Abstract
Lysobacter is a genus of Gram -negative gliding bacteria that are emerged as novel biocontrol agents and new sources of bioactive natural products. The bacteria are naturally resistant to many antibiotics commonly used in transformant selection, which has hampered the genetic manipulations. Here, we described a facile method for quick -and-easy identification of the target transformants from a large population of the wild type and non-target transformants. The method is based on a distinct yellow-to-black color change as a visual selection marker for site-specific integration of the gene of interest. Through transposon random mutagenesis, we identified a black-colored strain from the yellow-colored L. enzymogenes. The black strain was resulted from a disruption of hmgA, a gene required for tyrosine /phenylalanine metabolism. The disruption of hmgA led to accumulation of dark brown pigments. As proof of principle, we constructed a series of expression vectors for a regulator gene found within the WAP-8294A biosynthetic gene cluster. The yield of WAP-8294A in the black strains increased by 2 fold compared to the wild type. Interestingly, the yield of another antibiotic (HSAF) increased up to 7 fold in the black strains. WAP-8294A is a family of potent anti-MRSA antibiotics and is currently in clinical studies, and HSAF is an antifungal compound with distinct structural features and a novel mode of action. This work represents the first successful metabolic engineering in Lysobacter. The development of this facile method opens a way toward manipulating antibiotic production in the largely unexplored sources.
Keywords: Lysobacter, anti-MRSA antibiotics, WAP -8294A, nonribosomal peptides, metabolic engineering, site-specific gene integration
The genus Lysobacter is one of the most ubiquitous environmental microorganisms, belonging to the Xanthomonadaceae family within the gamma proteobacteria(1 –3). These Gram-negative gliding bacteria are drawing increased attention as novel biocontrol agents against pathogens of crop plants. For example, L. enzymogenes exhibits potent antifungal activity against a broad spectrum of pathogens, including Magnaporthe poae that causes Kentucky bluegrass summer patch, Pythium ultimum that causes sugar beet damping-off, Puccinia graminis that causes cereal crop rusts, Uromyces appendiculatus that causes bean plant rust, Bipolaris sorokiniana that causes turf grass leaf spots, and Rhizoctonia solani that causes brown patch on a wide range of plants (4–6). Our previous studies showed that L. enzymogenes produces a broad spectrum antifungal compound, HSAF (dihydromaltophilin), which is the critical factor of L. enzymogenes as a biocontrol agent against plant fungal diseases (7–10). In addition to the broad activities against fungal diseases, Lysobacter is emerging as a new source of bioactive natural products (11, 12). Several species of the genus are known to produce potent antibiotics, including the cyclic peptide lysobactin (13–15) and tripropeptins (16, 17), the cephem-typeβ-lactam antibiotic cephabacins (18–20), and the cyclic lipodepsi peptides WAP -8294A(21–24). Among them, the WAP-8294A family is particularly interesting because they exhibit a very potent activity against methicillin-resistant Staphylococcus aureus(MRSA), and one member of the family, WAP-8294A2, is now in clinical studies(25). However, WAP -8294 A compounds are produced by Lysobacter only under certain conditions with a very low yield (24). This is a bottleneck for structure-activity relationship studies and clinical tests.
As a relatively underexplored group of microorganisms, Lysobacter do not have many well developed genetic tools for gene manipulation. These bacteria are naturally resistant to most of the antibiotics commonly used in genetic selections, including ampicillin, kanamycin, streptomycin, tetracycline, and rifampin. This feature renders most of the selection marker genes useless when screening for Lysobacter transformants. Currently, gentamicin and chloramphenicol are the few antibiotics that can be used in Lysobacter transformant selection. Furthermore, no autonomous vector has been identified for Lysobacter, and chromosomal integration through homologous recombination is the only way for genetic engineering in these bacteria. A limiting step of this process is the initial selection of the putative transformants from a large number of colonies. Thus a facile method for quick- and-easy identifying the putative transformants is highly desirable in order to explore the potential of these prolific producers of antibiotics.
Here, we describe such a facile method for transformant identification. The method is based on a “yellow-to-black” color change for a visual selection of the putative transformants. We demonstrated the utility of this method through a site-specific integration of plasmid constructs that would express ORF8, a putative regulator gene found in the WAP -8294A gene cluster (24). ORF8 was predicted to encode a TonB -dependent receptor, which was also found in the HSAF gene cluster (26). Our results showed that the yield of WAP-8294A and HSAF in the ORF8-expressing strains increased 2 and 7 fold, respectively, compared to the wild type strain. This result not only demonstrated the usefulness of this new genetic tool, but also confirmed the positive regulatory role of the TonB-dependent receptor in the production of antibiotics in Lysobacter. The work also represents the first successful metabolic engineering of antibiotics in a Lysobacter strain.
RESULTS AND DISCUSSION
Identification and Characterization of the “Black Mutant” Generated by Transposon Random Mutagenesis
One of the characteristic features of L. enzymogenes OH11 is the yellowish appearance. Through transposon random mutagenesis(27, 28), we obtained a black (dark brown) mutant of the wild type strain. The subsequent sequencing of the transposon insertion site showed that the transposon was inserted into the hmgA gene, which is predicted to encode homogentisate 1, 2-dioxygenase. This dioxygenase catalyzes the oxidative cleavage of the aromatic ring of tyrosine/phenylalanine, which is a key step in aromatic amino acid degradation pathway (29, 30)(Figure 1). The disruption of hmgA gene blocks the oxidative cleavage reaction that leads to the accumulation of homogentisate. Further oxidations of homogentisate produce ochronotic pigments and render the black color in the organisms (29, 30).
Figure 1.
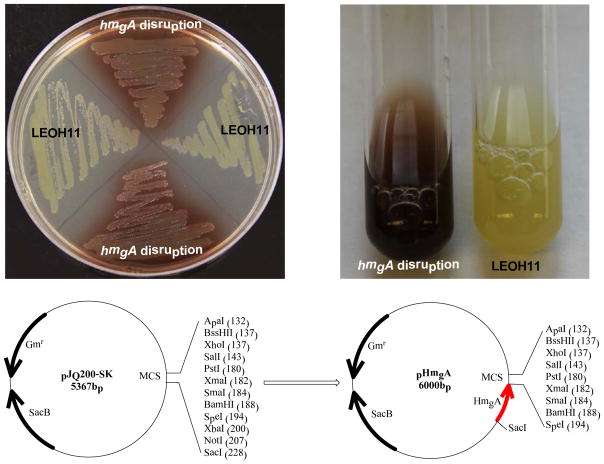
“Yellow-to-black” color change of Lysobacter enzymogenes OH11. The solid and liquid cultures of the wild type strain (LEOH11) and the hmgA integrated strains are shown at top, and the map of the plasmid construct containing the 633-bp fragment within hmgA that is cloned into pJQ200-SK is shown at bottom.
To verify this hmgA gene is responsible for the yellow-to-black color switch, we amplified a 633-bp fragment with in hmgA and cloned it into pJQ200 -SK(31), which generated the construct pHmgA. This construct was then introduced into L. enzymogenes OH11 by conjugation. The colonies in black color were picked up for PCR verification. The results showed that all the black colonies resulted from a disruption of the hmgA gene through homologous recombination between the pHmgA and the chromosomal copy of hmgA gene (Figure 1). As a control, the introduction of pJQ200-SK vector alone only produced yellow colonies. The results confirmed that hmgA is responsible for the color change. The black colonies did not show any obvious difference from the wild type in growth rate (approximately 1.2–1.4 of OD600nm in 24 hours) and other phenotypes. Thus, this pHmgA vector could be used for site-specific gene integration in L. enzymogenes, to facilitate a visual screening of putative transformants.
Construction of Broadly Adaptable Site-specific Gene Integration Vectors
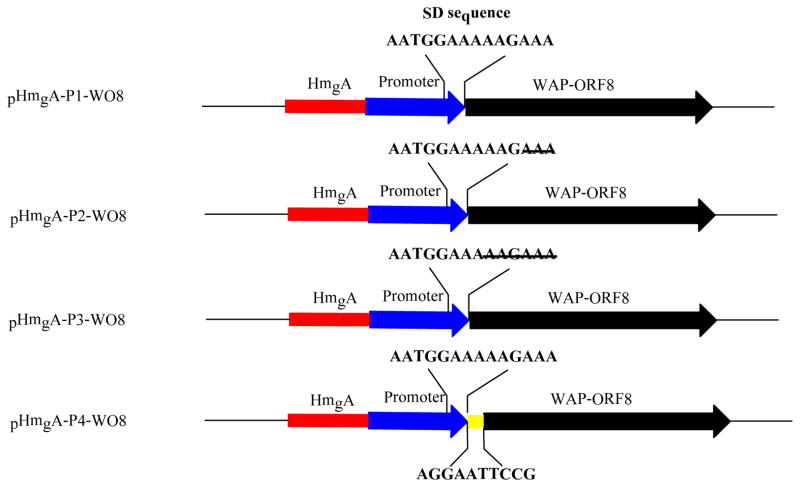
To demonstrate the utility of this quick-and-easy screening method, we constructed a series of vectors that are expected to express the target gene in Lysobacter. To do this, we first need to put a promoter into the pHmgA vector. Our previous study had indicated that the 5′-untranslated region of the key HSAF biosynthetic gene, encoding a polyketide synthase/nonribosomal peptide synthetase (PKS/NRPS), could be a good candidate(26). RT-PCR results showed that the PKS/NRPS gene was highly transcribed in the first three days of culture when the wild type strain was grown in 1/10 TSB medium, and the change of the transcription levels was consistent with the trend of HSAF production in the cultures. The bioinformatics analysis of the 5′-untranslated region suggested that this region may contain a putative promoter and a putative ribosomal binding site(Shine-Dalgarno sequence)(32). We subsequently amplified the 538-bp fragment and placed it behind hmgA in pHmgA vector, to generate pHmgA -P1 (Figure 2). This vector contains the natural SD sequence, AATGGAAAAAGAAA. In addition, we also generated a series of derivative vectors, with a modified SD sequence. pHmgA-P2 contains a shorter SD sequence, AATGGAAAAAG; pHmgA-P3 contains an even shorter SD sequence, AATGGAAA; pHmgA-P4 contains the putative SD sequence present in the 5′-untranslated region of ORF8, in addition to the promoter sequence of pHmgA -P1 (Figure 2). After sequencing the four vectors to confirm their identity, we individually transferred them into the wild type L. enzymogenes OH11. On the selection plates, approximately fifty percent of single colonies showed the black color. The phenotype is consistent with the expectation, because the se vectors, pHmgA -P1/P2/P3/P4, contain two homologous fragments, the hmgA fragment and the HSAF promoter fragment. Only the mutants resulted from the homologous recombination at the hmgA fragment would give the black color phenotype. The result showed all of the four vectors could be used for color screening.
Figure 2.
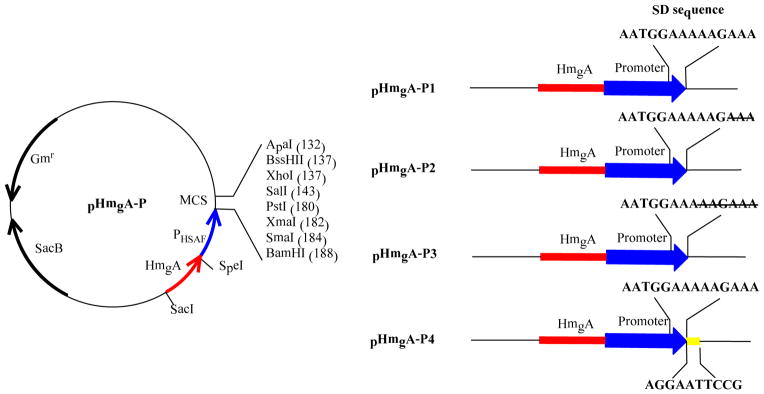
Construction of broadly adaptable vectors for site -specific gene integration in Lysobacter. The 538-bp promoter (PHSAF), from the highly expressed HSAF biosynthetic gene cluster (26), was placed immediately behind hmgA in pHmgA vector, to generate pHmgA -P1. This vector contains the natural SD sequence, AATGGAAAAAGAAA. Three pHmgA-P1 derived vectors, with a modified SD sequence, are also shown. Note that pHmgA-P4 contains the putative SD sequence of ORF8(24), in addition to the promoter sequence of pHmgA -P1.
Production of the Anti-MRSA WAP-8294A2 in Lysobacter enzymogenes Expressing a TonB-dependent Receptor
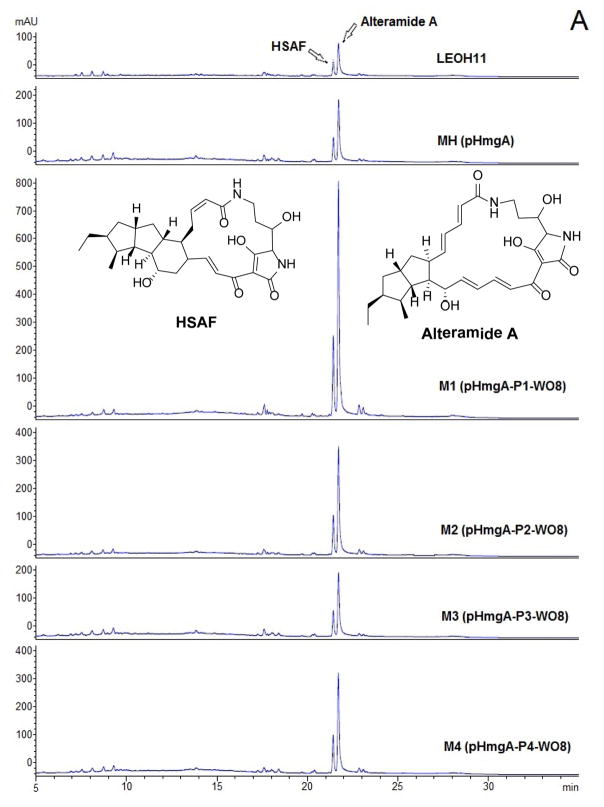
pHmgA -P1/P2/P3/P4 were constructed to express any target gene when it is placed under the control of the promoter sequence. To test the utility of the vectors and the color -based screening method, we placed the ORF8 gene of the WAP biosynthetic gene cluster behind the promoter in these vectors. ORF8 was predicted to encode a TonB-dependent receptor that positively regulates the production of WAP-8294A compounds (24). The gene was inserted behind the promoter of the pHmgA -P vectors to form pHmgA-P1-WO8, pHmgA-P2-WO8, pHmgA-P3-WO8, and pHmgA-P4-WO8, respectively (Figure 3). The vectors were introduced individually into the wild type through conjugation. Because there are three homologous fragments in each of the constructs, the vectors could be integrated into three potential sites. Mutant strain M1 (one black colony from approximately 60 yellow colonies) was obtained from the hmgA site integration of pHmgA-P1-WO8, strain M2 (one black colony from approximately 120 yellow colonies) from pHmgA -P2-WO8, strain M3 (two black colonies from approximately 60 yellow colonies) from pHmgA-P3-WO8, and strain M4 (one black colony from approximately 60 yellow colonies) from pHmgA-P4-WO8.
Figure 3.
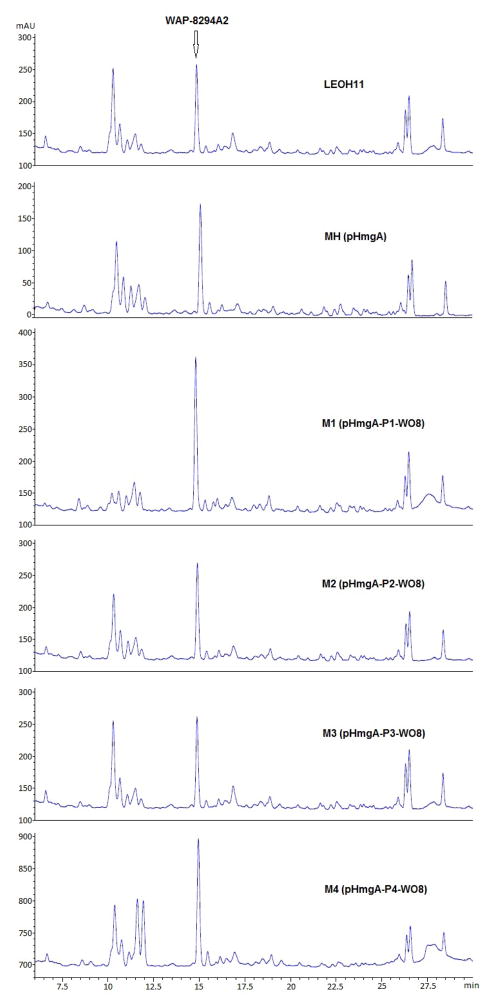
Construction of ORF8 expression vectors and integration of the constructs into the color-switching hmgA site of Lysobacter enzymogenes OH11 to improve the yield of the anti -MRSA antibiotics WAP-8294A. ORF8, the TonB-dependent receptor gene in the WAP-8294A biosynthetic gene cluster (24), was placed behind the promoter of vectors pHmgA -P1/P2/P3/P4, to form pHmgA-P1-WO8 (strain M1), pHmgA-P2-WO8(strain M2), pHmgA-P3-WO8(strain M3), and pHmgA-P4-WO8(strain M4), respectively. Black colonies (strain M1-M4) resulted from each of the constructs were selected, verified, cultured, and analyzed by HPLC and MS for WAP-8294A production. The wild type (LEOH11) and strain MH (pHmgA-alone integration) were used as controls.
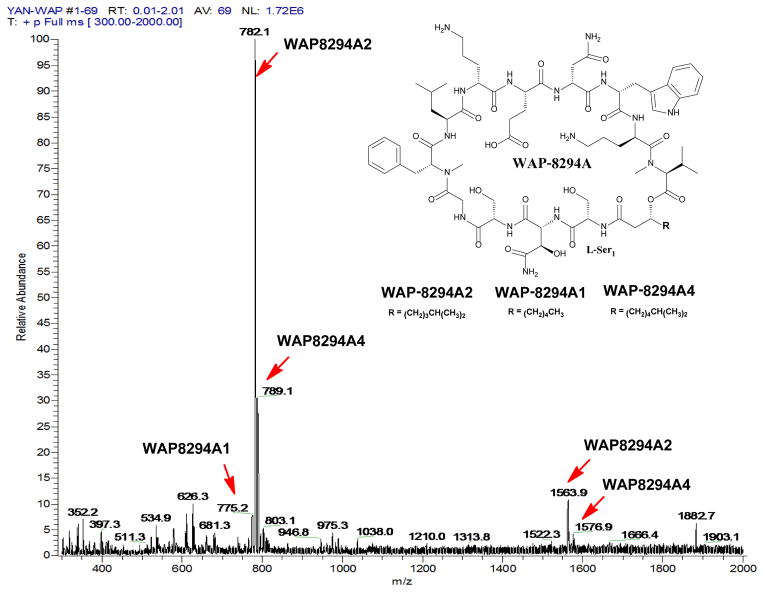
These black strains were analyzed for the production of WAP-8294A2. As a control, WAP-8294A2 produced in the wild type was first analyzed by HPLC. This compound had a retention time of 14.9 min (Figure 3). The strain containing HmgA-alone disruption (MH) produced a similar amount of WAP-8294A2 as the wild type. This indicates that the gene integration into the hmgA site does not impact the production of this antibiotic. The strain (M1) containing ORF8 under the control of the natural HSAF promoter, pHmgA -P1-WO8, produced 1.9-fold more WAP-8294A2 than the wild type (Figure 3). The strains (M2, M3, and M4) containing ORF8 under the control of a modified promoter also produced a slightly higher yield (1.2, 1.1, and 1.5 fold, respectively) than the wild type. The identity of WAP-8294A2 was confirmed by mass spectrometry and comparison with the standard compound. The results suggested that, among the tested, the natural HSAF promoter probably has the best effect on ORF8 gene expression in these Lysobacter strains and any modification, a shorter or extra SD sequence, would lead to an adversary effect.
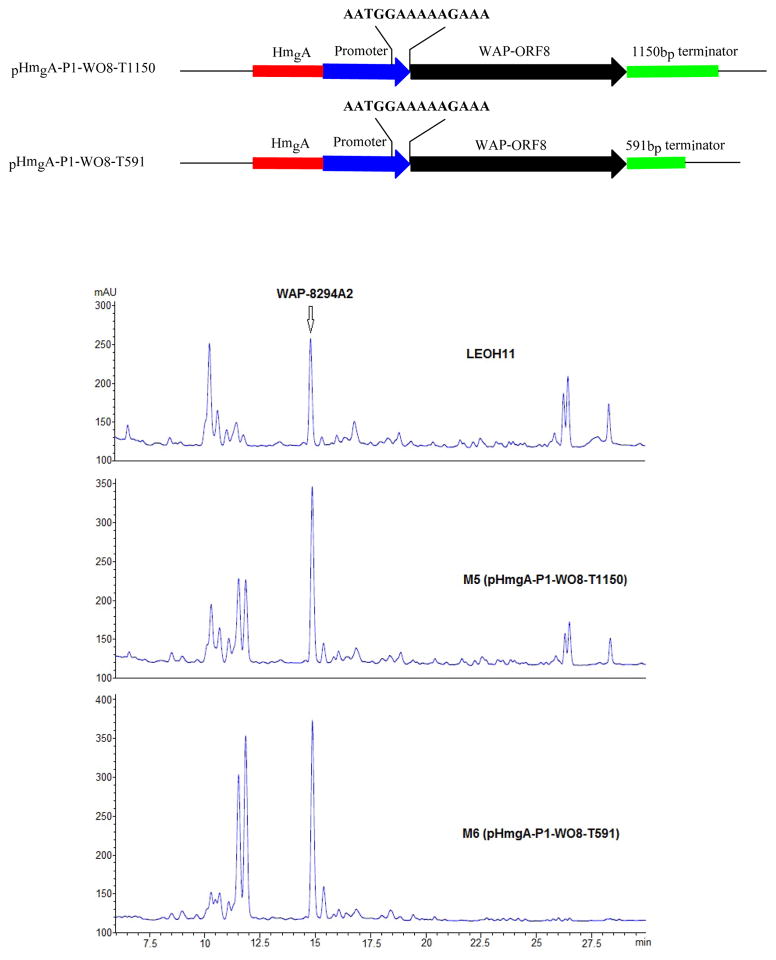
WAP-8294A Yield in Strains Expressing pHmgA-P1-WO8 with a Putative Terminator
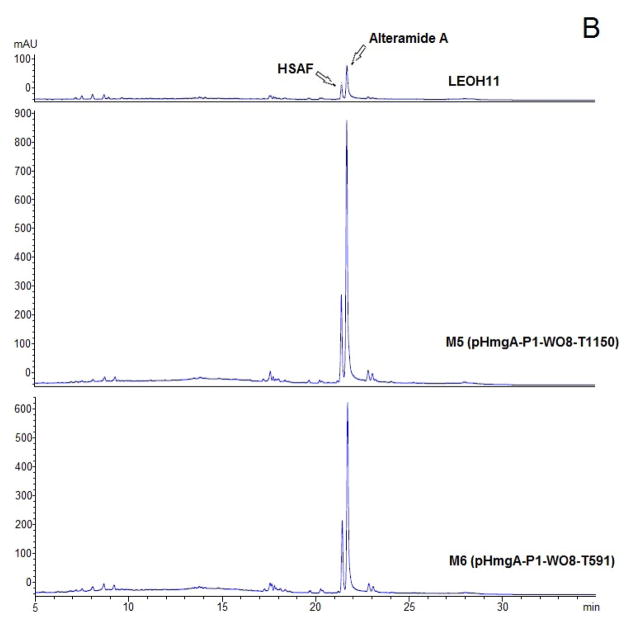
Since the strain containing ORF8 under the control of the natural HSAF promoter gave the best yield of WAP-8294A2, further optimization was based on this construct, pHmgA-P1-WO8. The DNA sequence at the end of a transcription unit could potentially contain controlling elements, such as a terminator, that are important for an optimized transcription. We subsequently generated two pHmgA-P1-WO8 derivative constructs, pHmgA-P1-WO8-T1150 and pHmgA-P1-WO8-T591, which contain a 1150-bp and 591-bp fragment, respectively, originated from the 3 ′-untranslated region of the HSAF biosynthetic genes. Each of the fragments was inserted immediately behind the ORF8 gene (Figure 4). The black colored strains resulted from the integration of the constructs, M5 (one black colony from approximately 80 yellow colonies) from pHmgA-P1-WO8-T1150 and M6 (three black colonies from approximately 80 yellow colonies) from pHmgA-P1-WO8-T591, were analyzed for WAP-8294A2 production. Strain M5 produced 1.9 times more than the wild type, while strain M6 produced 2.0 times more than the wild type (Figure 4).
Figure 4.
Optimization of ORF8 expression in Lysobacter enzymogenes OH11 to increase the production of WAP-8294A. pHmgA-P1-WO8-T1150 (strain M5) and pHmgA-P1-WO8-T591 (strain M6) were derived from pHmgA-P1-WO8 by inserting a 1150-bp and 591-bp fragment, respectively, of the 3′-untranslated region of the HSAF biosynthetic genes behind the ORF8 gene. The black colored strains resulted from the integration of the constructs into the hmgA site were analyzed for WAP-8294A production using HPLC.
HSAF Production in the ORF8-expressing Strains
A TonB -dependent receptor is also present in the HSAF biosynthesis gene cluster (26). To test whether the expression of the TonB-dependent receptor of the WAP biosynthetic gene cluster could also affect the production of HSAF, we analyzed the yield of HSAF in the ORF8-expressing strains. The wild type produced HSAF and its analog alteramide A (33), with the retention time of 21.5 and 21.8 min, respectively (Figure 5). Interestingly, the HmgA-alone disruption strain MH produced 2-fold more HSAF than the wild type. The ORF8-expressing strain pHmgA-P1-WO8 (M1) produced a significantly higher amount of HSAF, increased 6.1-fold, than the wild type. The ORF8-expressing strains with a modified SD sequence (M2, M3, and M4) had 3.1, 1.7, and 2.6-fold more HSAF. Adding the 1150-bp putative terminator to the ORF -8 expressing cassette resulted in an improved HSAF production, increased 7-fold in M5, but adding the 591-bp terminator (M6) did not have the same effect. The yield of HSAF in strain M6 was 5.2-fold of that in the wild type.
Figure 5.
Production of HSAF and its analog, alteramide A, in the ORF8 expression strains M1–M4 (A) and M5–M6 (B). The wild type (LEOH11) and strain MH (pHmgA-alone integration) were used as controls.
Summary and Concluding Remarks
The yellow-to-black color change provides a simple screening method for putative transformants. Recently, a color change-based method was also used to integrate genes into the fungus Aspergillus nidulans, which led to the discovery of cryptic polyketide metabolites (34). The pigment accumulation in Lysobacter did not have any obvious adversary effect on the organism. This is evident as the black colored strains showed the similar optical density as the yellow strains in the cultures and the engineered strains produced an increased amount of WAP-8294A2 and HSAF as expected. Thus, the hmgA locus provides a new means for site -specific gene integration in Lysobacter. Because of the naturally occurred resistance to many common antibiotics used in genetic selections, there are very few selection markers that can be used in genetic manipulation in Lysobacter. Our method is a visual screening that does not reply on an antibiotic selection marker gene. The color change in the colonies is very clear and distinct, which enables an easy identification of the target transformants even when the majority of the colonies are yellow (the wild type or transformants resulted from a random insertion of the construct). We demonstrated the utility of this method by constructing a series of vectors that express a regulator gene for the biosynthesis of WAP-8294A, a family of potent anti-MRSA antibiotics that are in clinical studies. In this proof of principle work, we were able to increase the yield of WAP-8294A2 by 2 times in strains that contain the regulator gene under the control of a promoter found in Lysobacter. In addition, we serendipitously found that the expression of the WAP TonB-dependent receptor led to a significant yield increase of another antibiotic, HSAF, which is produced by the same organism. One of the strains produced up to 7-fold more HSAF than the wild type. This finding suggests that the TonB-dependent regulatory mechanism may crosstalk between the WAP and the HSAF biosynthetic systems, as both gene clusters contain such a regulatory gene (24, 26).
Although species in Lysobacter exhibit a great potential as new sources of bioactive compounds, many of the compounds are produced only under very special conditions and often in a very low yield. The development of this facile screening method and the construction of the expression vectors open a way to ward solving these problems. These vectors contain varied promoters and multiple cloning sites, which facilitates further manipulations, such as the replacement with new promoters as well as new target genes.
METHODS
Bacterial Strains, Plasmids and General DNA Manipulations
Escherichia coli DH5α strain was used as the host for general DNA propagations. E. coli S17 –1 was used as the strain for conjugation. L. enzymogenes and other bacterial stains were grown in Luria Bertani (LB) broth or 1/10 strength tryptic soy broth (1/ 10 TSB, Sigma). Genomic DNA of L. enzymogenes was prepared as previously described (5). Plasmid mini-prep and DNA gel extraction were performed using kits from Qiagen. PCR primers were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). All other manipulations were carried out according to standard methods.
Construction of hmgA Site-specific Integration Vectors and Screening for hmgA Mutant
To construct the plasmid vectors for hmgA site -specific integration, a 633 -bp internal fragment was amplified, using the primer pairs HmgA-up and HmgA-down (Table 1), from the genomic DNA of the wild type L. enzymogenes OH11. The fragment was digested with SacI/SpeI and cloned into the site of the conjugation vector pJQ200SK to produce pHmgA. This construct was transferred into E. coli S17 –1, which was mated with L. enzymogenes OH11 for conjugal transfer of pHmgA. The black colonies grown on LB plates containing gentamicin (20 μg/ml) were picked up and inoculated into liquid cultures containing gentamicin. Genomic DNA was prepared from each of the black-colored cultures, and diagnostic PCR were performed to verify the mutants that resulted from homologous recombination.
Table 1.
The sequences of primers used in this study
| Primer | Sequence |
|---|---|
| HmgA-up | CGAGCTCAACGACTTCGGCGACGGC |
| HmgA-down | GGACTAGTGATCGACGGGTCGGGATG |
| p1-up | CGGGATCCTTTCTTTTTCCATTATCTTTCAG |
| p1-down | GGACTAGTCTGGGTGACTCTGCATGGA |
| p2-up | CGGGATCCCTTTTTCCATTATCTTTCAGA |
| p2-down | GGACTAGTCTGGGTGACTCTGCATGGA |
| p3-up | CGGGATCCTTTCCATTATCTTTCAGAAAATG |
| p3-down | GGACTAGTCTGGGTGACTCTGCATGGA |
| WO8-up | CGGGATCCATGAAGCAAACGCGCGGC |
| WO8-down | CCGCTCGAGTCAGAACTTGTAGCGCACCCC |
| WO8-p4-up | CGGGATCCAGGAATTCCGATGAAGCAAACGCGC |
| WO8-P4-down | CCGCTCGAGTCAGAACTTGTAGCGCACCCC |
| T591-up | CCGCTCGAGTCAGAACTTGTAGCGCACCCC |
| T591-down | CGGACCGGGCCCTCGCCCTTCTTCAGGTCG |
| T1150-up | CCGCTCGAGGCTGGCGAGACGAGTCACCA |
| T1150-down | CGGACCGGGCCCACCCCGAGCCTTGAGCGA |
| verify-up | TTCTCGCCGTTCGCCCAG |
| verify-down | CTTCCATGCAGAGTCACCCA |
Construction of Promoter-containing pHmgA-P Vectors
The promoter of the PKS/NRPS gene in HSAF biosynthetic gene cluster was amplified from the genomic DNA of the wild type strain. Three pairs of primers were used to generate three versions of promoter differing at the SD sequence. Primers p1-up and p1-down, p2-up and p2-down, and p3-up and p3-down (Table 1), were used to amplify the promoters to produce pHmgA -P1, pHmgA-P2, and pHmgA-P3, respectively. The PCR fragments were digested with SpeI/Bam HI and inserted into the same sites of vector pHmgA. This placed the promoter right behind the hmgA gene in pHmgA vector. The constructs were validated by DNA sequencing and transferred into L. enzymogenes OH11 by conjugation. The black colored colonies were selected and verified by diagnostic PCR.
Construction of WAP-ORF8 Expression Vectors
The coding region of WAP -ORF8 was amplified, using primers WO8-up and WO8-down (Table 1), from the genomic DNA of the wild type strain. The PCR product was digested with BamHI/XhoI and cloned into the same sites of the vectors pHmgA -P1, pHmgA-P2, and pHmgA-P3, respectively, to generate pHmgA-P1-WO8, pHmgA-P2-WO8, and pHmgA-P3-WO8. The PCR product amplified by primers WO8 -p4-up and WO8-p4-down was used to construct pHmgA-P4-WO8, which was generated by ligating the PCR fragment with the Bam HI/XhoI digested vector pHmgA -P1. In each the constructs, the coding region of ORF8 was placed right behind the respective promoter. To add the putative terminator into the expression constructs, primers T591-up and T591-down and primers T1150 -up and T1150-down were used to amplify a 591-bp and 1150-bp fragments, respectively, from the genomic DNA. The fragments were digested with XhoI/ApaI and cloned into the same sites of pHmgA -P1-WO8, to produce pHmgA-P1-WO8-T591 and pHmgA-P1-WO8-T1150. The constructs were validated and transferred into L. enzymogenes OH11 by conjugation. The black colored colonies were selected and verified by diagnostic PCR.
Extraction and Analysis of WAP-8294A
Various strains of L. enzymogenes OH11 were grown in 1/10 strength TSB medium for 24 hours, and an aliquot of 2 ml was transferred to a 250 ml sterile flask containing 50 ml of fermentation medium (2.5% glucose, 2% soybean, 0.4% soybean oil, 0.25% NaCl, 0.5% CaCO3, pH 7.2). The culture was incubated at 28°C for 3 days with shaking at 200 rpm. To extract WAP compounds, the 50 ml culture broth was centrifuged, and the pH of the supernatant was adjusted to 2.5 with 1N HCl. The treated supernatant was then extracted with n-butanol/ethyl acetate (1/1, vol) containing 0.05% TFA. The organic phase was dried using a Rotavapor, and the residues were re-dissolved in 2 ml methanol containing 0.05% TFA. A 50 μl aliquot of each extracts was analyzed by HPLC (Agilent, 1220 Infinity LC) using a reversed-phase column (Cosmosil, 4.6 ID × 250 mm). Water/0.05% TFA (solvent A) and acetonitrile/methanol (1/1, vol)/0.05% TFA (solvent B) were used as the mobile phases with a flow rate of 1.0 ml/min. HPLC program was as follows: 57% B in A in the first 5 min, 57–100% Bin 5 –32 min, 100% B in 32–40 min, back to 57% B at 41 min and maintained to 48 min. The metabolites were detected at 280 nm on a UV detector. LCQ-MS was used to verify the mass of the peak of WAP-8294A2 and analogs
Extraction and Analysis of HSAF
Various strains of L. enzymogenes OH11 were grown in 1/10 strength TSB medium for 24 hours, and an aliquot of 2 ml was transferred to a 250 ml sterile flask containing 50ml of fresh 1/10 strength TSB medium. The culture was incubated at 28°C for 3 days with shaking at 200 rpm. To extract HSAF, the 50 ml culture broth was centrifuged, and the pH of supernatant was adjusted to 2.5 with 1N HCl. The treated supernatant was then extracted with n-butanol/ethyl acetate (1/1, vol) containing 0.05% TFA. The organic phase was dried using a Rotavapor, and the residues were re-dissolved in 2 ml methanol containing 0.05% TFA. A 50 μl aliquot of each extracts was analyzed by HPLC (Agilent, 1220 Infinity LC) using a reversed-phase column (phenomenex, 150 ×4.6 mm, 3micron). Water/0.025% TFA (solvent A) and acetonitrile/0.025% TFA (solvent B) were used as the mobile phases with a flow rate of 1.0 ml/min. HPLC program was as follows: 5–25% B in A in the first 0–10 min, increased to 80% B at 25 min, to 100% at 26 min, and back to 5% B at 30 min. The metabolites were detected at 280 nm on a UV detector. LCQ-MS was used to verify the mass of the peak of HSAF and analogs.
Acknowledgments
This work was supported in part by the NIH (R01AI097260), Nebraska Research Initiatives, and National High Technology Research and Development Program of China (2011AA10A205). We thank Ron Cerny and Kurt Wulser for technique assistance.
References
- 1.Christensen P, Cook FD. Lysobacter, a new genus of non-fruiting, gliding bacteria with a high base ratio. Int J Syst Bact. 1978;28:367–393. [Google Scholar]
- 2.Sullivan RF, Holtman MA, Zylstra GJ, White JF, Kobayashi DY. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J Appl Microbiol. 2003;94:1079–1086. doi: 10.1046/j.1365-2672.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- 3.Reichenbach H. The genus Lysobacter. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. Prokaryotes. Springer; New York: 2006. pp. 939–957. [Google Scholar]
- 4.Kobayashi DY, Yuen GY. The role of clp-regulated factors in antagonism against Magnaporthe poae and biological control of summer patch disease of Kentucky bluegrass by Lysobacter enzymogenes C3. Can J Microbiol. 2005;51:719–723. doi: 10.1139/w05-056. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi DY, Reedy RM, Palumbo JD, Zhou JM, Yuen GY. A clp gene homologue belonging to the Crpgene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl Environ Microbiol. 2005;71:261–269. doi: 10.1128/AEM.71.1.261-269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuen GY, Steadman JR, Lindgren DT, Schaff D, Jochum CC. Bean rust biological control using bacterial agents. Crop Protection. 2001;20:395–402. [Google Scholar]
- 7.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother. 2007;51:64–72. doi: 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Jochum CC, Yu F, Zaleta-Rivera K, Du L, Harris SD, Yuen GY. An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopath. 2008;98:695–701. doi: 10.1094/PHYTO-98-6-0695. [DOI] [PubMed] [Google Scholar]
- 9.Yuen GY, Jochum CC, Lindgren DT. Evaluation of the antibiotic HSAF from Lysobacter enzymogenes C3 for bean rust control, 2005 and 2006. Plant Dis Manag Rep. 2007;1(162) [Google Scholar]
- 10.Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nett M, Konig GM. The chemistry of gliding bacteria. Nat Prod Rep. 2007;24:1245–1261. doi: 10.1039/b612668p. [DOI] [PubMed] [Google Scholar]
- 12.Xie Y, Wright S, Shen Y, Du L. Bioactive natural products from Lysobacter. Nat Prod Rep. 2012;19:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Sullivan J, McCullough JE, Tymiak AA, Kirsch DR, Trejo WH, Principe PA. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. I. Taxonomy, isolation and partial characterization. J Antibiot (Tokyo) 1988;41:1740–1744. doi: 10.7164/antibiotics.41.1740. [DOI] [PubMed] [Google Scholar]
- 14.Bonner DP, O’Sullivan J, Tanaka SK, Clark JM, Whitney RR. Lysobactin, a novel antibacterial agent produced by Lysobacter sp. II. Biological properties. J Antibiot (Tokyo) 1988;41:1745–1751. doi: 10.7164/antibiotics.41.1745. [DOI] [PubMed] [Google Scholar]
- 15.Hou J, Robbel L, Marahiel MA. Identification and characterization of the lysobactin biosynthetic gene cluster reveals mechanistic insights into an unusual termination module architecture. Chem Biol. 2011;18:655–664. doi: 10.1016/j.chembiol.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Hashizume H, Igarashi M, Hattori S, Hori M, Hamada M, Takeuchi T. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. I. Taxonomy, isolation and biological activities. J Antibiot (Tokyo) 2001;54:1054–1059. doi: 10.7164/antibiotics.54.1054. [DOI] [PubMed] [Google Scholar]
- 17.Hashizume H, Hirosawa S, Sawa R, Muraoka Y, Ikeda D, Naganawa H, Igarashi M. Tripropeptins, novel antimicrobial agents produced by Lysobacter sp. J Antibiot (Tokyo) 2004;57:52–58. doi: 10.7164/antibiotics.57.52. [DOI] [PubMed] [Google Scholar]
- 18.Ono H, Nozaki Y, Katayama N, Okazaki H. Cephabacins, new cephem antibiotics of bacterial origin. I. Discovery and taxonomy of the producing organisms and fermentation. J Antibiot (Tokyo) 1984;37:1528–1535. doi: 10.7164/antibiotics.37.1528. [DOI] [PubMed] [Google Scholar]
- 19.Harada S, Tsubotani S, Ono H, Okazaki H. Cephabacins, new cephem antibiotics of bacterial origin. II. Isolation and characterization. J Antibiot (Tokyo) 1984;37:1536–1545. doi: 10.7164/antibiotics.37.1536. [DOI] [PubMed] [Google Scholar]
- 20.Sohn YS, Nam DH, Ryu DD. Biosynthetic pathway of cephabacins in Lysobacter lactamgenus: molecular and biochemical characterization of the upstream region of the gene clusters for engineering of novel antibiotics. Metab Eng. 2001;3:380–392. doi: 10.1006/mben.2001.0200. [DOI] [PubMed] [Google Scholar]
- 21.Kato A, Nakaya S, Ohashi Y, Hirata H. WAP-8294A(2), a novel anti-MRSA antibiotic produced by Lysobacter sp. J Am Chem Soc. 1997;119:6680–6681. [Google Scholar]
- 22.Kato A, Nakaya S, Kokubo N, Aiba Y, Ohashi Y, Hirata H, Fujii K, Harada K. A new anti-MRSA antibiotic complex, WAP-8294A. I. Taxonomy, isolation and biological activities. J Antibiot (Tokyo) 1998;51:929–935. doi: 10.7164/antibiotics.51.929. [DOI] [PubMed] [Google Scholar]
- 23.Harad KI, Suzuki M, Kato A, Fujii K, Oka H, Ito Y. Separation of WAP-8294A components, a novel anti-methicillin-resistant Staphylococcus aureus antibiotic, using high-speed counter-current chromatography. J Chromatogr A. 2001;932:75–81. doi: 10.1016/s0021-9673(01)01235-3. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Li Y, Qian G, Wang Y, Chen H, Li YZ, Liu F, Shen Y, Du L. Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP -8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob Agents Chemother. 2011;55:5581–5589. doi: 10.1128/AAC.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirri G, Giuliani A, Nicoletto SF, Pizzuto L, Rinaldi AC. Lipopeptides as anti-infectives: a practical perspective. Centr Eur J Biol. 2009;4:258–273. [Google Scholar]
- 26.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiang SL, Mekalanos JJ. rfb mutations in Vibrio cholerae do not affect surface production of toxin-coregulated pili but still inhibit intestinal colonization. Infect Immun. 1999;67:976–980. doi: 10.1128/iai.67.2.976-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Qian G, Li Y, Wright S, Shen Y, Liu F, Du L. Biosynthetic mechanism for sunscreens of the biocontrol agent Lysobacter enzymogenes. PLoS One. 2013;8:e66633. doi: 10.1371/journal.pone.0066633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milcamps A, de Bruijn FJ. Identification of a novel nutrient-deprivation-induced Sinorhizobium melilotigene (hmgA) involved in the degradation of tyrosine. Microb. 1999;145(Pt 4):935–947. doi: 10.1099/13500872-145-4-935. [DOI] [PubMed] [Google Scholar]
- 30.Arias-Barrau E, Olivera ER, Luengo JM, Fernandez C, Galan B, Garcia JL, Diaz E, Minambres B. The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J Bacteriol. 2004;186:5062–5077. doi: 10.1128/JB.186.15.5062-5077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 32.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 33.Shigemori H, Bae MA, Yazawa K, Sasaki T, Kobayashi J. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonassp. associated with the marine sponge Halichondria okadai. J Org Chem. 1992;57:4317–4320. [Google Scholar]
- 34.Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, White TC, Tang Y. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth Biol ASA. 2013 doi: 10.1021/sb400048b. [DOI] [PMC free article] [PubMed] [Google Scholar]