Abstract
Background. Mutations in NPM1 and FLT3 genes represent the most frequent genetic alterations and important diagnostic and prognostic indicators in patients with acute myeloid leukemia (AML). Objective. We investigated the prevalence and clinical characteristics of NPM1 and FLT3 mutations in 161 patients of de novo AML including adults and children. Results. NPM1 mutation was found in 21% and FLT3 mutation in 25% of the AML patients. Thirteen (8%) samples were positive for both NPM1 and FLT3/ITD mutations. Adult patients had significantly higher frequency of NPM1 mutation than children (25.8% versus 8.8%; P = 0.02). Further, NPM1 mutation was found to be more frequent in patients above 45 years of age (P = 0.02). NPM1 mutation was significantly associated with higher platelet count (P = 0.05) and absence of hepatosplenomegaly (P = 0.01), while FLT3/ITD mutation was associated with higher white blood count (P = 0.01). Immunophenotypically, NPM1 mutation was associated with the lack of CD34 (P < 0.001) and HLD-DR expression (P < 0.001), while FLT3/ITD mutation was positively associated with the expression of CD7 (P = 0.04). No correlation was found between NPM1 mutation and fusion gene. Interestingly, FLT3/ITD mutation was found to be inversely associated with AML/ETO fusion gene (P = 0.04). Conclusions. The results suggest that distinct clinical and immunophenotypic characteristics of NPM1 and FLT3/ITD mutations present further insight into the molecular mechanism of leukemogenesis.
1. Introduction
Acute myeloid leukemia (AML) develops from malignant transformation of immature hematopoietic cells through a complex multistep process that requires cooperation of different types of genetic alterations [1]. Alterations associated with AML include recurrent fusion genes such as t(8; 21), inv(16), and t(15; 17) that affect transcription factors or components of the transcriptional coactivation complex, resulting in impaired differentiation and/or aberrant self-renewal capacity by hematopoietic progenitors. In addition to these, a group that comprises mutation in genes involved in the proliferation and survival potential of hematopoietic progenitors includes receptor tyrosine kinases (FLT3, KIT, and RAS), oncogenic transcription factor (CEBP/alpha), and nucleophosmin (NPM1) mutation. Many of these gene mutations figure importantly as a provisional clinicopathologic entity in the revised World Health Organization (WHO) 2008 classification of myeloid neoplasms [2].
Fms-like tyrosine kinase-3 (FLT3) is a class III receptor tyrosine kinase located on chromosome 13q12. Wild-type FLT3 mRNA is expressed on normal hematopoietic stem cells and in the majority of AML blast cells [3]. The most common mutation in the FLT3 gene is internal tandem duplication (FLT3/ITD) of the region coding for the juxtamembrane (JM) domain of the FLT3 receptor [4]. The mutated FLT3 gene is always transcribed in frame and coded mutant FLT3 with a long JM domain. Another mutation reported is missense mutation in the tyrosine kinase domain (TKD) of the FLT3 gene. This mutation also promotes constitutive phosphorylation of the receptor [5]. The most frequent mutations are point mutations and deletions of codons 835/836 in second TKD of the gene.
In addition, a frameshift mutation in exon 12 of the NPM1 that interferes with cell cycle regulation has also been reported to occur frequently in AML [6]. This mutation is an alternative leukemogenetic mechanism rather than chromosomal translocations that was discovered in 2005. Due to its unique molecular, genotypic, immunophenotypic, and prognostic features, AML with mutated NPM1 and another category of AML with CEBPA were included as two separate provisional entities in the 2008 WHO classification of myeloid neoplasm [2]. In leukemia and lymphoma, NPM1 is also a partner in chromosomal translocations, where the dislocated cytoplasmic nucleophosmin appears to contribute to oncogenesis. NPM1 is a nucleus-cytoplasm shuttling protein that is ubiquitously expressed and is highly conserved. The NPM1 protein has been shown to contribute to many basic cellular processes such as the regulation of centrosome function, biosynthesis of ribosomes, and preventing aggregation of proteins in the nucleolus, and it participates in (ARF-P53) tumor suppressor pathway [7].
To date, there are only few studies specifically investigating the prevalence and clinical characteristic of NPM1 and FLT3 mutations in AML in Indian population. Ahmad et al., 2009, reported lower frequency of NPM1 mutation in Indian population compared to western countries but did not study the clinical relevance of NPM1 mutation in AML patients [8]. Another study reported no difference in the CR rate of patients with and without NPM1 mutation though it found poor prognosis for FLT3/ITD mutation in AML patients [9]. Moreover, in our previous study on FLT3 mutation no association with the clinical outcome in AML patients with normal cytogenetic was reported [10]. The study presented here investigates the prevalence and clinical characteristics of the exon 12 NPM1 and FLT3 (ITD/D835) mutations in slightly different cohort of 161 de novo AML patients including adults and children. We also evaluated the association of these two gene mutations with immunophenotypic markers and other genetic alterations prevalent in AML.
2. Material and Method
2.1. Selection Criteria of Patients
One hundred sixty-one patients (116 adults and 45 children) with confirmed diagnosis of AML admitted to the Division of Hematology, Safdarjung Hospital, New Delhi, India, were consecutively included in the study from the year 2006 to the year 2010. Our earlier study was based on 133 patients (90 men and 43 women) who were screened for the detection of only FLT3 mutation [10]. This study included the overlapping cases of around 25 subjects (18 adults and 7 children) from the earlier study for whom we have both DNA and RNA available for the detection of mutation and translocation. The diagnosis of AML was made on routinely stained bone marrow aspiration/biopsies and peripheral blood smears and evaluated according to the French-American-British (FAB) criteria. Cytochemical stains were used to differentiate AML from ALL and to subclassify them into FAB subtypes. Written informed consent was obtained from all patients and all in vitro procedures were performed according to the protocol approved by the Ethical Committee of the Institute and Safdarjang Hospital. Clinical data, including age, gender, whole blood cell count (WBC), hemoglobin (Hb), platelet count, and presence of organomegaly, were collected at the time of diagnosis. Peripheral blood samples (4-5 mL) were obtained from all patients in heparinised vials (10–50 U/mL of blood) and transported to the laboratory on ice. In addition, bone marrow aspirates were collected in heparin containing phosphate buffered saline (PBS) when possible. All samples were processed on the same day. One mL of whole blood/bone marrow sample was used for immunophenotyping studies by flow cytometry and the remaining 3-4 mL of blood/bone marrow was used for separation of PBMC using Histopaque-1077 solution (Sigma, USA) for genetic studies.
2.2. Immunophenotyping
Immunophenotyping studies were carried out at the time of diagnosis as described previously [11]. The following monoclonal antibodies, all obtained from Becton-Dickinson Biosciences, USA, were included: CD3-FITC, CD5-FITC, CD7-FITC (T cell specific), CD10-PE (CALLA), CD19-FITC (B cell specific), CD13-PE, CD33-FITC, HLA-DR-FITC, MPO-FITC (myeloid cell specific), and TdT-FITC. CD45-PE and CD34-FITC antibodies were included in the panel to gate the blast cell population. For myeloperoxidase (MPO) antigen and TdT (intracellular), BD Cytofix/Cytoperm kit was used. Flow cytometric analysis was performed on a FACSCalibur Flow Cytometer (Becton-Dickinson, USA), and CellQuest software was used to acquire and analyze the data. Single color immunophenotyping was performed by collecting 10000 events/cells from each tube. Results were obtained by gating the blast cells using side scatter (SSC) versus forward light scatter (FSC) parameters and examining the associated FITC and PE fluorescence on FL1 and FL2 detectors, respectively. In addition, CD45 intensity versus side scatter (SSC) was used to detect blast cells and separate them from the normal populations. A case was considered as positive for specific antigen if the antigen was expressed in at least 20% of the leukemic cells of individual sample (20% cut-off level) and when the fluorescence intensity was clearly separated from negative controls (isotypic controls or negative cell population for the examined antigen). BD Calibur instrument settings were optimized by BD CaliBRITE (Catalog no. 340486) bead using FACSComp software version 2.0. To ensure data extraction from clinical samples, instrument settings were also optimized using a normal donor's peripheral blood samples (weekly) for FSC and SSC gains, FSC threshold, and fluorescence compensation levels.
2.3. Treatment Protocol and Response to Induction Chemotherapy
AML patients received the standard “3 + 7” induction chemotherapy. The details of the treatment protocol have been described previously [11]. Briefly, all patients included in the study were treated with the standard “3 + 7” induction chemotherapy or all-trans-retinoic acid (ATRA) therapy. Therapy-related AML cases and patients treated with other than the standard “3 + 7” induction chemotherapy were excluded from the study. All patients included in this study received the standard “3 + 7” induction chemotherapy protocol as described earlier: daunorubicin (45 mg/m2 per day) for 3 days and cytarabine (100 mg/m2 per day as a continuous 24 h intravenous infusion) for 7 days followed by G-CSF (5 μg/kg) daily from day 8 till counts are recovered (neutrophils > 1.5 × 109/L and platelet count > 100 × 109/L). Consolidation is comprised of three courses of high-dose cytosine arabinoside (3 g/m2 every 12 h on days 1, 3, and 5; total 18 g/m2) followed by G-CSF (5 μg/kg) daily from day 8 till counts are recovered. Bone marrow aspiration was done 28 days after initiation of chemotherapy. A complete remission was defined as the absence of abnormal clinical symptoms and presence of less than 5% of myeloblasts in the bone marrow. In this study, clinical response of the patients was evaluated after completion of induction chemotherapy only. Complete response (CR) to induction chemotherapy was defined clinically by disappearance of abnormal physical findings attributable to the leukemia and by the of return of the patient to good physical health and hematologically by the return of the peripheral blood to normal values with respect to hemoglobin, total and differential white blood cell count and platelets, and the presence of less than 5% morphologically normal blast cells in a marrow preparation of normal cellularity. Patients who did not meet the criteria of CR after induction therapy were considered as noncomplete response (NR).
2.4. Detection of NPM1 and FLT3 Mutations
NPM1 exon 12 mutation was detected by PCR and DNA sequencing. Briefly, genomic DNA from AML patients was extracted by using Himedia kit (Mumbai, India). NPM1 exon 12 was amplified by genomic PCR using primers reported by Gale et al., 2008 [12]. The reaction mixture contained 10 pmol of each primer, 100 ng of genomic DNA, 200 μM dNTPs, 2.0 mM Mgcl2, and 1 unit of Taq DNA polymerase (MBI Fermentas, UK) in the buffer provided by the manufacturer. Amplification was performed in a thermal cycler (PTC 200; MJ Research, USA), at an initial denaturation at 95°C for 5 minutes, followed by 30 cycles of 95°C for 2 minutes, 62°C for 45 seconds, 72°C for 45 seconds and followed by a hold at 72°C for 10 minutes and a cooldown. The 6-FAM-labeled PCR products were diluted tenfold in water, and 0.5 μL was mixed with 9.5 μL of HiDi formamide (Applied Biosystems, Inc., USA) and 0.25 μL of GeneScan Liz 250 internal size standards (Applied Biosystems, Inc., USA) and heated to 95°C for 2 minutes snap chill down on ice. The samples were run on an ABI 3130xl Genetic Analyzer using 16 cm capillaries. Raw data were analyzed with GeneMapper v4.0 software (Applied Biosystems, Inc., USA). For simplicity, rounded base pair values are used in the text. WT product size was 198 bp. For FLT3 mutation analysis, polymerase chain reaction (PCR) and DNA sequencing method were used to detect FLT3/ITD and FLT3/D835 mutations in AML samples as described previously [10]. To validate the result from capillary gel electrophoresis, PCR products larger than the wild-type product were sequenced to identify the type and extent of duplication/insertion. Amplified fragments were cut and extracted from agarose gels using Qiagen MinElute kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Approximately 100 ng purified PCR product was directly sequenced with 3.3 pmol primers as described above with the Big Dye Terminator Cycle Sequencing kit v3.1. Sequence analysis was performed on an ABI 3130xl Sequence Detection System (Applied Biosystems, Inc., USA).
2.5. Detection of Fusion Genes
Association of NPM1 and FLT3 mutations with commonly occurring translocation was also studied in 120 patients (91 adults and 29 children) in whom RNA samples were available. The reverse transcription polymerase chain reaction (RT-PCR) was performed for the detection of fusion genes BCR/ABL, PML/RAR, and AML/ETO [11]. AML/ETO and PML/RARα fusion genes were detected by simple RT-PCR using primers and conditions according to the BIOMED-1 Concerted Action protocol. The presence of the fusion gene BCR/ABL was detected by multiplex RT-PCR.
2.6. Statistical Analysis
For the analysis of categorical variables, either Fischer's exact or Pearson's X2 test was applied. Mann-Whitney rank sum test was used for the differences in the median variable in hemoglobin level, WBC/platelet count, and peripheral blast percentage. A P-value of ≤0.05 was considered statistically significant. All the analysis was performed using SPSS software (version 19).
3. Results
3.1. Patients Characteristics
The main clinical and hematologic characteristics of the patients included in the study are summarized in Table 1. Of 161 de novo AML patients included in the study, 116 were adults (median age: 32; range 16–85) and 45 were children (median age: 10; range 1–15). M2 (60/161, 37%) subtype was found to be the most common FAB subtype followed by M5 (26/161, 16%) and M0/M1 (24/161, 15%) subtype. Three patients were diagnosed with biphenotypic acute leukemia (BAL). Hepatosplenomegaly (HSM) was very common and seen in 92 patients (57.1%). Lymphadenopathy (LAP) was present in 46 patients (28.5%).
Table 1.
Clinical characteristics of AML patients with NPM1 and FLT3 mutations.
| Total (161) | No mutation % (100) | NPM1 % (34) | FLT3/ITD % (35) | FLT3/D835 % (5) | FLT3 + NPM1 % (13) | |
|---|---|---|---|---|---|---|
| Male : Female | 110 : 51 | 69 : 31 | 20 : 14 | 24 : 11 | 4 : 1 | 7 : 6 |
| Children : Adult | 45 : 116 | 33 : 67 | *4 : 30 | 8 : 27 | 2 : 3 | 2 : 11 |
| M0/M1 | 24 | 15 (15) | 12 (4) | 17 (6) | 40 (2) | 23 (3) |
| M2 | 60 | 36 (36) | 47 (16) | 37 (13) | 20 (1) | 46 (6) |
| M3 | 9 | 5 (5) | 6 (2) | 6 (2) | 0 (0) | 0 |
| M4 | 18 | 14 (14) | 6 (2) | 6 (2) | 20 (1) | 8 (1) |
| M5 | 26 | 16 (16) | 15 (5) | 17 (6) | 20 (1) | 15 (2) |
| M6 | 1 | 1 (1) | 0 (0) | 0 (0) | 0 (0) | 0 |
| BAL | 3 | 3 (3) | 0 (0) | 0 (0) | 0 (0) | 0 |
| AML (NC) | 20 | 10 (10) | 15 (5) | 17 (6) | 0 (0) | 8 (1) |
| MPO (+) stain | 121 | 86 (63/73) | 96 (24/25) (P = 0.28) | 93 (28/30) (P = 0.50) | 100 (2/2) (P = 1.0) | 100 (9/9) (P = 0.59) |
| AR present | 79 | 46 (46) | 56 (19) (P = 0.33) | 51 (18) (P = 0.69) | 20 (1) (P = 0.36) | 38 (5) (P = 0.76) |
| LAP | 46 | 26 (26) | 23 (8) (P = 0.82) | 40 (14) (P = 0.13) | 40 (2) (P = 0.62) | 31 (4) (P = 0.74) |
| HSM | 92 | 64 (64) | 38 (13) (P = 0.01) | 46 (16) (P = 0.07) | 60 (3) (P = 1.0) | 31 (4) (P = 0.03) |
| Hb level (g/dL) | 6.2 (2–13.8) | 6.2 (2.6–13.8) | 6.6 (3–12.2) (P = 0.73) | 6.2 (2–10.4) (P = 0.57) | 5.3 (3.6–10.0) (P = 0.49) | 6.2 (3.6–10.1) (P = 0.99) |
| TLC (109/L) | 31.5 (1–428) | 27.4 (1–290) | 34.7 (1.6–380)(P = 0.78) | 60.2 (1.4–428) (P = 0.01) | 41.6 (2.5–256) (P = 0.94) | 48.7 (19–312) (P = 0.04) |
| Platelet (109/L) | 22 (1–343) | 20 (1–270) | 46.5 (9–343) (P = 0.05) | 23 (2–126) (P = 0.86) | 20 (2–191) (P = 0.75) | 37 (9–191) (P = 0.39) |
| Blast (%) | 69 (10–98) | 67.5 (17–98) | 75 (10–95) (P = 0.28) | 70 (29–96) (P = 0.50) | 80 (50–90) (P = 0.56) | 60 (29–92) (P = 0.73) |
| CR | 83 (24/29) | 52 (15/29) | 73 (8/11) (P = 0.28) | 29 (2/7) (P = 0.42) | 100 (1) (P = 1.0) | 50 (1/2) (P = 1.0) |
P values were obtained on comparing each of FLT3 (ITD/D835), NPM1, and FLT3 + NPM1 mutation groups with the no mutation group (column 2). Mann-Whitney rank sum test was used for the difference in median variable of Hb, Plt, WBC, and blast parameters. For Hb, Plt, WBC, and blast, numbers represent median values while, numbers in parenthesis indicate range. For other categorical variables, either Fischer's exact or Pearson's χ2 test is used. P indicated P value. CR indicated complete remission after induction chemotherapy. Abbreviations: AML (NC): AML not classified, BAL: biphenotypic acute leukemia, AR: auer rod, MPO: myeloperoxidase, LAP: lymphadenopathy, and HSM: hepatosplenomegaly. *Bold significant P value = 0.02.
3.2. NPM1 and FLT3 (ITD/D835) Mutations
NPM1 mutation was detected in 34 out of 161 cases (21%). Eight samples were randomly selected from 34 positive samples for sequence analysis of NPM1 mutation. Six different sequence variants were observed, all leading to a frame shift in the region encoding the C-terminal of the NPM1 protein. All mutations consisted of insertion of 4 bp insertions either between nucleotides 960-961 or between nucleotides 964-965 (see Supplementary Figure 1 available online at http://dx.doi.org/10.1155/2013/582569). The most frequent mutation (Type A mutation) was the duplication of a TCTG tetranucleotide found in 3 cases (Table 2). Alterations in FLT3 gene were detected in 25% (40/161) patients. These aberrations include FLT3/ITD in 35 (22%) cases and FLT3/D835 mutation in 5 cases (3%). None of the samples showed combination of both FLT3/ITD and FLT3/D835 mutations. Thirteen (38%) of the 34 samples with NPM1 mutation had also FLT3/ITD mutation.
Table 2.
Sequencing analysis of different patterns of NPM1 mutation in AML patients.
| Sample ID | DNA sequence of exon 12 of NPM1 gene |
|---|---|
| Wild type | GATCTCTG —GCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH542 | GATCTCTGTCTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH545 | GATCTCTGCATGGCAGTGGAGGAAGTCTCTTTAAGAAAATAG |
| UH533 | GATCTCTG TTTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH581 | GATC CGCTTCTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH591 | GATC TATGTCTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH595 | GATCTCTGTCTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH576 | GATCTCTGTCTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
| UH567 | GATCTCTGTGTGGCAG TGGAGGAAGTCTCTTTAAGAAAATAG |
Sequence highlighted in bold color represents inserted nucleotides in NPM1 gene. TCTG is the most common 4 bp nucleotide insertion in exon 12 of NPM1 gene.
3.3. Relationship of NPM1 and FLT3 Mutations with Clinical Characteristics
NPM1 Mutation. Baseline characteristics of patients with NPM1 and FLT3 (ITD and D835) mutations are summarized in Table 1. NPM1 mutation was found more often in females (27%, 14/51) than males (18%, 20/110) (P = 0.24). The NPM1 mutation was found more frequently in adults as compared to children (26% versus 9%; P = 0.01). Further stratification of age showed that frequency of NPM1 mutation increases with age, 38% (12/32) being the highest frequency observed in AML patients above 45 yrs of age (P = 0.02) (Figure 1). Patients with NPM1 mutation had significantly higher platelet count (P = 0.05) as compared to patients without any NPM1 and FLT3 mutations. Further, platelet count was significantly higher in NPM1 mutation group than in no mutation group in adult patients (56 × 109/L versus 20 × 109/L; P = 0.02) and in children (25 × 109/L (20–50 × 109/L); P = 0.62). Data are not shown. Moreover, NPM1 mutation was found to be significantly associated with the absence of extramedullary diseases such as HSM (P = 0.01).
Figure 1.
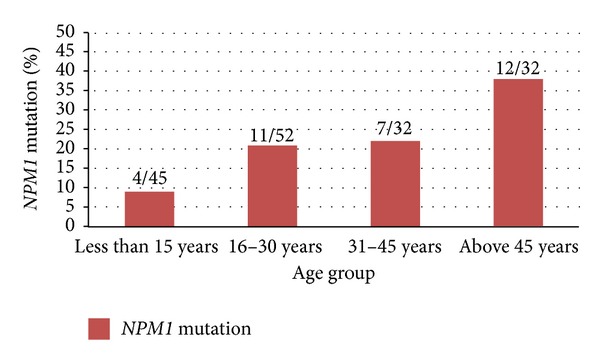
Incidence of NPM1 mutation according to age.
FLT3 Mutation. No significant difference in age or gender was found in patients with FLT3/ITD (P = 1.0 and P = 0.28) and FLT/D835 mutation as compared to those patients without any mutations. Similar to NPM1 mutation, FLT3/ITD mutation was found more often in adults (23%) as compared to children (18%); however, this difference was not statistically significant (P = 0.29). Other clinical parameters except WBC count showed no association with either FLT3/ITD or FLT3/D835 mutations. Patients with FLT3/ITD mutation had significantly higher WBC count 60.2 × 109/L (range 1.4–428 × 109/L) compared to patients without mutations 27.4 × 109/L (range 1–290 × 109/L) (P = 0.01). Furthermore, adult patients (>15 years) had significantly higher median WBC count in FLT3 mutation group compared to no mutation group (56.9 × 109/L versus 27.6 × 109/L; P = 0.03). However, there was no significant difference in the WBC count in children between FLT3/ITD mutation and no mutation groups (113 × 109/L versus 25.6 × 109/L; P = 0.21). Though the median count was much higher in cases with FLT3/ITD mutation but not significant, this can perhaps be accounted for by the small numbers of patients with FLT3/ITD mutations in children.
Patients with both NPM1 and FLT3/ITD mutations had higher WBC count as compared to patients without mutation (P = 0.04). This group similar to NPM1 mutation was also associated with the absence of HSM (P = 0.03). However, other clinical parameters were not significantly different between patients with both NPM1 and FLT3/ITD mutations and without mutations. NPM1 and FLT3/ITD mutations status did not affect significantly the induction of CR rate in the study probably due to the smaller number of patients (29) in the clinical outcome category.
3.4. Immunophenotypic Characteristics of NPM1 and FLT3 Mutations
Immunophenotypically, NPM1 mutation was found to be associated with the lower positivity of CD34 (29% versus 70%; P < 0.001) and HLA-DR expression (29% versus 81%; P = 0.001) compared to the group without NPM1 and FLT3/ITD mutations. In contrast to this, FLT3/ITD mutation was associated with the higher positivity of CD7 marker (33% versus 14%; P = 0.04). Further, there was no difference in the positivity of CD34 in the group with FLT3/ITD mutation compared to those without FLT3/ITD mutation (61% versus 70%; P = 0.38). However, the group with both NPM1 and FLT3/ITD mutations was associated with the lack of CD34 expression (P = 0.03). Other markers such as CD3, CD5, CD10, CD19, CD14, CD13, CD33, TdT, HLA-DR, and MPO were not significantly associated with NPM1 and FLT3/ITD mutations status (Table 3).
Table 3.
Immunophenotypic characteristics of AML patients with NPM1 and FLT3/ITD mutations.
| Antigen marker | Total (161) | No mutation (100) | NPM1 (34) | FLT3/ITD (35) | NPM1 + FLT3 (13) |
|---|---|---|---|---|---|
| CD3 | 3% | 4% | 0% | 0% | 0% |
| CD5 | 3% | 4% | 0% | 0% | 0% |
| CD7 | 17% | 14% | 9% | 33%* | 11% |
| CD10 | 8% | 11% | 3% | 0% | 0% |
| CD13# | 86% | 90% | 71% | 94% | 100% |
| CD14 | 20% | 22% | 18% | 15% | 22% |
| CD19 | 22% | 29% | 12% | 12% | 8% |
| CD33 | 88% | 85% | 94% | 94% | 100% |
| CD34 | 60% | 70% | 29%* | 61% | 38%* |
| MPO# | 67% | 67% | 77% | 56% | 60% |
| HLA-DR# | 71% | 81% | 29%* | 81% | 60% |
| TdT# | 10% | 12% | 0 % | 11% | 0% |
P values were obtained on comparing each of FLT3, NPM1, and FLT3 + NPM1 mutation groups with the no mutation group (column 2). Numbers in the table represent the percentage of positive cases for that antigen marker. *Significant P-value <0.05. #Markers not done in all cases.
3.5. Correlation of NPM1 and FLT3 Mutations with Translocations
AML/ETO was the most frequent translocation detected (23%, 28/120) in AML patients (Table 4). PML/RARα translocation was detected in 4% (5/120) while 9% (11/120) of samples were positive for BCR/ABL translocation. All the five positive cases of PML/RAR were of typical M3 AML morphology with highly granulated blast cells, nuclear size irregular and highly variable. Majority of the promyelocytes were with bilobed and irregular nuclei. Faggot cells were also seen in the three cases. All the cases were strongly MPO positive and PAS negative. Correlation with gene mutation data in the patients showed that AML/ETO translocation was inversely associated with the FLT3/ITD mutation (P = 0.04). Frequency of AML/ETO translocation was low in FLT3/ITD mutation group compared to FLT3/ITD wild group (12% versus 28%; P = 0.04). No association of NPM1 and FLT3/ITD mutations was observed with other translocation due to smaller number of cases positive for the translocation. BCR/ABL translocation was detected in two patients positive for FLT3/ITD mutation while in one patient with NPM1 mutation. PML/RARα translocation was detected only in one case of both FLT3 and NPM1 mutations.
Table 4.
Relationship of FLT3 and NPM1 mutations with translocations.
| Translocation (n) | NPM1 mut. % (22) | NPM1 wild % (98) | P value | FLT3 mut. % (31) | FLT3 wild % (89) | P value |
|---|---|---|---|---|---|---|
| BCR/ABL (11) | 4 (1) | 10 (10) | 0.68 | 6 (2) | 10 (9) | 0.72 |
| PML/RARα (5) | 4 (1) | 4 (4) | 1.0 | 3 (1) | 5 (4) | 1.0 |
| AML/ETO (28) | 14 (3) | 25 (25) | 0.27 | 11 (3) | 28 (25) | 0.04* |
*Bold significant P-value = 0.02.
4. Discussion
Genetic alterations in AML are known to be major determinants of the patient response to therapy and outcome besides their role in the pathogenesis of the disease. In addition to the aberrations that are revealed by conventional karyotyping, submicroscopic lesions such as FLT3 and NPM1 mutations which are undetectable at chromosome level have been described in AML [1, 6]. FLT3 and NPM1 mutations have been shown to be the most prevalent somatic alterations in AML, particularly in cytogenetically normal AML. These mutations play significant role in the diagnosis, risk assessment, and guidance of therapy [2, 13]. In the current study, the frequency of NPM1 and FLT3/ITD mutations was 26% (30/116) and 23% (27/116) in adults, respectively, similar to the frequency reported in Japanese (24.9% NPM1, 22.6% FLT3/ITD) [14] and in German adult AML population (27.4% NPM1, 21% FLT3/ITD) [15]. When compared to patients from Southeast Asian region, the frequency of NPM1 mutation was equivalent to our study (26%); however, the frequency of FLT3/ITD mutations was higher in Southeast Asian region as compared to our study (33% versus 23%) [16].
Adult patients usually have a higher frequency of NPM1 mutation than children. The frequency of NPM1 mutation was 9% in children (≤15 years) and 26% in adult patients (≥15 years) in the current study (P = 0.02). The frequency of NPM1 mutation in adult is nearly comparable to reports from other countries including Japan, Germany, and England (25–41%) [12, 14, 15]. Similarly, the frequency of NPM1 mutations was 7.5% overall in pediatric AML patients (range 0–12%) [17–19]. Observed variation in the frequency of NPM1 mutations might be due to the differences in the sample size of the study, ethnicity, and geographical distribution of the population studied. Moreover, we observed a striking relationship between incidence of NPM1 mutation and age of the AML patients in adults. The incidence of NPM1 mutation appears to increase with age such that patients between 30 and 45 years have frequency of 22% and patients older than 45 years of age have frequency of 38%. Similar age-dependent increase in the incidence of NPM1 mutation has also been reported earlier [6, 17]. Ahmad et al., 2009, have also reported a higher frequency of NPM1 mutation in adults as compared to children in Indian population (23.6 versus 2.5%) [8].
Certain associations between the two gene mutations and clinical characteristics have been found in the current study, for instance, a higher platelet count (P = 0.05) and absence of hepatosplenomegaly (P = 0.01) with NPM1 mutation and high WBC count with FLT3/ITD mutation. Thiede et al., 2006, also observed that blasts with NPM1 mutation may retain a certain capacity for thrombocytic differentiation as demonstrated by in vitro experiments of Hsu et al., 2003 [15, 20]. Further, platelet count was significantly higher in NPM1 mutation group than in no mutation group in adults only not in children. In concurrence with our report, Falini et al., 2005, and Döhner et al., 2005 also found correlation of mutated NPM1 with high platelet counts in adult AML patients [6, 21]. Similarly in line with our study, Braoudaki et al., 2010, too did not find association of NPM1 mutation with platelet count in pediatric AML patients [22].
We found a significantly increased WBC count in patients with FLT3/ITD mutation (P = 0.01) but not with FLT3/D835. FLT3/ITD mutation leads to constitutively activated FLT3 protein which causes proliferation, inhibits apoptosis, and suppresses differentiation of leukemic cells. It is reported that ligand independent constitutive activation of FLT3 induced by ITD mutation could activate some downstream signal molecules including mitogen activated protein (MAP) kinase, signal transducer and activator of transcription 5 (STAT5), and serine threonine kinases Akt, which contribute to cell proliferation and survival advantages [4, 23, 24]. This may be the reason for the higher WBC count in these cases. However, this significant association was observed only in adult not in pediatric patients. This supports the observation reported in earlier studies that FLT3/ITD but not the FLT3/D835 mutation is associated with higher WBC count in adult AML patients [25, 26]. However, median WBC count in pediatric patients with FLT3/ITD mutation was higher than in the patient without mutation but was not statistically significant, although others disagree with this [27].
Immunophenotyping analysis showed that NPM1 mutation was associated with the lower positivity of CD34 expression (29 versus 70%; P < 0.001) and lack of HLA-DR expression (29 versus 81%; P < 0.05). In several European studies on AML, NPM1 mutation is inversely associated with the expression of CD34 marker [15, 21]. Similar report of lack of CD34 expression in NPM1 mutation positive has also been reported from South Asian countries [16, 17]. All these studies including ours are in agreement with the fact that the lack of CD34 expression in NPM1 mutated cases could be an indicator for good response to induction chemotherapy. Though FLT3/ITD patients did not display a unique immunophenotype, a significant association of FLT3/ITD mutation with aberrant expression of CD7 was found (P = 0.04). An earlier study by us and Rausei-Mills et al., 2008 has also shown the association of coexpression of CD7 antigen with FLT3/ITD mutation [10, 28].
In concurrence with other reports, there was no significant association of NPM1 mutation with the AML/ETO, PML/RARα, and BCR/ABL translocations in the current study [14, 17, 22]. However, AML/ETO translocation was found to be negatively associated with the FLT3/ITD mutation (P = 0.04). This is quite contrast to the study of Schessl et al., (2005) which reports that AML/ETO collaborates with FLT3-LM (length mutation) in inducing acute leukemia in a murine BM transplantation model [29]. Of note, both myeloblastic and lymphoblastic leukemias of B cell and T cell types developed in this model of AML/ETO and FLT3-LM cooperation suggest that additional mutations in AML/ETO positive progenitors are necessary for the transformation into leukemia-initiating cells.
In summary, the frequency of NPM1 and FLT3 mutations was 21% and 25% in de novo AML patients comparable to other reports despite geographic and ethnic differences. Distinct clinical and immunophenotypic characteristics associated with the NPM1 and FLT3/ITD mutation were found which seems to characterize a distinct entity of AML. FLT3/ITD was found to be inversely associated with the presence of AML/ETO translocation, indicating requirement of other mutations to cause AML. Further, comprehensive studies on the biological effects of NPM1 and FLT3/ITD mutations and their interactions with other genetic and epigenetic alterations are needed to gain insight into the molecular mechanism of these mutations involved in the pathogenesis of AML.
Supplementary Material
Supplementary figure 1(a) shows the capillary electrophoresis of NPM1 mutation in AML samples run on the ABI 3130xl Genetic Analyzer (16-cm capillaries). Fluorescent PCR product analysis of exon 12 showed one wild type NPM1 peak at 198 bp and additional peak at 202 bp in patients with NPM1 mutation. Figure 1(b) shows the sequence electropherogram of NPM1 mutation generated by direct sequencing of PCR product from normal and NPM1 mutant samples. Reverse PCR primer was used as a sequencing primer. The boxed tetranucleotides shown in the mutant sequence are the 4 bp inserted between nucleotide 960 and 961 from the transcription start site
Conflict of Interests
The authors declare that there is no conflict of interests.
Acknowledgment
This work was supported by a Grant under the Indian Council of Medical Research (ICMR) Fellowship (IOP/RA/2011-2012/2347).
References
- 1.Gary Gilliland D, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100(5):1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 2.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 3.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10(12):1911–1918. [PubMed] [Google Scholar]
- 4.Hayakawa F, Towatari M, Kiyoi H, et al. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19(5):624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 5.Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT. Identification of novel FLT-3 Asp835 mutations in adult acute myeloid leukaemia. British Journal of Haematology. 2001;113(4):983–988. doi: 10.1046/j.1365-2141.2001.02850.x. [DOI] [PubMed] [Google Scholar]
- 6.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. The New England Journal of Medicine. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 7.Colombo E, Marine J-C, Danovi D, Falini B, Pelicci PG. Nucleophosmin regulates the stability and transcriptional activity of p53. Nature Cell Biology. 2002;4(7):529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad F, Mandava S, Das BR. Mutations of NPM1 gene in de novo acute myeloid leukaemia: determination of incidence, distribution pattern and identification of two novel mutations in Indian population. Hematological Oncology. 2009;27(2):90–97. doi: 10.1002/hon.883. [DOI] [PubMed] [Google Scholar]
- 9.Dunna NR, Rajappa S, Digumarti R, et al. Fms like tyrosine kinase (FLT3) and nucleophosmin 1 (NPM1) mutations in De Novo normal karyotype acute myeloid leukemia (AML) Asian Pacific Journal of Cancer Prevention. 2010;11(6):1811–1816. [PubMed] [Google Scholar]
- 10.Chauhan PS, Bhushan B, Mishra AK, et al. Mutation of FLT3 gene in acute myeloid leukemia with normal cytogenetics and its association with clinical and immunophenotypic features. Medical Oncology. 2011;28(2):544–551. doi: 10.1007/s12032-010-9485-4. [DOI] [PubMed] [Google Scholar]
- 11.Chauhan PS, Bhushan B, Singh LC, et al. Expression of genes related to multiple drug resistance and apoptosis in acute leukemia: response to induction chemotherapy. Experimental and Molecular Pathology. 2012;92(1):44–49. doi: 10.1016/j.yexmp.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Gale RE, Green C, Allen C, et al. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111(5):2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 13.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T, Kiyoi H, Ozeki K, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106(8):2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 15.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107(10):4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 16.Boonthimat C, Thongnoppakhun W, Auewarakul CU. Nucleophosmin mutation in Southeast Asian acute myeloid leukemia: eight novel variants, FLT3 coexistence and prognostic impact of NPM1/FLT3 mutations. Haematologica. 2008;93(10):1565–1569. doi: 10.3324/haematol.12937. [DOI] [PubMed] [Google Scholar]
- 17.Chou W-C, Tang J-L, Lin L-I, et al. Nucleophosmin mutations in de novo acute myeloid leukemia: the age-dependent incidences and the stability during disease evolution. Cancer Research. 2006;66(6):3310–3316. doi: 10.1158/0008-5472.CAN-05-4316. [DOI] [PubMed] [Google Scholar]
- 18.Shimada A, Taki T, Kubota C, et al. No nucleophosmin mutations in pediatric acute myeloid leukemia with normal karyotype: a study of the Japanese Childhood AML Cooperative Study Group. Leukemia. 2007;21(6, article 1307) doi: 10.1038/sj.leu.2404625. [DOI] [PubMed] [Google Scholar]
- 19.Thiede C, Creutzig E, Reinhardt D, Ehninger G, Creutzig U. Different types of NPM1 mutations in children and adults: evidence for an effect of patient age on the prevalence of the TCTG-tandem duplication in NPM1-exon 12. Leukemia. 2007;21(2):366–367. doi: 10.1038/sj.leu.2404519. [DOI] [PubMed] [Google Scholar]
- 20.Hsu CY, Yung B. Involvement of nucleophosmin/B23 in TPA-induced megakaryocytic differentiation of K562 cells. British Journal of Cancer. 2003;89(7):1320–1326. doi: 10.1038/sj.bjc.6601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 22.Braoudaki M, Papathanassiou C, Katsibardi K, Tourkadoni N, Karamolegou K, Tzortzatou-Stathopoulou F. The frequency of NPM1 mutations in childhood acute myeloid leukemia. Journal of Hematology and Oncology. 2010;3, article 41 doi: 10.1186/1756-8722-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiyoi H, Ohno R, Ueda R, Saito H, Naoe T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene. 2002;21(16):2555–2563. doi: 10.1038/sj.onc.1205332. [DOI] [PubMed] [Google Scholar]
- 24.Rocnik JL, Okabe R, Yu J-C, et al. Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood. 2006;108(4):1339–1345. doi: 10.1182/blood-2005-11-011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Lin D, Zhang X, Chen S, Wang M, Wang J. Analysis of FLT3 internal tandem duplication and D835 mutations in Chinese acute leukemia patients. Leukemia Research. 2005;29(12):1393–1398. doi: 10.1016/j.leukres.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 26.Colovic N, Tosic N, Aveic S, et al. Importance of early detection and follow-up of FLT3 mutations in patients with acute myeloid leukemia. Annals of Hematology. 2007;86(10):741–747. doi: 10.1007/s00277-007-0325-3. [DOI] [PubMed] [Google Scholar]
- 27.Meshinchi S, Alonzo TA, Stirewalt DL, et al. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108(12):3654–3661. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rausei-Mills V, Chang KL, Gaal KK, Weiss LM, Huang Q. Aberrant expression of CD7 in myeloblasts is highly associated with de novo acute myeloid leukemias with FLT3/ITD mutation. American Journal of Clinical Pathology. 2008;129(4):624–629. doi: 10.1309/NRTX9AKXHR5JBT93. [DOI] [PubMed] [Google Scholar]
- 29.Schessl C, Rawat VPS, Cusan M, et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. Journal of Clinical Investigation. 2005;115(8):2159–2168. doi: 10.1172/JCI24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure 1(a) shows the capillary electrophoresis of NPM1 mutation in AML samples run on the ABI 3130xl Genetic Analyzer (16-cm capillaries). Fluorescent PCR product analysis of exon 12 showed one wild type NPM1 peak at 198 bp and additional peak at 202 bp in patients with NPM1 mutation. Figure 1(b) shows the sequence electropherogram of NPM1 mutation generated by direct sequencing of PCR product from normal and NPM1 mutant samples. Reverse PCR primer was used as a sequencing primer. The boxed tetranucleotides shown in the mutant sequence are the 4 bp inserted between nucleotide 960 and 961 from the transcription start site


