Abstract
To search for an alternative to using protein conjugated aflatoxin as a coating antigen in aflatoxin detection by an ELISA method, a random-8-peptide library was constructed and used as a source of peptides that mimic aflatoxins (termed as mimotopes). Five mimotope peptides were obtained by panning-elution from the library and were successfully used in an indirect competitive ELISA for analyzing total aflatoxin concentration. The assay exhibited an IC50 value of 14 µg/kg in samples (with 1 in 7 dilution of sample extract) for aflatoxins. The linear range is 4–24 µg/kg. Further validation indicated relatively good recovery (60–120%) in peanut, rice and corn. Natural contaminated samples (peanut and feedstuff) were analyzed for aflatoxin concentration by both conventional ELISA and phage ELISA. The results showed good correlation. It can be concluded that the mimotope preparation is an effective substitute for the aflatoxin based coating antigen in ELISA and can be used in real sample analysis.
Keywords: Aflatoxin, phage-displayed peptide, phage peptide library, mimotope, matrix effect
INTRODUCTION
Aflatoxins are a group of extremely toxic metabolites produced mainly by Aspergillus species namely A. flavus, A. parasiticus and the rare A. nomius.1 More than 20 aflatoxins have been identified,2 among which aflatoxin B1, B2, G1, and G2 are most commonly found in nature.3 In addition, aflatoxin M1 is found in animal tissues and fluids (milk and urine) as a metabolic product of aflatoxin B1.4, 5 Aflatoxins are potent carcinogens, mutagens and teratogens, thus pose a significant threat to both humans and animals.3, 6 In 1987, the International Agency for Research on Cancer (IARC) of the World Health Organization (WHO) accepted that aflatoxin should be classified as a Group 1 carcinogen (carcinogenic to humans). Then, in 1993, WHO-IARC classified AFB1 as Group 1.7 As a result, governments in many countries have regulatory limits for levels of aflatoxins in agricultural products. There are a variety of analytical methods available for aflatoxin detection among which ELISA is the most widely used method due to its simplicity, sensitivity, specificity and facile sample preparation.8, 9 Aflatoxin is a low molecular weight, non-proteinaceous toxin, so it is necessary to use an aflatoxin-protein conjugate to raise antibodies. It is also necessary to use it as a reporter or competing antigen in assay development. In some cases, the synthesis of small molecules as competing ligands can be difficult, expensive, and in the case of aflatoxins, potentially hazardous. It is also common to synthesize a library of coating antigens. For optimum performance of the assay, one needs a reporter ligand that binds to the antibody only slightly less well than the target analyte.
One possible alternative to using aflatoxin as immunochemical reagents is to develop protein or peptide mimics that serve the same function.10 One way to realize this is to develop anti-idiotype antibodies.11–14 However, these methods are time-consuming and costly. It has been found that analyte peptidomimetic isolated from phage display libraries can be used as surrogate competing haptens in both monoclonal antibody-based immunoassays15, 16 and polyclonal antibody-based immunoassays.17 This concept has a high potential to be of practical use in the development of heterologous immunoassays. Phage-displayed peptide libraries have been used in a number of applications, including epitope mapping,18, 19 identifying peptide ligands,20, 21 and defining protein-protein interaction.22–24 Also, researchers have used the technology to select peptide mimics of non-proteinaceous chemicals like deoxynivalenol,10 aflatoxin B1, 9, 25 zearalenone 26 and ochratoxin.27 Monoclonal antibody 1C11 is an anti-aflatoxin antibody developed in our laboratory. It is the most sensitive antibody for all four major aflatoxins described so far.28 In this study, we constructed and characterized a phage peptide library with a random 8-amino-acid sequence with which to identify a peptide that can bind MAb 1C11. The peptides which can be used as a substitute of the coating antigen were tested as competitors with aflatoxin B1, B2, G1, G2 and M1. One of these mimotopes was applied in an ELISA for quantitative estimation of aflatoxins in samples.
MATERIALS AND METHODS
Reagents
All reagents were of analytical grade unless otherwise specified. Anti-aflatoxin monoclonal antibody 1C11 was produced in our laboratory.28 Aflatoxin B1, B2, G1, G2, M1 standards, bovine serum albumin (BSA), goat anti-mouse IgG-peroxidase antibody, hapten AFB1 conjugated with BSA, polyethylene glycol 8000 (PEG 8000), Tween 20, and 3, 3′, 5, 5′-tetramethylbenzidine (TMB) were obtained from Sigma (St. Louis, MO, USA). E. coli ER2738 host strain, M13KE gIII and 96 gIII sequencing primer were purchased from New England Biolabs (Ipswich, MA, USA). Mouse anti-M13 MAb-HRP was purchased from GE Healthcare (Piscataway, NJ, USA).
Construction of the 8-amino-acid random peptide library
The 8-amino-acid random peptide library was constructed according to the manufacturer’s instructions. The library was made by ligating a synthetic 42-bp fragment into the M13KE vector and transfecting E. coli cells with the ligation product by electroporation. Within the fragment was the degenerate coding sequence 5′-GCTTGT (NNK)8 TGCGGTGGAGGT-3′, where N stands for an equimolar mixture of G, A, T, and C while K is an equimolar mixture of G and T. The single strand degenerate oligonucleotide is converted to double strand by polymerase chain reaction (PCR) using the extension primer 5′-CATGCCCGGGTACCTTTCTATTCTC-3′ (New England Biolabs Inc., Ipswich, MA). The PCR reaction was performed (heat to 95 °C, then cool slowly to less than 37 °C; 10 min, 37 °C; 15 min, 65 °C) with 5µg of template, 3 molar equivalents of extension primer and 15 units of DNA polymerase (Klenow fragment) (New England Biolabs Inc., Ipswich, MA).
A 196 µL aliquot of the PCR-amplified oligonucleotide mixture was digested (3 h, 37 °C ) with 5 µL EagI (10 U/µL) (New England Biolabs Inc., Ipswich, MA) and 5 µL Acc65I (10 U/µL) (New England Biolabs Inc., Ipswich, MA) in a total volume of 400 µL. The digested duplex was purified by phenol/chloroform extraction and ligated into a digested M13KE vector at 16 °C overnight.
Electroporation of 1 µL of ligation product into electrocompetent ER2738 cells (100 µL) was performed by discharging a 25 µF capacitor charged to 2.5 kV across a 0.2 cm gap in parallel with a 200 S resistance. After 30 min incubation in SOC medium (LB medium with 0.01 M MgCl2, 0.02 M glucose) at 37 °C, serial dilution of the cells was mixed with 3 mL top agar and 200 µL a mid-log culture of ER2738 and pooled on LB/IPTG/Xgal plate. The remaining cells were added to one liter of early-log cells (OD600 0.01–0.05) and grown at 37 °C with shaking for amplification. After 5-h shaking, the culture was centrifuged at 5000 g for 20 min at 4 °C. The supernatant was collected and 1/6 volume of 20% PEG/2.5 M NaCl solution was added. Phage virions were collected by centrifugation as above. The final phage library was suspended in TBS (50 mM Tris-HCl, 150 mM NaCl, pH 7.5) containing 0.02% NaN3.
Phage selection by panning-elution
Three wells of one Nunc-Immuno plate were coated with purified 1C11 MAb (10 µg/mL) in 100 µL of phosphate-buffered saline (PBS) at 4 °C overnight. Nonspecific binding was blocked by incubation with 300 µL PBS containing 3% BSA for 1 h at 37 °C. To eliminate nonspecific binding of phage with BSA, another plate coated with 100 µL 3% BSA-PBS was used for pre-absorption. For the panning-elution procedure, the phage library (1×1010 pfu/mL) diluted with PBS was first added to the pre-absorption plate and incubated at 37 °C for 1 h. Then the supernatant was transferred to the plate containing MAb and incubated with shaking at room temperature for 2 h. The wells were washed 10 times by PBS containing 0.1% (v/v) Tween 20 (PBST). To elute the bound phage, 100 µL AFB1 (100 ng/mL in 10% methanol, competitive elution) was added to each well with shaking for 30 min to compete the binding phage from the coating antibody, or 100 µL 0.2 M glycine-HCl (pH 2.2, acidic elution), 1 mg/mL BSA was added with gently rocking for no more than 20 min and neutralized with 15 µL of 1 M Tris-HCl (pH 9.1). The elution solution was then collected and used to infect E. coli ER2738 for amplification and titration. The amplified phage was used for a subsequent round of panning. In the second and third rounds of panning, the concentration of coating antibody was reduced to 5 µg/mL and 1 µg/mL, while the elution buffer was 10 ng/mL and 1 ng/mL AFB1, respectively. After three rounds of panning-elution selection, individual plaques were picked up from LB/IPTG/Xgal plates and tested for their ability to bind to the MAb by phage ELISA. Positive clones were further selected by titration and submitted for DNA sequencing using the primer 96gIII (CCCTCATAGTTAGCGTAACG) (Division of Biological Sciences, Automated DNA Sequencing Facility, University of California, Davis).
Screening of phage eluate for positive clones by phage ELISA
After three rounds of panning, 200 µL of ER2738 cell culture (mid-log phase, OD600=0.5 AU) was resuspended with 10 µL of diluted phage eluates. Then the infected cells were transferred to culture tubes containing 45 °C top agar and poured on a LB/IPTG/Xgal plate. The plates were incubated overnight at 37 °C. A total of 20 clones were picked, transferred to diluted ER2738 culture and grown at 37 °C with shaking for 4.5 h. Cells were pelleted by centrifugation at 10000 rpm for 10 min and the supernatants were collected for phage ELISA.
To select the positive clones, a Nunc-Immuno plate was coated with anti-aflatoxin MAb and blocked as described above for panning-elution selection. Wells coated with 3% BSA were used to detect the nonspecific binding of each clone. Fifty microliters of phage supernatant of each clone was mixed with 50 µL of 100 ng/mL AFB1 in 10% methanol-PBS or pure dilution buffer. The mixtures were added to the wells and the preparations were incubated at room temperature for 2 h with shaking. After the wells were washed six times with 0.1% PBST, 100 µL of anti-M13 phage antibody conjugated with HRP (1:5000 dilution in PBS) was added. After a 1-h incubation and six-time washing, amounts of bound enzyme were determined by adding 100 µL of peroxidase substrate (25 mL of 0.1 M citrate acetate buffer [pH 5.5], 0.4 mL of 6 mg/mL dimethyl sulfoxide [DMSO] solution of TMB, and 0.1 mL of 1% H2O2). The absorbance at 450nm was determined after the reaction was stopped by adding 50 µL of 2 N H2SO4 per well.
Competitive ELISA performed with phage-displayed peptide
A Nunc-Immuno plate was coated with 100 µL of MAb 1C11 at a concentration of 5 µg/mL in PBS by incubation overnight at 4 °C and blocked with 3% skimmed milk in PBS for 1 h at 37 °C. Various concentrations of standard AFB1 (0 to 30 ng/mL in 10% methanol-PBS) or sample extract were mixed with equal volumes of phage-displayed peptide (diluted 1:1000 in PBS). The mixtures were added to MAb 1C11 coated wells (0.5 µg /well) followed by 2-h incubation at room temperature with shaking. The plate was washed 6 times with 0.05% PBST and 100 µL of anti-M13 phage antibody conjugated with HRP was added. After 1-h incubation and washing, the color was developed as above.
Sample preparation
Peanuts, rice and corn samples used for spike-and-recovery assessment were purchased from a local supermarket in California. Natural contaminated samples were collected from farmers in China. Five gram of ground sample was extracted using 15 mL of methanol-water (80:20, v/v) by shaking on a shaker at 250 rpm for 20 min. The mixture was then centrifuged at 4000 rpm for 30 min. The supernatant was diluted 1 in 7 with PBS containing 2% or 4% BSA prior to analysis (the final concentration of methonal in the sample solution is 10%).
Conventional ELISA
To compare the effect of phage ELISA with conventional ELISA, an indirect conventional ELISA was used to detect the natural contaminated samples. The plate was coated with 0.02 µg AFB1-BSA in PBS by incubation overnight at 4 °C. The next day, plate was blocked with 3% skimmed milk in PBS at 37 °C for 1 h. A mixture of 50 µL 1 µg/mL MAb 1C11 and 50 µL serial dilution of AFB1 or sample extract was added to the plate and incubated for 1 h at 37 °C. Then 100 µL goat anti-mouse antibody conjugated with HRP (1:4000) was added to the plate. After 1-h incubation, TMB substrate was added and the OD value was detected at 450nm with a microplate reader. Samples were extracted in the same way as for phage ELISA, but diluted 1 in 7 with pure PBS.
RESULTS AND DISCUSSION
Characterization of the phage library
PⅢ library is a powerful tool to select high affinity proteins or peptides that bind to particular target. Capacity and diversity is critical to the success of biopanning. The greater the diversity of the library, the more chances to obtain the target ligands. We constructed a loop-constrained octapeptide library. The transducing unit (TU) of the library was determined by infecting E coli ER 2738 cells and pooled on LB/IPTG/Xgal plates. The calculated TU was 5.52×109 pfu/µg. To identify the diversity of peptide sequences in the library, we selected 20 clones from the library for sequencing. The results showed that there were no identical inserted sequences among them. The titer of the library was 3.4×1013.
Panning-elution selection of specific phages
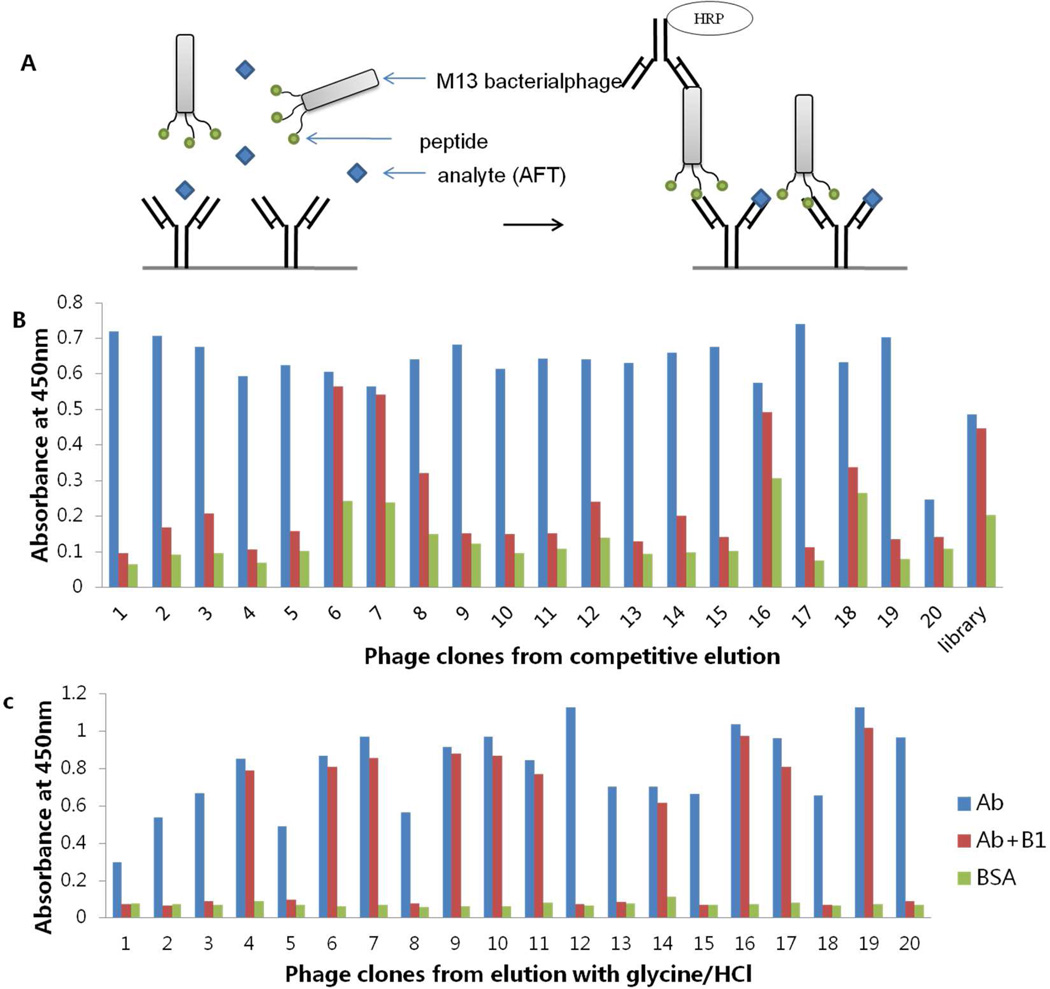
Figure 1A is a schematic diagram of the competitive ELISA using a phage displayed mimotope. This method requires a phage-borne peptide that mimics the epitope to be recognized by antibody rather than nonspecific binding to the surface of the antibody molecule outside the antigen binding pocket. After each round of panning, the number of eluted phage was determined. Enrichment for specific phage with affinity to MAb 1C11 was observed after two rounds of panning (data not shown).
Figure 1.
Schematic diagram of (A) phage peptide based competitive ELISA, (B) screening of positive clones by phage ELISA from competitive elution and (C) screening of positive clones by phage ELISA from acidic elution. Library without panning was used as negative control.
Randomly chosen clones (20 clones) derived from the third round of panning were analyzed by phage ELISA in the presence (100 ng/mL) or absence of AFB1. Bound phages were eluted by two distinct strategies: nonspecific disruption by incubation with 0.2 M glycine-HCl (pH 2.2) and competitive elution by incubation with high concentration of aflatoxin B1. Figure 2B and C show the results of phage ELISA with two elution methods. Eighteen clones out of 20 showed big signal differences with or without AFB1 in the assay using competitive elution, while 10 out of 20 were positive with acidic elution. The percentage of isolated phage peptides that bound desired epitope of the coating antibody dramatically increased when soluble aflatoxin B1 was used for specific removal of phage that preferentially bound to the plate.
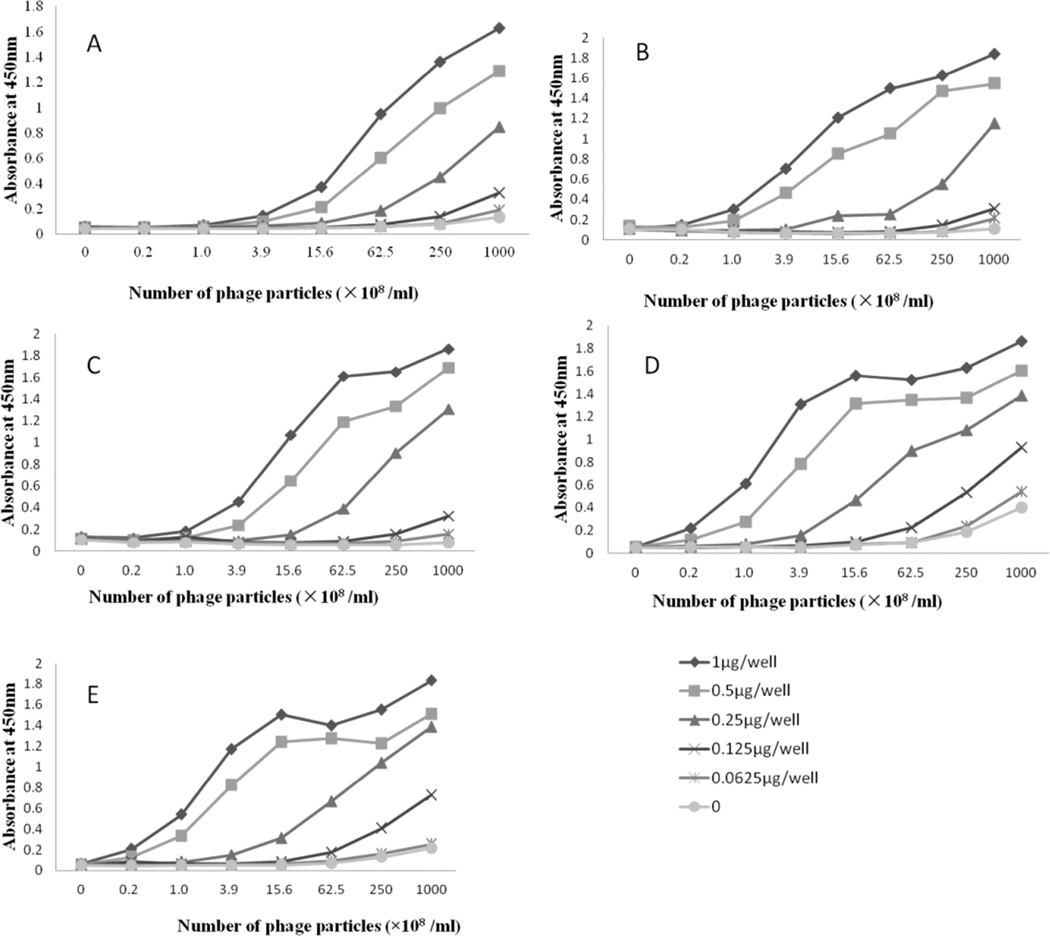
Figure 2.
Checkerboard results of each phage peptide. (A) CM2, (B) CM4, (C) CM8, (D) PM13 and (E) PM23.
Phage single-stranded DNA from the positive clones were isolated, and the nucleotide sequence of each of them was determined. Only five different sequences (designated as CM2, CM4, CM8, PM13, PM23) were identified (Table 1). Each sample was sequenced twice. CM2 and CM8 had the consensus sequence of ‘-PHPWNP-’. CM4 and PM13 showed the consensus peptide sequence ‘-T-HRNW-‘. In addition, each amino acid sequence of the mimotope peptides contained one or more aromatic amino acids (tyrosine, tryptophan and phenylalanine). With a different monoclonal antibody and phage-displayed library, Liu et al.25 obtained an insert amino acid sequence rich in histidine (H) and tryptophan (W). Thirumala-Devi et al. 9 searched mimotope with two monoclonal antibodies (MAb 13 and MAb 24). Peptides binding MAb 24 (specific for AFB1 and AFG1) contained the sequence of “YMD”, and those bound MAb 13 (specific for aflatoxin B1, B2, G2, G2) contained “PW”. The most common amino acids proline (P), tryptophan (W) and histidine (H) in those publications reflect a similarity interaction of different anti-aflatoxin antibodies with aflatoxins.
Table 1.
Amino Acid Sequences of Mimotope Peptides
| Clone name | Sequence |
|---|---|
| CM2 | FPHPWNPP (3) |
| CM4 | HTSHRNWD (4) |
| CM8 | YPHPWNPT (3) |
| PM13 | QTTHRNWA (8) |
| PM23 | PRYLPWFP (2) |
Note. A total of 20 clones were sequenced. The numbers of isolates bearing the same sequence are indicated in parenthesis.
Sensitivity and specificity test
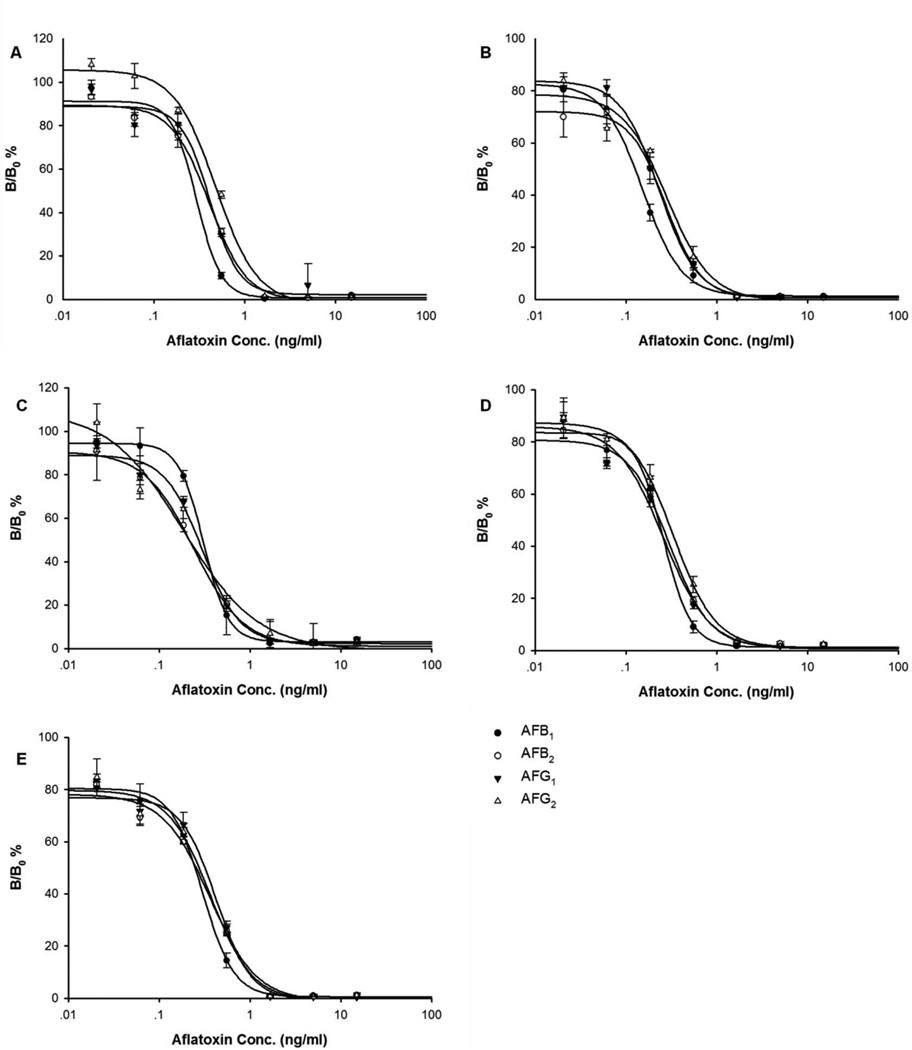
Unlike pⅧ library displaying peptides in a high copy number, pⅢ library has only five insert peptide copies on the phage. Instead of using it as coating antigen, we established a different ELISA format with antibody coated on the plate (Figure 1A). Five clones with different sequences were amplified and purified. The optimal concentrations of coating antibody and phage peptide were checked by a checkerboard procedure. Serial dilutions of phage particles were added to the wells of the plates coated with four different concentrations of antibody (10, 5, 2.5 and 1.25 µg/mL). The best results (Figure 2) were obtained with antibody concentrations of 0.5 µg/well and phage concentrations of 2.5×1010 pfu/mL for clone CM2, 6.25×109 pfu/mL for clone CM4 and CM8, 1.56×109 pfu/ml for PM13 and PM23. Under these conditions, the indirect competitive ELISA was conducted with each mimotope towards aflatoxin B1, B2, G1 and G2 (Figure 3). For each mimotope, the competitive difference among the four toxins was not significant. However, for most of the mimotopes, the homologous toxin (AFB1) was the most competitive. The specificity test towards aflatoxin M1 was also performed. The sensitivity and cross reactivity of the assay are presented in Table 2. These results further confirm that the strongest competition was with aflatoxin B1, and the weakest competition was with aflatoxin M1. The phage peptide-based competitive ELISA showed good cross reactivity with aflatoxin B2, G1, G2 and M1. We chose mimotope CM8, which has high cross reactivity towards four different aflatoxins, for the following sample analysis. The linear range is 0.09–0.5 ng/mL and the half inhibition concentration (IC50) is 0.29 ng/mL.
Figure 3.
Competition between pure aflatoxin (●) B1, (○) B2, (▼) G1 and (△) G2 with phage mimotope (A) CM2, (B) CM4, (C) CM8, (D) PM13 and (E) PM23 for binding to MAb 1C11 in an indirect competitive ELISA. Results are the average of three independent experiments.
Table 2.
Results of Sensitivity and Cross-Reactivity of the Phage Peptide-Based ELISA with Five Mimotopes
| Aflatoxins | Clones | ||||
|---|---|---|---|---|---|
| PM13 | PM23 | CM2 | CM4 | CM8 | |
| Sensitivity (ng/mL) | |||||
| B1 | 0.238 | 0.261 | 0.285 | 0.121 | 0.300 |
| B2 | 0.252 | 0.293 | 0.370 | 0.236 | 0.235 |
| G1 | 0.217 | 0.354 | 0.378 | 0.183 | 0.294 |
| G2 | 0.289 | 0.325 | 0.456 | 0.162 | 0.271 |
| M1 | 0.331 | 0.339 | 0.516 | 0.358 | 0.383 |
| Cross-reactivity (%)* | |||||
| B1 | 100 | 100 | 100 | 100 | 100 |
| B2 | 94.4 | 89.1 | 77.0 | 51.3 | 127.7 |
| G1 | 109.7 | 73.7 | 75.4 | 66.1 | 102.0 |
| G2 | 82.3 | 80.3 | 62.5 | 74.7 | 110.7 |
| M1 | 71.9 | 77.0 | 55.2 | 33.8 | 78.3 |
cross-reactivity was calculated as: % cross-reactivity= (IC50AFB1/IC50analyte) × 100
Optimization of phage peptide-based competitive ELISA
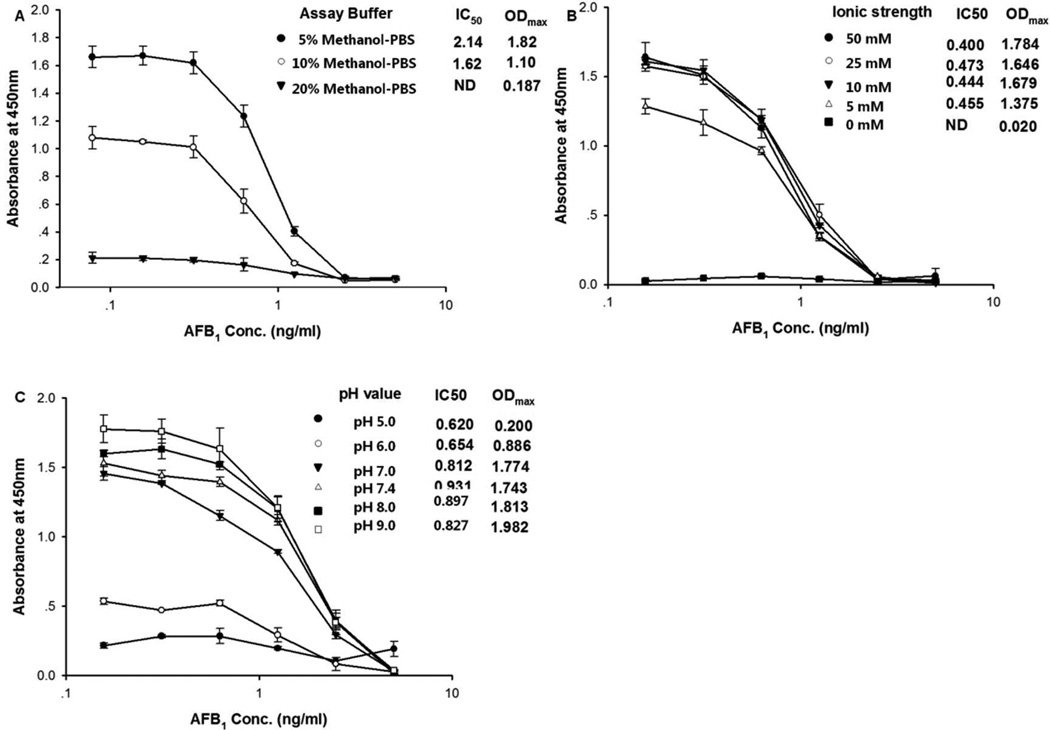
Aflatoxins are highly lipophilic, so the addition of solvent to the assay buffer is necessary to the assay performance. Methanol is the most commonly used solvent in aflatoxin analysis. A high tolerance to methanol may allow the analysis of samples without extensive dilution of the sample extract decreasing the limit of quantification. For these reasons, we tested the effect of methanol using PBS containing three different concentrations of methanol (5%, 10%, 20%) (Figure. 4A). High concentrations of methanol resulted in significant decrease of the maximum signal in the assay, which indicated that activity of the phage peptide can be influenced by the solvent. The lowest IC50 was observed at 10% methanol. Thus, we used the assay buffer containing final 10% methanol throughout this study.
Figure 4.
Influence of (A) methanol concentration, (B) ionic strength and (C) pH on the performance of the assay. Each value represents the mean value of four replicates.
The pH and buffer ionic strength were also investigated. The pH value is a key point influencing the assay performance. From Figure 4B we can see that the maximum signal decreased when we lowered the pH of the buffer. A similar result was obtained with low ionic strength buffer, but there was little difference when concentration was over 0.01 M. Finally, a pH of 7.0 and 0.01 M PBS buffer was selected as the optimum for the assay giving a favorable IC50 value.
Matrix effect and sample analysis
Matrix effects, caused by constituents in the specimen, are an important interference effect for immunoassays, influencing the measurement of a target analyte.29 It may cause false positive or negative results, reducing the sensitivity and specificity of the method. Dilution of the sample extracts is a good way to reduce matrix effects, however, high dilution also results in lower sensitivity. Peanut, corn and rice were chosen as test samples to study matrix interference. Aflatoxin concentration in these samples has been tested to be lower than 0.1 ppb by conventional ELISA.28 Sample extracts were diluted by 8, 20 and 50 times with PBS or 10% methanol-PBS and spiked with a series of concentrations of AFB1. Table 3 shows the % color reduction compared with that from no matrix containing assay buffer and IC50 of matrix assay. Color development was inhibited significantly by sample co-extractants, however, the sensitivity was not affected. This suggested the matrix interferes mainly with the enzyme activity not the antibody binding activity. The degree of interference varied with different samples.
Table 3.
Matrix Effect Measured as % Color Reduction and IC50 Values
| 8 times dilution | 20 times dilution | 50 times dilution | ||||
|---|---|---|---|---|---|---|
| Samples | % color reduction |
IC50 value (ppb) |
% color reduction |
IC50 value (ppb) |
% color reduction |
IC50 value (ppb) |
| Peanut | 47.9% | 0.481 | 47.4% | 0.515 | 35.3% | 0.515 |
| Rice | 74.4% | 0.393 | 60.1% | 0.42 | 31.7% | 0.418 |
| Corn | 75.1% | 0.5 | 71.6% | 0.444 | 58.3% | 0.46 |
For the standard assay, 10% methanol-PBS was used. The sample extract was diluted 1 in 7 with PBS (the final methanol concentration in the test solution is 10%), 1 in 19 with 10% Methanol-PBS (the final concentration of methanol in the extract is 13.5% Methanol-PBS) and 1 in 49 with 10% methanol-PBS (final concentration of methonal is 11.4%) for matrix assay. % color reduction was calculated as (1-Asample/Amethanol)×100, where Asample is the absorbance (450 nm) of test sample extract without aflatoxin and Amethanol is the absorbance of 10% methanol without aflatoxin.
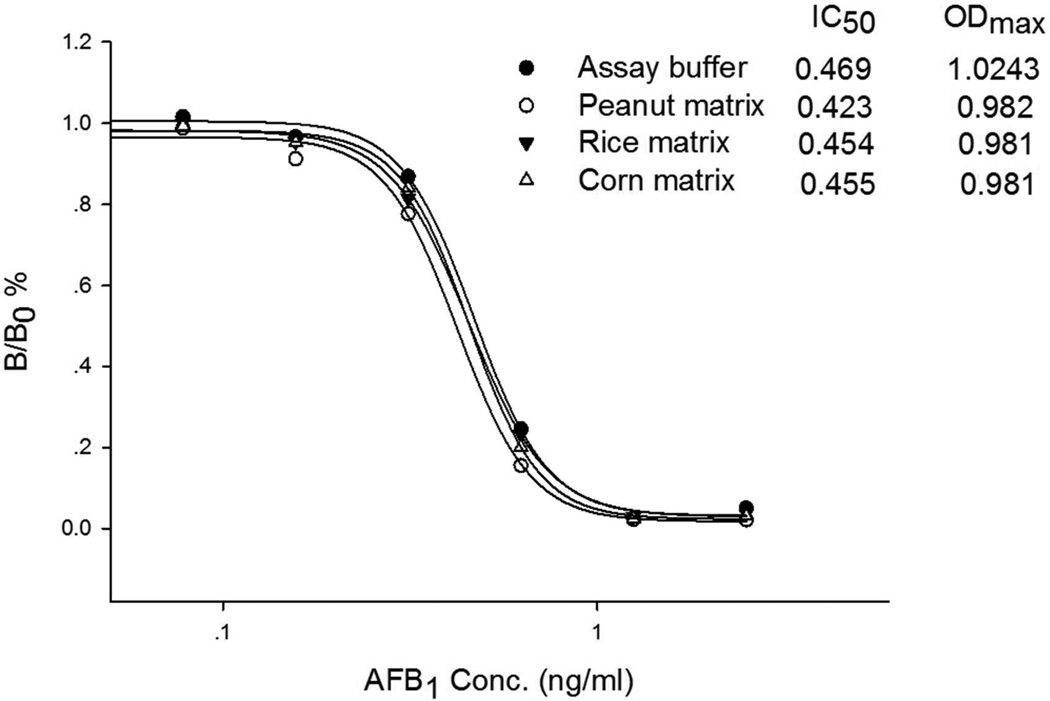
For corn and rice, the problem was overcome by diluting the extract using 2% BSA-PBS instead of PBS. However, for peanut, we optimized the concentration of BSA in the dilution buffer, and finally found 4% BSA is effective to reduce color reduction less than 10%. The protein in the dilution buffer seems to be a stabilizer in the assay to protect phage from interferences30. No significant % color reduction and inhibition was observed from Figure 5. Due to the dilution, the final sensitivity of the assay is 14 µg/kg with a linear range of 4–24 µg/kg, which is suitable for monitoring total aflatoxin concentration under current regulatory limits of aflatoxin in most countries (20 µg/kg).
Figure 5.
The assay standard curve in 10% methanol-PBS (●), peanut (○), rice (▼)and corn (㖳) matrix.
Validation studies
To test the effectiveness of the phage ELISA, we carried out tests with samples spiked with known concentrations of aflatoxins. Before the test, all samples were verified to have aflatoxin less than 0.1 ppb by conventional ELISA. The results are presented in Table 4. The overall recoveries ranged from 60%–120%.
Table 4.
Recovery Analysis of Aflatoxin by Phage Peptide-Based Competitive ELISA
| Sample type | Spike level (µg/kg) | Mean ±SD | Average recovery (%) |
|---|---|---|---|
| AFB1 5 | 4.7±0.99 | 94.35 | |
| AFB1 10 | 9.6±1.1 | 95.97 | |
| Peanut | AFB1 20 | 19.2±1.6 | 96.12 |
| Total aflatoxinsa 5 | 4.3±0.9 | 85.71 | |
| Total aflatoxins 10 | 11.3±3.2 | 112.71 | |
| AFB1 5 | 5.1±0.9 | 102.9 | |
| AFB1 10 | 6.2±1.4 | 62.5 | |
| Rice | AFB1 20 | 20.9±3.1 | 100.5 |
| Total aflatoxins 5 | 4.5±0.9 | 89.5 | |
| Total aflatoxins 10 | 11.5±4.0 | 114.7 | |
| AFB1 5 | 4.0±1.0 | 81.1 | |
| AFB1 10 | 9.7±1.4 | 97.4 | |
| Corn | AFB1 20 | 20.7±0.5 | 103.5 |
| Total aflatoxins 5 | 4.4±1.4 | 87.3 | |
| Total aflatoxins 10 | 11.4±1.4 | 114.4 |
Mixed aflatoxin standard was prepared with aflatoxin B1, B2, G1 and G2 at a ratio of 2:1:1:1. Each sample was assayed with four replicates.
Nine samples with unknown concentration of aflatoxin were tested using the developed phage ELISA and conventional ELISA. From Table 5, when contamination is between 4–20 µg/kg, the results of phage ELISA were in good agreement with those of conventional ELISA.
Table 5.
Detection Results of Conventional ELISA and Phage ELISA in Peanuts and Feedstuff Samples
| Samples | Conventional ELISA (µg/kg±SD, n=3) |
Phage ELISA (µg/kg±SD, n=3) |
|---|---|---|
| Peanut | ||
| 1 | 10.4±1.5 | 9.2±1.9 |
| 2 | 0.76±0.13 | ND |
| 3 | 12.9±4.6 | 15.5±1.8 |
| 4 | 3.9±2.2 | 2.3±0.07 |
| 5 | 15.6±1.9 | 15.5±5.4 |
| 6 | 29.8±8.9 | 24.8 |
| Feedstuff | ||
| 1 | 31.4±3.1 | ND |
| 2 | 8.1±1.2 | 7.6±0.5 |
| 3 | 12.6±3.1 | 11.2±2.9 |
ND, not detectable.
Safety
Aflatoxins are powerful hepatotoxins and carcinogens, so great care should be taken to avoid personal exposure and potential laboratory contamination. All items coming in contact with aflatoxins (glassware, vials, tubes, ELISA plates, et al.) were immersed in a 10% bleach solution for 1–2h before they were discarded.
Acknowledgments
Funding
This work was supported by the Project of National Science & Technology Pillar Plan (2012BAB19B09), the Special Fund for Agro-scientific Research in the Public Interest (201203094), and the Key Project of the Ministry of Agriculture (2011-G5). The research is also supported in part by the National Institute of Environmental Health Sciences Superfund Basic Research Program P42 ES04699, the National Institute of Occupational Safety and Health Western Center for Agricultural Health and Safety U50 OH07550 and the Counter ACT Program, National Institutes of Health Office of the Director, and the National Institute of Neurological Disorders and Stroke, Grant U54 NS079202.
REFERENCES
- 1.Creppy EE. Update of survey, regulation and toxic effects of mycotoxins in Europe. Toxicol. Lett. 2002;127:19–28. doi: 10.1016/s0378-4274(01)00479-9. [DOI] [PubMed] [Google Scholar]
- 2.Delmulle BS, De Saeger S, Sibanda L, Barna-Vertro I, Van Peteghem CH. Development of an immunoassay-based lateral flow dipstick for the rapid detection of aflatoxin B1 in pig feed. J. Agric. Food Chem. 2005;53:3364–3368. doi: 10.1021/jf0404804. [DOI] [PubMed] [Google Scholar]
- 3.Cho S, Lee C, Jang M, Son Y, Lee S, Choi I, Kim S, Kim D. Aflatoxins contamination in spices and processed spice products commercialized in Korea. Food Chem. 2008;107:1283–1288. [Google Scholar]
- 4.Allcroft R, Roberts BA, Lloyd MK. Excretion of aflatoxin in a lactating cow. Food Cosmet. Toxicol. 1968;6:619–625. doi: 10.1016/0015-6264(68)90311-8. [DOI] [PubMed] [Google Scholar]
- 5.Guan D, Li P, Zhang Q, Zhang W, Zhang D, Jiang J. An ultra-sensitive monoclonal antibody-based competitive enzyme immunoassay for aflatoxin M1 in milk and infant milk products. Food Chem. 2011;125:1359–1364. [Google Scholar]
- 6.Blesa J, Soriano JM, Moltó JC, Mañes J. Limited survey for the presence of aflatoxins in foods from local markets and supermarkets in Valencia, Spain. Food Addit. Contam. 2004;21:165–171. doi: 10.1080/0265203031000. [DOI] [PubMed] [Google Scholar]
- 7.Li P, Zhang Q, Zhang W. Immunoassays for aflatoxins. Trends Anal. Chem. 2009;28:1115–1126. [Google Scholar]
- 8.Chu FS, Fan TS, Zhang GS, Xu YC, Faust S, Mcmahon PL. Improved enzyme-linked immunosorbent assay for aflatoxin B1 in agricultural commodities. J. Assoc. Off. Anal. Chem. 1987;70:854–857. [PubMed] [Google Scholar]
- 9.Thirumala-Devi K, Miller JS, Reddy G, Reddy DVR, Mayo MA. Phage-displayed peptides that mimic aflatoxin B1. J. Appl. Microbiol. 2001;90:330–336. doi: 10.1046/j.1365-2672.2001.01249.x. [DOI] [PubMed] [Google Scholar]
- 10.Yuan Q, Pestka JJ, Hespenheide BM, Kuhn LA, Linz JE, Hart LP. Identification of mimotope peptides which bind to the mycotoxin deoxynivalenol-specific monoclonal antibody. Appl. Environ. Microbiol. 1999;65:3279–3286. doi: 10.1128/aem.65.8.3279-3286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu FY, Chu FS. Production and characterization of a monoclonal anti-anti-idiotype antibody against fumonisin B1. J. Agric. Food Chem. 1999;47:4815–4820. doi: 10.1021/jf990185x. [DOI] [PubMed] [Google Scholar]
- 12.Becker MI, Aguayo JE, Jamett A, Juica F, Yudelevich A, Foradori A, De Ioannes AE. An alternative ELISA for T4 determination based on idiotype anti-idiotype interaction and a latex method for anti-idiotype monoclonal antibody selection. J. Immunol. Methods. 1996;192:73–85. doi: 10.1016/0022-1759(96)00023-3. [DOI] [PubMed] [Google Scholar]
- 13.Edwards AJ, Durst RA. Flow-injection liposome immunoanalysis (FILIA) with electrochemical detection. Electroanalysis. 1995;7:838–845. [Google Scholar]
- 14.Guan D, Li P, Cui Y, Zhang Q, Zhang W. A competitive immunoassay with a surrogate calibrator curve for aflatoxin M1 in milk. Anal. Chim. Acta. 2011;703:64–69. doi: 10.1016/j.aca.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Felici F, Luzzago A, Folgori A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides. II. Selection of clones recognized by a protective monoclonal antibody against the Bordetella pertussis toxin from phage peptide libraries. Gene. 1993;128:21–27. doi: 10.1016/0378-1119(93)90148-v. [DOI] [PubMed] [Google Scholar]
- 16.Cardozo S, González-Techera A, Last JA, Hammock BD, González-Sapienza GG. Analyte peptidomimetics selected from phage display peptide libraries: a systematic strategy for the development of environmental immunoassays. Environ. Sci. Technol. 2005;39:4234–4241. doi: 10.1021/es047931l. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, González-Techera A, González-Sapienza GG, Ahn KC, Gee SJ, Hammock BD. Phage-borne peptidomimetics accelerate the development of polyclonal antibody-based heterologous immunoassays for the detection of pesticide metabolites. Environ. Sci. Technol. 2008;42:2047–2053. doi: 10.1021/es702219a. [DOI] [PubMed] [Google Scholar]
- 18.Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. Proc. Nati. Acad. Sci. U. S. A. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamie KS, Scott GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 20.Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat. Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 21.Stricker NL, Christopherson KS, Yi BA, Schatz PJ, Raab RW, Dawes G, Bassett DE, Bredt DS, Li M. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat. Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- 22.Ryan ST, Chi-Rosso G, Bonnycastle LL, Scott JK, Koteliansky V, Pollard S, Gotwals PJ. Epitope mapping of a function-blocking beta 1 integrin antibody by phage display. Cell Adhes. Commun. 1998;5:75–82. doi: 10.3109/15419069809005600. [DOI] [PubMed] [Google Scholar]
- 23.Burks EA, Chen G, Georgiou G, Iverson BL. In vitro scanning saturation mutagenesis of an antibody binding pocket. Proc. Nati. Acad. SciUSA. 1997;94:412–417. doi: 10.1073/pnas.94.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright RM, Gram H, Vattay A, Byme S, Lake P, Dottavio D. Binding epitope of somatostatin defined by phage-displayed peptide libraries. Biotechnology. 1995;13:165–169. doi: 10.1038/nbt0295-165. [DOI] [PubMed] [Google Scholar]
- 25.Liu R, Xu L, Qiu X, Chen X, Deng S, Lai W, Xu Y. An Immunoassay for determining aflatoxin B1 using a recombinant phage as a nontoxic coating conjugate. J. Food Safety. 2012;32:318–325. [Google Scholar]
- 26.He Q, Xu Y, Huang Y, Liu R, Huang Z, Li Y. Phage-displayed peptides that mimic zearalenone and its application in immunoassay. Food Chem. 2011;126:1312–1315. [Google Scholar]
- 27.Liu R, Yu Z, He Q, Xu Y. An immunoassay for ochratoxin A without the mycotoxin. Food Control. 2007;18:872–877. [Google Scholar]
- 28.Zhang D, Li P, Zhang Q, Zhang W, Huang Y, Ding X, Jiang J. Production of ultrasensitive generic monoclonal antibodies against major aflatoxins using a modified two-step screening procedure. Anal. Chim. Acta. 2009;636:63–69. doi: 10.1016/j.aca.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Wood WG. “Matrix effects” in immunoassays. Scand. J. Clin. Lab. Invest. Suppi. 1991;205:105–112. [PubMed] [Google Scholar]
- 30.Lee NA, Wang S, Allan RD, Kennedy IR. A rapid aflatoxin B1 ELISA: development and validation with reduced matrix effects for peanuts, corn, pistachio, and Soybeans. J. Agric. Food Chem. 2004;52:2746–2755. doi: 10.1021/jf0354038. [DOI] [PubMed] [Google Scholar]