Abstract
The Bam machine assembles β-barrel membrane proteins into the outer membranes of Gram-negative bacteria. The central component of the Bam complex, BamA, is a β-barrel that is conserved in prokaryotes and eukaryotes. We have previously reported an in vitro assay for studying the assembly of β-barrel proteins by the Bam complex and now apply this assay to identify the specific components that are required for BamA assembly. We establish that BamB and BamD, two lipoprotein components of the complex, bind to the unfolded BamA substrate and are sufficient to accelerate its assembly into the membrane.
The outer membranes (OMs) of Gram-negative bacteria contain transmembrane proteins with β-barrel structure. These proteins are synthesized in the cytoplasm with N-terminal signal sequences, which target them for secretion across the inner membrane (IM) via the Sec machine.1–3 They are then transported in complex with chaperones across the aqueous periplasmic compartment between the membranes and are finally assembled in the OM by the β-barrel assembly machine (Bam).4 The E. coli Bam complex contains two proteins that are essential for cell viability: an integral membrane β-barrel, BamA, and an OM lipoprotein, BamD, which is anchored to the membrane by an N-terminal lipid and which binds to the soluble region of BamA that extends into the periplasm.5–7 Three other lipoproteins, BamB, C, and E, associate with these two proteins but are not essential.5, 8–10 The mechanism of β-barrel assembly is believed to be highly conserved because orthologs of BamA are found in all organisms that contain β-barrels.11–16 However, very little is known about how that mechanism proceeds; it is thought to involve multiple steps, including substrate recognition, folding, and membrane insertion, but it is not clear how the components of the Bam complex accomplish those steps.
We have reconstituted the process of β-barrel assembly in vitro from purified components and now make use of this system to dissect the Bam complex and observe the effects of its individual components.17, 18 We chose to study the assembly of BamA because it is an essential outer membrane protein (OMP), and given that its function is to assemble other OMPs, we hypothesized that its assembly mechanism might reveal or reflect aspects of how it functions in the more general OMP assembly process. Through this analysis, we have determined that BamB and BamD bind to unfolded substrates.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The methods used to express and purify the proteins used in this study are described in the supporting information.
Proteoliposome Preparation
Proteoliposomes containing the Bam complex and Bam subcomplexes were prepared by the detergent dilution methods described previously.18 Briefly, E. coli phospholipids (40 μL of a 20 mg/mL sonicated aqueous suspension) were added to the purified Bam complexes (200 μL of 10 μM solutions) in TBS (pH 8), 0.03% DDM, 1 mM TCEP and incubated on ice for 5 min. These phospholipid, detergent, protein complex mixtures were then diluted with 8 mL of TBS (pH 8) and incubated on ice for 30 min. They were then ultracentrifuged at 300,000 × g for 2 hours at 4 °C. The pelleted proteoliposomes were resuspended in 200 μL of TBS (pH 8). Empty liposomes were prepared in parallel with these proteoliposomes by the same detergent dilution method, simply omitting the Bam proteins. Liposomes and proteoliposomes that were not used immediately were flash frozen in liquid nitrogen and stored at −80 °C.
Folding Assays
Folding into Bam Proteoliposomes
The unfolded FLAG-BamA or FLAG-OmpA substrate was prepared at a concentration of 5 μM in 8 M urea and then diluted ten-fold into solutions containing empty liposomes or the Bam proteoliposomes. The proteoliposomes were also diluted four-fold from their stock concentrations into these reactions. A typical reaction contained 2.5 μL liposomes or proteoliposomes, 6.5 μL TBS (pH 8), and 1 μL of 5 μM substrate such that the final concentrations of the substrate and Bam complex were 0.5 μM and ~2.5 μM, respectively. If the experiment included a preincubation, the substrate was first diluted ten-fold from a 50 μM solution in 8 M urea into a solution of TBS (pH 8) or purified SurA in TBS (pH 8) and incubated at 25 °C for 10 min. These preincubated solutions were then diluted ten-fold into the proteoliposomes. Unless noted otherwise in the figures, the concentrations of SurA and the substrates were, respectively, 50 μM and 5 μM in the preincubation, and 5 μM and 0.5 μM in the final reactions. Reactions were stopped after 60 min of incubation at 25 °C (unless noted otherwise in the figures) by adding ice cold 2x SDS sample loading buffer (125 mM Tris, pH6.8, 4% SDS, 30% glycerol, 0.005% bromophenol blue, 5% β-mercaptoethanol). For the time course experiments, aliquots of the reactions were removed at the indicated time points, quenched by the same method, and kept on ice. All quenched samples were applied to SDS-PAGE (4–20% gel), and run at 150 V for 110 min at 4 °C. The proteins were transferred from the gel to a PVDF membrane by semi-dry transfer in 25 mM Tris-HCl, 192 mM glycine (pH 8.3) at 10 V for one hour. The products of the reaction were detected by immunoblotting with FLAG-HRP antibodies (used at a dilution of 1:200,000). The blot images were scanned, and ImageQuant TL was used to calculate the densities of the observed bands. The percent yields of folded protein were determined by comparing the densities of the unfolded and folded bands in each lane.
Folding in Detergent
FLAG-tagged substrate proteins were prepared at a concentration of 5 μM in 8 M urea. They were then diluted ten-fold into a solution of TBS (pH 8), 0.5% LDAO and incubated at 25 °C for one hour. The folding reactions were stopped with 2x SDS sample loading buffer. The quenched samples were run on SDS-PAGE and immunoblotted as described in the previous section.
Folded Chimeric BamA Affinity Purifications
FLAG-tagged wild-type and mutant BamA substrates were prepared at a concentration of 100 μM in 8 M urea. These substrates were then diluted 10-fold into TBS (pH 8), 0.5% LDAO and incubated at 25 °C for 60 min to allow their β-barrels to fold. Concentrated, purified BamCDE-His6 complex was then added to each of the folded substrates to a final concentration of 10 μM. Aliquots of these mixtures were removed and used as “input” samples for SDS-PAGE analysis. The remainder of each mixture was subjected to Ni-NTA affinity chromatography in TBS (pH 8), 0.05% DDM. Proteins in the eluates were precipitated with 10% trichloroacetic acid and incubated on ice for 30 min. These samples were then centrifuged at 18,000 × g for 10 min at 4 °C, and the pellets were resuspended in 1 M Tris (pH 8) and 2x SDS sample loading buffer. The “input” and these “eluate” samples were subjected to SDS-PAGE on a 4–20% gradient gel at 200 V for 45 min. The proteins were then detected by staining with Coomassie blue.
Unfolded Substrate Affinity Purifications
Urea-denatured FLAG-BamA and FLAG-OmpA were prepared at a concentration of 100 μM and subsequently diluted ten-fold into a solution of soluble BamB-His6, BamD-His6, or BamE-His6 and incubated a room temperature for 10 min. The final concentrations of the unfolded OMP and the soluble Bam proteins were 10 μM and 100 μM, respectively. A small aliquot of each of these mixtures was removed for use as an “input” sample. The remainder of the mixture was subjected to Ni-NTA affinity purification; after loading the material on the column, it was washed with TBS (pH 8) with 20 mM imidazole and the bound proteins were eluted in TBS (pH 8) with 200 mM imidazole. Proteins in the eluates were precipitated with 10% trichloroacetic acid and resuspended in 1 M Tris (pH 8) and 2x SDS sample loading buffer as described in the previous section. The proteins in the “input” and these “eluate” samples were separated by SDS-PAGE and stained with Coomassie Blue.
RESULTS
We began by identifying the minimal set of components that are required to assemble a BamA substrate in vitro. The fact that the OMP chaperones and some of the Bam proteins are non-essential suggests that they are not required for the assembly of all OMPs. We found that BamA can be assembled in vitro without a chaperone (Figures 1A and 1B). We diluted urea-denatured, FLAG-tagged BamA with or without SurA, a major periplasmic chaperone,19–21 into proteoliposomes containing the Bam complex.17, 18 The reaction products were separated on semi-native SDS-PAGE, and the folded and unfolded forms of the proteins were then visualized by immunoblotting with anti-FLAG antibodies. (β-barrels do not unfold in SDS unless they are boiled, so their folded and unfolded forms have different mobilities on SDS-PAGE, and the FLAG-tag on the substrate BamA distinguishes it from the untagged BamA in the complex.) The substrate did become less foldable in the absence of solubilizing factors (i.e. in tris-buffered saline), and SurA maintained its folding-competent state. However, SurA could be functionally replaced by urea (Figure S1). These results are consistent with in vivo measurements demonstrating that BamA levels do not decrease when surA is deleted.22–25 Therefore, the BamA substrate can be delivered to the Bam complex in different ways without affecting its assembly on the machine. Accordingly, we proceeded with our in vitro analysis of the direct effects of the Bam complex components on this substrate in the absence of a chaperone.
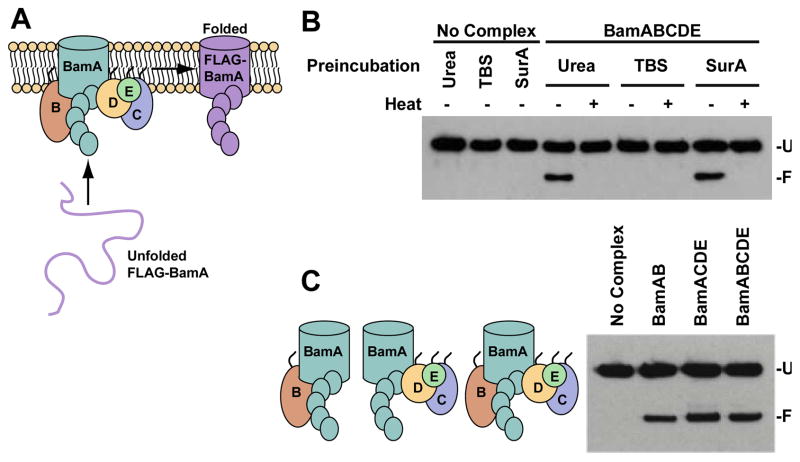
Figure 1.
BamA can be folded by a minimal set of OMP assembly components. A. Schematic of the experimental design. Purified Bam complex is incorporated into liposomes composed of E. coli phospholipids, and unfolded FLAG-tagged BamA is added to these proteoliposomes with or without a chaperone. B. Urea and SurA maintain the folding-competence of BamA equally well. FLAG-BamA was prepared in 8 M urea and then diluted directly into empty liposomes or proteoliposomes containing the Bam complex, or the denatured substrate was first incubated in solutions of tris-buffered saline (TBS) or SurA and then added. The final concentrations of the substrate and SurA were 0.5 μM and 5 μM, respectively. The reactions were stopped after 60 minutes and analyzed by SDS-PAGE and immunoblotting with anti-FLAG antibodies. C. Bam subcomplexes lacking the lipoproteins demonstrate activity equal to that of the complete complex in assembling full-length FLAG-BamA. (U: unfolded FLAG-BamA, F: folded FLAG-BamA)
We found that no specific Bam lipoprotein is required to assemble BamA in vitro; BamAB and BamACDE subcomplexes both assembled BamA into proteoliposomes (Figure 1C and S2A).17 The BamAB subcomplex is less effective, but it appears that BamB can at least partially substitute for BamCDE. Clearly, BamD is essential in vivo while BamB is not; therefore, these proteins must have additional non-redundant functions that may relate to the assembly of other OMP substrates. Given that BamA is the only common component in the subcomplexes, we examined whether the functions of the lipoproteins are critical in the assembly mechanism or whether BamA alone is capable of assembling more BamA. We compared the activities of proteoliposomes containing just BamA or BamCDE to that of BamACDE proteoliposomes to determine if BamA functions cooperatively with the lipoproteins (Figures 2A and S2B). Surprisingly, unfolded BamA substrate assembled into proteoliposomes containing only the BamCDE lipoproteins more efficiently than into proteoliposomes containing only BamA. Therefore, the assembly of an unfolded BamA molecule does not require a preassembled BamA molecule in the membrane.
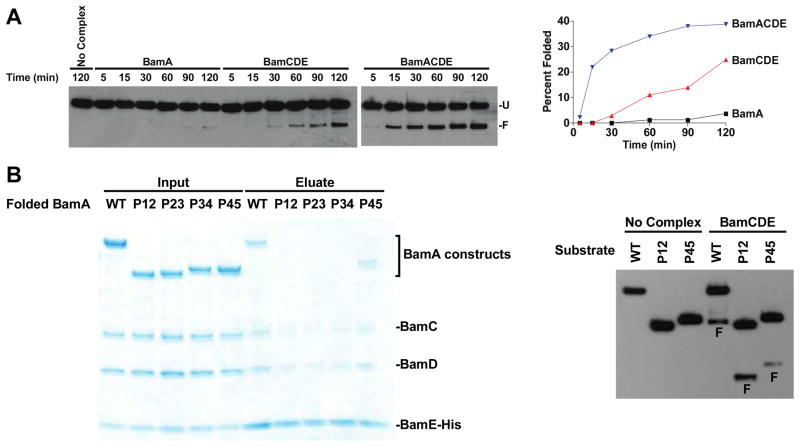
Figure 2.
Bam lipoproteins are sufficient to facilitate BamA assembly. A. The BamCDE lipoproteins facilitate FLAG-BamA assembly more effectively than BamA alone, but BamA and BamCDE function most effectively as a complex. FLAG-BamA assembly into proteoliposomes containing BamA, BamCDE, or BamACDE was monitored over the course of two hours. Reactions were stopped at the indicated time points. Folding yields were determined by comparing the densities of the folded and unfolded bands and plotted. B. The chimeric BamAP12 substrate does not bind to BamCDE after it is folded (left panel), but these lipoproteins do facilitate its assembly (right panel). The FLAG-tagged wild-type, chimeric (P12, P23, P34), and truncated (P45) BamA substrates were folded in detergent (0.5% LDAO) for 60 minutes; purified BamCDE-His6 lipoproteins were then added, and the complexes were isolated by Ni-NTA affinity chromatography (left). Unfolded FLAG-tagged BamA substrates were added to empty liposomes or BamCDE proteoliposomes, and the folding reactions were stopped after 120 minutes (right). (F: folded FLAG-BamA construct)
We considered an alternate explanation for the observed folding in the BamCDE proteoliposomes in which the folded BamA product might form a complex with BamCDE and thereby produce a more active assembly machine. We discounted this hypothesis because a BamA substrate that cannot bind to the lipoproteins after it is folded is assembled equally well by them (Figures 2B and S3). The periplasmic region of BamA contains five polypeptide transport associated (POTRA) domains. The most C-terminal of these, P5, is adjacent to the β-barrel and known to bind to BamCDE.7 We generated chimeric and truncated BamA substrates containing two POTRA domains and the β-barrel domain. When the truncated substrate containing POTRA domains 4 and 5 is folded in detergent and then mixed with the BamCDE lipoproteins, it can be co-purified with the lipoproteins; the chimeric BamA proteins containing other pairs of POTRA domains (P12, P23, or P34) do not co-purify with the lipoproteins. Nevertheless, the BamCDE lipoproteins facilitate the assembly of the chimeric BamAP12. We therefore favor a model in which the lipoproteins directly and independently facilitate the assembly of the unfolded BamA substrate (vide infra).
In vivo experiments have indicated that BamA assembly is facilitated by the Bam complex.7, 24 Our results do not contradict those studies; the fact that the BamACDE complex was more active than BamCDE clearly demonstrates that when BamA is preassembled in a complex with the lipoproteins it facilitates the assembly of more BamA. Given the difference in the kinetics of the BamABCDE and BamCDE catalyzed processes, the latter is unlikely to occur in a wild-type cell. However, by dissecting the Bam complex in vitro we were able to observe the independent function of the BamCDE lipoproteins. It seemed possible that the lipoproteins might perform this same function in the context of the complete complex, and we therefore began to characterize how they assemble BamA in order to understand their mechanistic role.
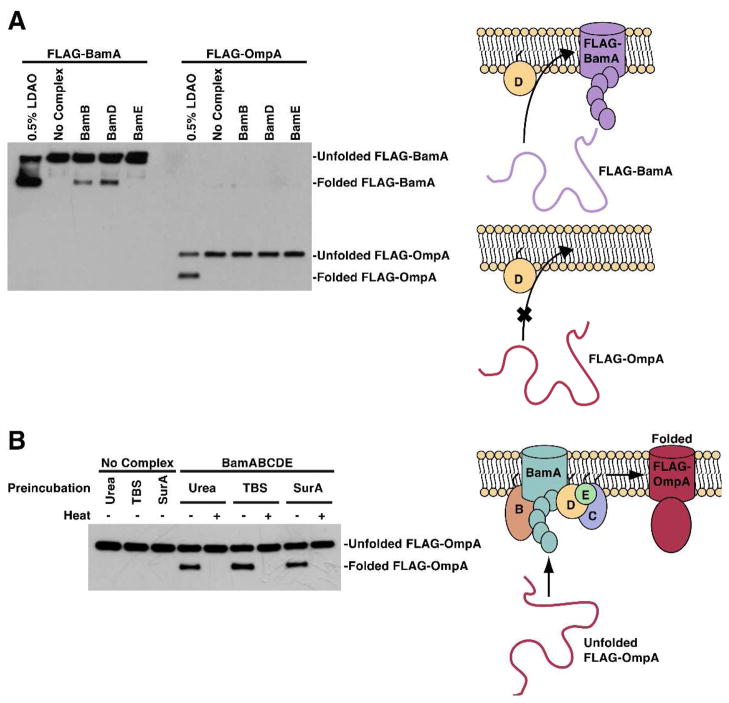
We examined whether other OMPs can also be assembled by the lipoproteins alone or if BamA is unusual in this regard. We compared how the individual Bam lipoproteins affect the folding of BamA and OmpA, an abundant, but non-essential OMP (Figure 3). Individually BamB and BamD both facilitated BamA assembly in vitro, but BamE did not. (BamC could not be purified individually in a stable form and consequently was not examined.) Clearly, the lipoproteins must have an important, direct effect because BamA does not assemble efficiently into empty liposomes or into BamE proteoliposomes. BamB and BamD must share a common function that has a specific effect on the BamA substrate. In contrast, none of the lipoproteins was sufficient to fold OmpA, but this substrate was assembled if the Bam complex was present in the membrane (Figures 3 and S4).26 Therefore, OmpA is not inherently unstable in the membranes used here, and we attribute the difference the ability of BamA and OmpA to assemble to a difference in the properties of the substrates. BamB and BamD have a clear function in the folding process, but it is not sufficient to assemble OmpA.
Figure 3.
BamB and BamD are individually sufficient to facilitate BamA’s self-assembly, but are not sufficient to assemble OmpA. A. Unfolded FLAG-BamA or FLAG-OmpA was added to a detergent solution or to proteoliposomes containing BamB, BamD, or BamE. Reactions were stopped after 120 minutes. B. OmpA is assembled by the complete Bam complex without a chaperone. FLAG-OmpA was added directly to proteoliposomes or preincubated in TBS or a ten-fold excess of SurA as in the experiment in Figure 1B.
We hypothesized that the common function of BamB and BamD relates to binding substrates at the OM as has been suggested by some crystal structures and cross-linking experiments.27–33 We examined whether the lipoproteins interact directly with unfolded OMPs; urea-denatured BamA or OmpA was mixed with an excess of soluble, His-tagged BamB, D, or E and then affinity purified. Soluble constructs of BamB, D, and E, which lack their N-terminal lipids, were used so that detergents could be omitted from the experiment to prevent folding of the substrates. BamA co-purified with BamD-His, to a lesser extent with BamB-His, and not at all with BamE-His (Figure 4). Therefore, the lipoproteins likely facilitate the assembly of the BamA substrate by binding to its unfolded state. In that respect, they may act like other enzymes by stabilizing a transient or intermediate state in the reaction pathway, and accordingly, their function in assembling the substrate does not involve or require binding to the final product (the folded state of BamA) as indicated by the assembly of the BamAP12 substrate described above.
Figure 4.
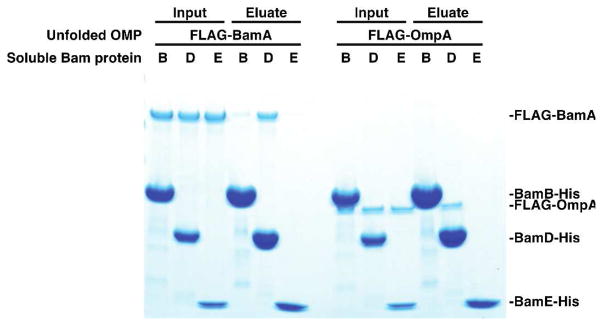
Certain Bam lipoproteins bind unfolded BamA and OmpA. Urea-denatured FLAG-BamA or FLAG-OmpA was diluted into a detergent-free solution containing a 10-fold molar excess of the indicated soluble, His-tagged Bam protein (lacking its N-terminal lipid anchor). Binding of the unfolded OMP to the Bam protein was then assessed by its ability to co-purify by Ni-NTA affinity chromatography.
However, this direct interaction between the unfolded substrate and the lipoproteins is not sufficient to produce folding on its own. When soluble BamB or BamD was added to empty liposomes, no folding of BamA was observed (Figure S5)—implying that the membrane localization or orientation of the lipoproteins matters. Furthermore, BamD bound unfolded OmpA, but, in this case, binding was not sufficient to produce folding of the substrate OMP (Figures 3A and 4). OmpA does not partition into the membrane even if it is bound near it. BamB and BamD are not capable of completing OmpA’s assembly alone, but simply binding unfolded BamA in proximity to the membrane is sufficient to facilitate this substrate’s assembly. BamA may be unusual or unique in its ability to assemble independent of other components. We were able to exploit this property of BamA to observe the individual functions of BamB and BamD in vitro. This is an advantage of our in vitro system in that it enables dissection of an essential machine; we do not have to contend with the pleiotropic effects of mutations and deletions in vivo and can isolate the effects of individual components.
DISCUSSION
Here we have shown that BamB and BamD can bind unfolded substrates and that this function facilitates the assembly of BamA. These lipoproteins likely interact with the unfolded substrate in different ways, but both are capable of facilitating BamA assembly by localizing the unfolded substrate to the membrane. Although BamD can also bind unfolded OmpA, it is not sufficient to catalyze the assembly of this substrate in vitro. We attribute this difference in assembly requirements to the function of BamA. BamA may be able to assemble aided only by the lipoproteins because it performs some of the OMP assembly mechanism; the fact that it is conserved in all organisms may reflect its role in the later folding and insertion steps of β-barrel assembly. BamD alone cannot assemble OmpA because this substrate relies on a preassembled BamA to complete the later steps of its assembly.
Many OMPs have been shown to assemble spontaneously into lipid bilayers in vitro, but their ability to do so depends strongly on the lipid content of the artificial membranes.26, 34, 35 In vivo, however, all OMPs must assemble into the same membrane. The inability of OmpA to assemble under the same conditions as BamA (i.e. into a membrane containing only BamB or BamD) suggests that the BamA substrate may possess some additional or unusual features. We propose that the structure of BamA in some way facilitates its own assembly such that it is less reliant on a preassembled Bam complex than other OMPs. This spontaneous assembly process is clearly much less efficient than the Bam complex catalyzed process, but it provides an intriguing solution to the “chicken and egg problem.” Perhaps in a primitive organism, an ancestral BamA protein assembled itself, and the other complex components later evolved to adapt BamA to assemble more and different types of other OMPs. In turn, BamA became more reliant on the other Bam components for its assembly and the spontaneous process became comparatively less efficient and important. By creating an efficient catalyst for β-barrel assembly, it also became possible to segregate β-barrels to a single membrane by making the rate of their assembly into the membrane containing the Bam complex dramatically faster than into an empty membrane.
By dissecting the Bam complex in vitro, we have identified a substrate binding interaction that appears to be important in BamA’s assembly. Our next step is to establish whether inhibition of this binding event is sufficient to inhibit OMP assembly.
Supplementary Material
Acknowledgments
Funding Sources
This work is supported by NIH grant AI081059.
ABBREVIATIONS
- Bam
β-barrel assembly machine
- OM
outer membrane
- IM
inner membrane
- OMP
outer membrane protein
- DDM
n-dodecyl-β-D-maltopyranoside
- TCEP
tris(2-carboxyethyl)phosphine
- SDS
sodium dodecylsulfate
- LDAO
lauryldimethylamine-N-oxide
- POTRA
polypeptide transport associated
Footnotes
The authors have no competing financial interests.
Experimental procedures and five additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 3.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 4.Hagan CL, Silhavy TJ, Kahne D. β-Barrel Membrane Protein Assembly by the Bam Complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 5.Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 8.Eggert US, Ruiz N, Falcone BV, Branstrom AA, Goldman RC, Silhavy TJ, Kahne D. Genetic basis for activity differences between vancomycin and glycolipid derivatives of vancomycin. Science. 2001;294:361–364. doi: 10.1126/science.1063611. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reumann S, Davila-Aponte J, Keegstra K. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: identification of a cyanobacterial homolog. Proc Natl Acad Sci USA. 1999;96:784–789. doi: 10.1073/pnas.96.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 13.Wiedemann N, Kozjak V, Chacinska A, Schonfisch B, Rospert S, Ryan MT, Pfanner N, Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- 14.Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- 15.Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel R, Hsu SC, Bedard J, Inoue K, Jarvis P. The Omp85-related chloroplast outer envelope protein OEP80 is essential for viability in Arabidopsis. Plant Physiol. 2008;148:235–245. doi: 10.1104/pp.108.122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagan CL, Kahne D. The reconstituted Escherichia coli Bam complex catalyzes multiple rounds of β-barrel assembly. Biochemistry. 2011;50:7444–7446. doi: 10.1021/bi2010784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouviere PE, Gross CA. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 21.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics. 2009;9:2432–2443. doi: 10.1002/pmic.200800794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tellez R, Jr, Misra R. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a bamB bamE strain of Escherichia coli. J Bacteriol. 2011;194:317–324. doi: 10.1128/JB.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denoncin K, Schwalm J, Vertommen D, Silhavy TJ, Collet JF. Dissecting the Escherichia coli periplasmic chaperone network using differential proteomics. Proteomics. 2012;12:1391–1401. doi: 10.1002/pmic.201100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel GJ, Kleinschmidt JH. The Lipid Bilayer-Inserted Membrane Protein BamA of Escherichia coli Facilitates Insertion and Folding of Outer Membrane Protein A from Its Complex with Skp. Biochemistry. 2013 doi: 10.1021/bi400103t. [DOI] [PubMed] [Google Scholar]
- 27.Noinaj N, Fairman JW, Buchanan SK. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J Mol Biol. 2011;407:248–260. doi: 10.1016/j.jmb.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Albrecht R, Zeth K. Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem. 2011;286:27792–27803. doi: 10.1074/jbc.M111.238931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heuck A, Schleiffer A, Clausen T. Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J Mol Biol. 2011;406:659–666. doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal Structure of BamD: An Essential Component of the β-Barrel Assembly Machinery of Gram-Negative Bacteria. J Mol Biol. 2011;409:348–357. doi: 10.1016/j.jmb.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J Mol Biol. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 32.Kim KH, Aulakh S, Paetzel M. Crystal structure of β-barrel assembly machinery BamCD protein complex. J Biol Chem. 2011;286:39116–39121. doi: 10.1074/jbc.M111.298166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter β domain. Proc Natl Acad Sci U S A. 2011;108:E383–391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinschmidt JH. Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers. Chem Phys Lipids. 2006;141:30–47. doi: 10.1016/j.chemphyslip.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 35.Burgess NK, Dao TP, Stanley AM, Fleming KG. β-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J Biol Chem. 2008;283:26748–26758. doi: 10.1074/jbc.M802754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.