2.0) | 1 (0.5) | - | - | 8.8 |
| 2 | GCTAGCXGGTCC | 172 | 144 (8.1) | 6 (3.5) | 6 (3.5) | 3 (1.7) | - | 16.8 |
| 3 | GCTAGAXGGTCC | 209 | 95 (4.3) | 1 (0.5) | 4 (1.9) | 1 (0.5) | - | 7.2 |
| 4 | GCTAGGXGGTCC | 194 | 10 (5.2) | 1 (0.5) | 4 (2.1) | 14 (7.2) | 2 (1.0) | 16.0 |
| 5 | GCTAGTXGGTCC | 231 | 12 (5.2) | 1 (0.4) | - | 14 (6.1) | - | 11.7 |
| 6 | GCTAATXGGTCC | 244 | 12 (4.9) | 1 (0.4) | 121 (5.3) | 6 (2.5) | - | 13.1 |
| 7 | GCTAGTXTGTCC | 238 | 23 (9.7) | 4 (1.7) | 3 (1.3) | 7 (2.9) | - | 15.6 |
| 8 | GCTAGTXAGTCC | 229 | 272 (11.8) | 2 (0.9) | 4 (1.7) | 2 (0.9) | - | 15.3 |
| 9 | GCTAGTXCGTCC | 215 | 293 (13.5) | 3 (1.4) | 12 (5.6) | 2 (0.9) | - | 21.4 |
2 out of 14 G to A transitions were accompanied by a G to C transversion at the 3'-neighboring site (TXG → TAC);
2 out of 27 G to T transversions were accompanied by a G to T transversion at the site two nucleotides downstream of the adduct (GTXA → TTTA);
1 out of 29 G to T transversions was accompanied by deletion at the site two nucleotides upstream of the adduct (TXCGT → TTC-T);
2 out of 14 G to T transversions were accompanied by an additional mutation: G to T transversion at the 3'-neighboring site (CXG → CTT) or a tandem base substitution downstream of the adduct (GCXG → AGTG);
1 out of 9 G to T transversions was accompanied by a G to T transversion at the 3'-neighboring site (AXG → ATT).
Figure 2.
Mutagenic bypass of MeFapy-dG: proposed primer-template misalignment models. With sequences 5–9, a subset of G to T transversions, the commonly observed base substitutions, could be generated by a primer-template misalignment-realignment (model I). Additional mechanism leading to G to T transversions could involve a non-instructive dA insertion opposite the lesion site and is not shown in this scheme. In the template slippage model (II-b), alternative slippage intermediates are possible.
MeFapy-dG-induced mutagenesis in the 5'-NXG-3' sequence context
The spectrum of MeFapy-dG-induced mutations in the above 5'-CXA-3' sequence context was different from those predicted from the data of the replication bypass assays with individual DNA polymerases.9 Thus, adducted oligodeoxynucleotides were redesigned to match the local sequence contexts of the templates that were used in the previous in vitro analyses, and additional sequences were investigated to assess the nearest neighbor effects on the mutagenic outcome of replication past MeFapy-dG. The first set of vectors contained the lesion in the 5'-NXG-3' sequences. Replication of DNAs carrying the lesion in the 5'-CXG-3' sequence gave a complex spectrum of mutations with an overall frequency of ~17% (Table 1, sequence 2). The mutations included G to T (8.1%), G to C (3.5%), and G to A (3.5%) base substitutions and single-nucleotide deletions of a dG (1.7%), (Table 2). The level of G to C transversions was relatively high, suggesting that as in sequence 1, the 5'-dC of the template could instruct incorporation of dG in a subset of encounters (Figure 2, model I). Deletions could also be generated via similar mechanism, except that after dG was incorporated via the dNTP-stabilized misalignment, the primer did not realign prior to extension (Figure 2, model II-a). Alternatively, deletions could be formed as result of the primer-template slippage, similar to that described by a standard Streisinger model17 (Figure 2, model II-b).
Table 2.
Types of deletions and insertions.
| Sequence | Number | Type | Sequence | |
|---|---|---|---|---|
| 1 | GCTAGCXAGTCC | - | - | |
| 2 | GCTAGCXGGTCC | 3 | deletion of dG | GCTAGCGGTCC |
| 3 | GCTAGAXGGTCC | 1 | single nucleotide deletion accompanied by a base substitution | GCTAGTGGTCC |
| 4 | GCTAGGXGGTCC | 14 | deletion of dG | GCTAGGGGTCC |
| 1 | insertion of dG | GCTAGGGGGGTCC | ||
| 1 | insertion of dT | GCTAGGTGGGTCC | ||
| 5 | GCTAGTXGGTCC | 12 | deletion of downstream dT | GCTAGGGGTCC |
| 1 | dinucleotide deletion | GCTAGGGTCC | ||
| 1 | deletion of dG | GCTAGTGGTCC | ||
| 6 | GCTAATXGGTCC | 5 | deletion of dG | GCTAATGGTCC |
| 1 | dG deletion accompanied by a G to T transversion | GCTAATTGTCC | ||
| 7 | GCTAGTXTGTCC | 5 | dinucleotide deletion | GCTAGTGTCC |
| 1 | deletion of downstream dT | GCTAGGTGTCC | ||
| 1 | dG deletion accompanied by a G to T transversion | GCTATTTGTCC | ||
| 8 | GCTAGTXAGTCC | 2 | deletion of dG | GCTAGTAGTCC |
| 9 | GCTAGTXCGTCC | 1 | deletion of dG | GCTAGTCGTCC |
| 1 | deletion of dG accompanied by a G to T transversion | GCTAGTCTTCC | ||
Relative to the 5'-CXG-3' context (sequence 2), the 5'-AXG-3' context (sequence 3) resulted in overall few mutations (~7%), but still favored G to T transversions (4.3%) with minimal numbers of G to A transitions (1.9%) and a single G to C transversion (Table 1). The only deletion found was a single-nucleotide deletion at either the 5'-dA or the lesion site which was accompanied by G to T transversion (Table 2).
When the mutagenic potential of MeFapy-dG was tested in the 5'-GXG-3' context (sequence 4), all types of base substitutions were observed with G to T transversions being most frequent (5.2%); the occurrence of G to A transitions was less common (2.1%), and only a single G to C transversion was detected (Table 1). Relative to sequences 2 and 3, replication in this context yielded the highest level of single-nucleotide deletions with an overall frequency of 7.2% (Table 1 and 2). In addition, insertions of either dG or dT were found, but each of these was represented by only a single clone. The precise site where deletions or insertion of dG occurred was not possible to determine because the greater sequence context changed from 5'-GGXGG-3' to 5'-GGGG-3' or 5'-GGGGGG-3', respectively; however, we rationalize that these frameshift mutations most likely were generated via a Streisinger mechanism.
The frequency of base substitutions in the 5'-TXG-3' context (sequence 5) was relatively low (Table 1). As with all other sequences, G to T transversions predominated (5.2%). Of the 231 colonies analyzed, only one G to C and no G to A mutations were observed. In addition, deletions were detected in this context at a 6.1% frequency, with the major type being an unusual single-nucleotide deletion that occurred immediately downstream of the MeFapy-dG adduct site (12 out of 14), (Table 2). We hypothesized that after dC incorporation opposite the lesion in this sequence, the 5'-dT had sufficient conformational freedom to rotate to an extrahelical position and in this conformation, the nucleotide was skipped by a DNA polymerase. Further, we hypothesized that formation of such a structure could be facilitated by a transient base-pairing between the 3'-terminal dC of the primer and the downstream dG (+2 site) (Figure 2, model III-a). Primer extension from this pair would result in dinucleotide deletion, and this mutation was observed in the 5'-TXG-3' sequence, but only as a rare event (Table 2). It seems that more often, the 3'-terminal dC would realign back opposite MeFapy-dG, while leaving the 5'-dT out of the helix. This primer-template conformation would be different from the initial structure that was formed after dC incorporation opposite the lesion, since the 5'-dT would now be moved out of the template site. A similar structure could also be generated following dC incorporation opposite the +2 dG from the −1 primer (Figure 2, model III-b). Interestingly, single-nucleotide deletions downstream of the adduct were commonly observed only when dT was present at that site in the 5'-NXG-3' sequences, probably because of all DNA bases, T has the lowest stacking ability.
In order to elucidate a possible role of the downstream dG in generating the unusual dT deletions, we modified sequence 5 (5'-GTXG-3') by placing dA instead of dG at the +2 site (5'-ATXG-3', sequence 6). The overall frequency of mutations was ~13% in this sequence, with G to A transitions and G to T transversions being the major types (5.3 and 4.9%, respectively), (Table 1). In addition, deletions of dG (Table 2) were observed at low frequency (2.5%), which are hypothesized to be generated via either the dNTP-stabilized misalignment (Figure 2, model II-a) or the template slippage (Figure 2, model II-b). Consistent with our prediction however, no deletions of the 5'-dT were found in the absence of the downstream dG.
MeFapy-dG-induced mutagenesis in the 5'-TXN-3' sequence context
The next set of experiments was designed to assess the effect of the 3' neighbor on the mutagenic properties of the MeFapy-dG adduct. The vectors were constructed to contain the lesion in either the 5'-TXT-3', 5'-TXA-3', or 5'-TXC-3' sequence context, and mutagenesis was evaluated as described above. For the 5'-TXT-3' context (sequence 7), the overall mutation frequency was determined to be ~16% with the spectra being dominated by G to T transversions (9.7%), (Table 1). G to C transversions and G to A transitions were observed at 1.7% and 1.3%, respectively. Single and dinucleotide deletions were also generated (2.9%) with dinucleotide deletions being more common (5 out of 7), (Table 2). The precise site of these deletions could not be determined, since the original sequence had three 5'-GT-3' repeats, with the middle repeat containing the adduct. We hypothesize that this repetitive sequence was prone to undergo a Streisinger-like primer-template isomerization, which explains the origin of dinucleotide deletions. With respect to the single-nucleotide deletions, one of these occurred immediately downstream of, but not including, the site of the MeFapy-dG adduct, while another occurred either at the adduct site or at the dG two nucleotides downstream. Since a G to T mutation accompanied the latter deletion, the exact sites of these mutations are uncertain.
The MeFapy-dG adduct caused considerable levels of deletions in the 5'-TXG-3' context (sequences 5 and 6), probably because in both cases the lesion was positioned in a 5'-G3-3' repeat. In contrast, deletions were rare when the context was either 5'-TXA-3' (sequence 8) or 5'-TXC-3' (sequence 9), (Tables 1 and 2). Specifically, single-nucleotide deletions were observed at the site of the lesion at ~0.9% frequency in each sequence context. The most common mutations were base substitutions; G to T transversions predominated and these were detected at ~12 and 14% frequencies in the 5'-TXA-3' and 5'-TXC-3' sequences, respectively.
Thus, the 5'-TXG-3' sequence was specifically prone to the dT deletions downstream of the MeFapy-dG adduct relative to any other 5'-TXN-3' sequence. We speculate that a number of factors may be responsible for this phenomenon, including strong stacking interactions between the MeFapy-dG and the upstream dG and a high melting energy barrier of the 3' neighboring pair (dG:dC). These factors could favor stabilization of the adducted base inside the helix. However, the presence of dG at the +2 site and the run of three dCs at the primer terminus that can be formed following dC incorporation opposite the lesion are probably the major structural elements involved in a primer-template isomerization that resulted in exclusion of the 5'-dT from the template, but not the MeFapy-dG.
Additional mutations
In addition to the previously described mutations, the majority of the sequences produced at least one progeny plasmid with a base substitution two nucleotides upstream or one to six nucleotides downstream of the MeFapy-dG adduct (Table 3). These sequence alterations occurred in 11 out of 1937 progeny plasmids examined and were formed more frequently downstream of the lesion site. In contrast, no mutations were found in 725 clones originated from non-damaged vectors. Thus, at least a subset of these additional mutations could be generated by a low-fidelity DNA polymerase recruited to bypass the adduct.
Table 3.
Additional mutations.
| Sequence | Mutation | |
|---|---|---|
| 1 | GCTAGCXAGTCC | - |
| 2 | GCTAGCXGGTCC | GGTAGCGGGTCC |
| 3 | GCTAGAXGGTCC | GCTATAGGGTCC, TCTAGAGGGTCC |
| 4 | GCTAGGXGGTCC | TATTGGGGGTCC |
| 5 | GCTAGTXGGTCC | - |
| 6 | GCTAATXGGTCC | GTTAATGGGTCC |
| 7 | GCTAGTXTGTCC | GCTATTGTGTCC |
| 8 | GCTAGTXAGTCC | GCTAATGAGTCC, GCTAGAGAGTCC, CCTAGTGAGTCC, GCTATTGAATCC |
| 9 | GCTAGTXCGTCC | GCTAGTGCTGTCC |
Discussion
In order to understand the long-term biological effect that may result following exposures to electrophilic alkylating agents, it is critical to identify those lesions that are both sufficiently long-lived to be still present during genome replication and have the capacity to be miscoding for either replicative or TLS DNA polymerases. For alkylating agents that modify both N-and O-atoms, it is well established that the relatively minor O6-Me-dG and O4-Me-dT adducts disproportionately contribute to mutagenesis and cytotoxicity.18–20 Further, N7-dG adducts are generally regarded as neither mutagenic nor cytotoxic, but can serve as a biomarker of exposure [reviewed21]. However, the cationic N7-Me-dG species can undergo a secondary chemical reactions involving C8-addition of hydroxide ion and ring-opening to form a stable MeFapy-dG adduct.
This study highlights the complex mutagenic spectra that can be generated by a DNA lesion in different sequence contexts. Previous investigations of the mutagenic properties of the unsubstituted (Figure 1, R=H) Fapy-dG adduct (also using the pMS2/COS-7 system) have been reported by Kalam et al.22 to show sequence context effects and mutation spectra that are similar to, but significantly less complex than those found in this current study. The mutagenesis of Fapy-dG was examined in two sequence contexts, 5'-TXT-3' and 5'-TXA-3', in which the predominant mutations were G to T transversions. Specifically, the 5'-TXT-3' context yielded 29.6% G to T transversions and 1.1% G to C transitions, while only G to T transversions were observed at a frequency of 8.2% in the 5'-TXA-3' context. Results from the current study using the 5'-TXT-3' context (sequence 7), but with the MeFapy-dG adduct revealed an overall lower frequency, but with greater complexity, including G to A transitions and dinucleotide deletions; for the 5'-TXA-3' context (sequence 8), both the absolute frequency and complexity of the mutation spectra were increased (Tables 1 and 2). However, the data obtained for Fapy-dG22 and MeFapy-dG (this report) should not be directly compared, since the larger sequence contexts of lesion-containing DNAs was different and as evident from the current study, this can influence the mutagenic outcome of replication bypass.
There are strong general trends that are evident throughout the mutagenesis data. First is that in all sequence contexts examined, the error-free bypass of the MeFapy-dG adduct predominated. Based on the data of the replication bypass assays in vitro,9 this could be achieved by a combined action of multiple DNA polymerases such that pol δ, Rev1, pol η, or pol κ will insert a dC opposite the lesion, while pol ζ, pol η, or pol κ will extend the primer beyond this site. Despite the existence of mechanisms for the error-free bypass of MeFapy-dG, replication of DNA containing this lesion resulted in mutation frequencies between 8–22%. These are comparable or higher than that observed for DNA adducts induced by polycyclic aromatic hydrocarbons,23–25 butadiene,26 bifunctional aldehydes,11–14 reactive oxygen species,22,27,28 ionizing radiation,29 and UV light.30,31 Since the majority of these agents are known mutagens and carcinogens, concluding the potential biological importance of the MeFapy-dG adducts is self-evident.
A second generalization is that G to T transversions occurred in every sequence context with mutational frequencies between 4.3–13.5%, while G to A transitions were measured in 8 out of 9 contexts at 0.5–5.6% frequencies. G to C transversions were also observed in all contexts, but with relatively low frequencies (0.4–3.5%). The levels of G to C transversions were slightly elevated in the 5'-CXN-3' sequences (sequence 1 and 2). The highest frequencies of G to T transversions were observed in three out of five 5'-TXN-3' sequences tested (sequence 7, 8, and 9). Thus, conventional models for the primer-template misalignment-realignment (Figure 2, model I) can explain a fraction of base substitutions.
Other possible mechanisms that explain the MeFapy-dG-induced base substitutions are less apparent. Hypothetically, the major mutation, the G to T transversion, could be formed because of default dA insertion by DNA polymerase(s) that follows the “A-rule”. If this was the case, DNA polymerases of the B family, such as pol α, pol δ, pol ε, and pol ζ, would be the primary suspects, since the members of the B family and not the Y family have been shown to preferentially incorporate dA opposite abasic sites in vitro.32 Furthermore, at least one polymerase of the B family was required to replicate past abasic sites in mammalian cells and this bypass generally proceeded via incorporation of dA opposite the lesion.33 The MeFapy-dG lesion exists partially in the unnatural α-anomeric configuration,10,34,35 and it is possible that this is recognized by the polymerases as an abasic site. Insertion of dA opposite the lesion site was found in bypass products generated by pol η or pol κ in the 5'-TXT-3' template, but not the 5'-TXG-3' template.9 The predominant replication errors made by these polymerases in vitro were single-nucleotide deletions at the 5'-dT in the 5'-TXT-3' context and misinsertions of dT in the 5'-TXG-3' context. Thus, pol η and pol κ are unlikely to be responsible for all base substitutions formed as result of intracellular bypass of MeFapy-dG based on the biochemical data.
The final general observation is that single and dinucleotide deletions were generated in the majority of the sequence contexts that were investigated. Although the frequencies and types of deletions differed depending on the sequence context, consistent patterns were observed. The highest levels of deletions were found in the repetitive sequences. These included sequence 4 (run of five dGs), 5 and 6 (run of three dG’s) that yielded single-nucleotide deletions (7.2, 5.6, and 2.5%, respectively) and sequence 7 (run of three GT repeats) that was specifically prone to dinucleotide deletions (2.1%). Single dG deletions were observed in 6 out of 9 sequence contexts. The mechanism(s) that generate deletions may conform to conventional models, such as template slippage17 or dNTP-stabilized misalignment,15,16 and the nature of the structures that stabilize the generation of such replication intermediates await structural analysis. Nevertheless, the fact that frameshift mutations are frequent products of replication bypass of Me-Fapy-dG will have considerable biological importance.
Current investigations are designed to investigate the structural basis of these mismatches and single and dinucleotide deletions using a combination of NMR and X-ray crystallographic techniques. In the case of the Fapy-dG adducts, it was hypothesized that differential stabilities in base stacking of the syn conformation of the lesion in the two sequence contexts might account for some of the mispairing properties, while the anti conformation yielded no differences in the various contexts. Our future studies on the MeFapy-dG adduct will likely generate additional new hypotheses based on structural data (versus molecular modeling) for the miscoding properties of this lesion in different sequence contexts. Additionally, it would be interesting to determine if Fapy-dG also produce deletions in repetitive sequences given its importance as a major lesion for oxidative damage.36,37
Acknowledgments
Funding. This work was supported by NIH Grants P01 ES05355 (C.J.R. & R.S.L.), P01 CA160032 (C.J.R. & R.S.L.), P30 ES00267, Center in Molecular Toxicology; P30 CA068485, Vanderbilt-Ingram Cancer Center.
Abbreviations
- (MeFapy-dG)
N6-(2-deoxy-D-erythro-pentofuranosyl)-2-6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine
- (pol)
polymerase
- TLS
translesion DNA synthesis.
Footnotes
The authors declare no competing financial interest.
References
- 1.Dahlmann HA, Vaidyanathan VG, Sturla SJ. Investigating the biochemical impact of DNA damage with structure-based probes: Abasic sites, photodimers, alkylation adducts, and oxidative lesions. Biochemistry. 2009;48:9347–9359. doi: 10.1021/bi901059k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrivastav N, Li D, Essigmann JM. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis. 2010;31:59–70. doi: 10.1093/carcin/bgp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem. Res. Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 4.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J. Biochem. Mol. Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 5.Beranek DT, Weis CC, Evans FE, Chetsanga CJ, Kadlubar FF. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem. Biophys. Res. Commun. 1983;110:625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- 6.Kadlubar FF, Beranek DT, Weis CC, Evans FE, Cox R, Irving CC. Characterization of the purine ring-opened 7-methylguanine and its persistence in rat bladder epithelial DNA after treatment with the carcinogen N-methylnitrosourea. Carcinogenesis. 1984;5:587–592. doi: 10.1093/carcin/5.5.587. [DOI] [PubMed] [Google Scholar]
- 7.Christov PP, Angel KC, Guengerich FP, Rizzo CJ. Replication past the N5-methyl-formamidopyrimidine lesion of deoxyguanosine by DNA polymerases and an improved procedure for sequence analysis of in vitro bypass products by mass spectrometry. Chem. Res. Toxicol. 2009;22:1086–1095. doi: 10.1021/tx900047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christov PP, Brown KL, Kozekov ID, Stone MP, Harris TM, Rizzo CJ. Site-specific synthesis and characterization of oligonucleotides containing an N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem. Res. Toxicol. 2008;21:2324–2333. doi: 10.1021/tx800352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christov PP, Yamanaka K, Choi JY, Takata K, Wood RD, Guengerich FP, Lloyd RS, Rizzo CJ. Replication of the 2,6-diamino-4-hydroxy-N5-(methyl)-formamidopyrimidine (MeFapy-dGuo) adduct by eukaryotic DNA polymerases. Chem. Res. Toxicol. 2012;25:1652–1661. doi: 10.1021/tx300113e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christov PP, Banerjee S, Stone MP, Rizzo CJ. Selective incision of the α-N5-methyl formamidopyrimidine anomer by Escherichia coli Endonuclease IV. J. Nucleic Acids. 2010 doi: 10.4061/2010/850234. Article ID 850234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. Error prone translesion synthesis past γ-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J. Biol. Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- 12.Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat. Res. 2008;637:161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes PH, Kanuri M, Nechev LV, Harris TM, Lloyd RS. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ. Mol. Mutagen. 2005;45:455–459. doi: 10.1002/em.20117. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes PH, Wang H, Rizzo CJ, Lloyd RS. Site-specific mutagenicity of stereochemically defined 1,N2-deoxyguanosine adducts of trans-4-hydroxynonenal in mammalian cells. Environ. Mol. Mutagen. 2003;42:68–74. doi: 10.1002/em.10174. [DOI] [PubMed] [Google Scholar]
- 15.Bloom LB, Chen X, Fygenson DK, Turner J, O'Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase III holoenzyme. The effects of β, γ complex processivity proteins and ε proofreading exonuclease on nucleotide misincorporation efficiencies. J. Biol. Chem. 1997;272:27919–27930. doi: 10.1074/jbc.272.44.27919. [DOI] [PubMed] [Google Scholar]
- 16.Efrati E, Tocco G, Eritja R, Wilson SH, Goodman MF. Abasic translesion synthesis by DNA polymerase β violates the "A-rule". Novel types of nucleotide incorporation by human DNA polymerase β at an abasic lesion in different sequence contexts. J. Biol. Chem. 1997;272:2559–2569. doi: 10.1074/jbc.272.4.2559. [DOI] [PubMed] [Google Scholar]
- 17.Streisinger G, Okada Y, Emrich J, Newton J, Tsugita A, Terzaghi E, Inouye M. Frameshift mutations and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- 18.Newbold RF, Warren W, Medcalf AS, Amos J. Mutagenicity of carcinogenic methylating agents is associated with a specific DNA modification. Nature. 1980;283:596–599. doi: 10.1038/283596a0. [DOI] [PubMed] [Google Scholar]
- 19.Mitra G, Pauly GT, Kumar R, Pei GK, Hughes SH, Moschel RC, Barbacid M. Molecular analysis of O6-substituted guanine-induced mutagenesis of ras as oncogenes. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8650–8654. doi: 10.1073/pnas.86.22.8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Basu AK, Essigmann JM. Site-specifically alkylated oligodeoxynucleotides: probes for mutagenesis, DNA repair and the structural effects of DNA damage. Mutat. Res. 1990;233:189–201. doi: 10.1016/0027-5107(90)90162-w. [DOI] [PubMed] [Google Scholar]
- 21.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. The formation and biological significance of N7-guanine adducts. Mutat. Res. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriya M, Spiegel S, Fernandes A, Amin S, Liu T, Geacintov N, Grollman AP. Fidelity of translesional synthesis past benzo[a]pyrene diol epoxide 2'-deoxyguanosine DNA adducts: marked effects of host cell, sequence context, and chirality. Biochemistry. 1996;35:16646–16651. doi: 10.1021/bi9608875. [DOI] [PubMed] [Google Scholar]
- 24.Khalili H, Zhang FJ, Harvey RG, Dipple A. Mutagenicity of benzo[a]pyrene-deoxyadenosine adducts in a sequence context derived from the p53 gene. Mutat. Res. 2000;465:39–44. doi: 10.1016/s1383-5718(99)00203-x. [DOI] [PubMed] [Google Scholar]
- 25.Dong H, Bonala RR, Suzuki N, Johnson F, Grollman AP, Shibutani S. Mutagenic potential of benzo[a]pyrene-derived DNA adducts positioned in codon 273 of the human P53 gene. Biochemistry. 2004;43:15922–15928. doi: 10.1021/bi0482194. [DOI] [PubMed] [Google Scholar]
- 26.Kanuri M, Nechev LV, Tamura PJ, Harris CM, Harris TM, Lloyd RS. Mutagenic spectrum of butadiene-derived-N1-deoxyinosine adducts and N6, N6-deoxyadenosine intrastrand cross-links in mammalian cells. Chem. Res. Toxicol. 2002;15:1572–1580. doi: 10.1021/tx025591g. [DOI] [PubMed] [Google Scholar]
- 27.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C-->T.A transversions in simian kidney cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon JH, Bhatia G, Prakash S, Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases κ and ζ in human cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14116–14121. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colis LC, Raychaudhury P, Basu AK. Mutational specificity of γ-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase η. Biochemistry. 2008;47:8070–8079. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon JH, Prakash L, Prakash S. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase ζ in mouse and human cells. Genes Dev. 2010;24:123–128. doi: 10.1101/gad.1872810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi JY, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Avkin S, Adar S, Blander G, Livneh Z. Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:3764–3769. doi: 10.1073/pnas.062038699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patro JN, Haraguchi K, Delaney MO, Greenberg MM. Probing the configurations of formamidopyrimidine lesions Fapy•dA and Fapy•dG in DNA using endonuclease IV. Biochemistry. 2004;43:13397–13403. doi: 10.1021/bi049035s. [DOI] [PubMed] [Google Scholar]
- 35.Lukin M, Minetti CA, Remeta DP, Attaluri S, Johnson F, Breslauer KJ, de Los Santos C. Novel post-synthetic generation, isomeric resolution, and characterization of Fapy-dG within oligodeoxynucleotides: Differential anomeric impacts on DNA duplex properties. Nucleic Acids Res. 2011;39:5776–5789. doi: 10.1093/nar/gkr082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: Mechanisms of formation, repair, and biological effects. Free Radic. Biol. Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Greenberg MM. The formamidopyrimidines: Purine lesions formed in competition with 8-oxopurines from oxidative stress. Acc. Chem. Res. 2012;45:588–597. doi: 10.1021/ar2002182. [DOI] [PMC free article] [PubMed] [Google Scholar]