Abstract
The allosteric effect of fluconazole (effector) on the formation of 1’-hydroxymidazolam (1’-OH-MDZ) and 4-hydroxymidazolam (4-OH-MDZ) from the CYP3A4/5 substrate, midazolam (MDZ), was examined in healthy volunteers. Following pre-treatment of fluconazole, AUC4-OH/AUCMDZ increased 35–62%, while AUC1’-OH/AUCMDZ decreased 5–37%; AUC1’-OH/AUC4-OH ratio decreased 46–58% by fluconazole and had no association with CYP3A5 genotype. 1’-OH-MDZ formation in vitro was more susceptible than 4-OH-MDZ formation to inhibition by fluconazole. Fluconazole decreased the intrinsic formation clearance ratio of 1’-OH-MDZ/4-OH-MDZ to an extent that was quantitatively comparable to in vivo observations. The elimination clearance of midazolam metabolites appeared unaffected by fluconazole. This study demonstrated that fluconazole alters midazolam product formation both in vivo and in vitro in a manner consistent with an allosteric interaction. The 1'-OH-MDZ/4-OH-MDZ ratio may serve as a biomarker of such interactions between midazolam, CYP3A4/5 and other putative effectors.
INTRODUCTION
Non-Michaelis–Menten kinetics are observed commonly in vitro in cytochrome P450 (CYP) catalyzed biotransformation reactions and are believed to be a consequence of homotropic or heterotropic cooperative (allosteric) interactions between substrate, effector and enzyme (1–3). Interest in investigating allosterism with CYP enzymes includes a need to recover an accurate in vitro intrinsic clearance for prediction of in vivo clearance and drug-drug-interactions (DDIs), which can alter total clearance and metabolite exposure. Studies to date have focused primarily on one CYP subfamily, CYP3A (mainly CYP3A4 and CYP3A5), the principal CYP subfamily in humans (4).
Unlike enzyme inhibition- and induction-mediated DDIs, which have been studied extensively and generally have well-established in vitro–in vivo correlations, CYP allosterism rarely has been investigated in vivo. This characteristic has hindered the understanding of clinical relevance of CYP allosterism. Previous in vivo studies in monkeys and CYP3A4 humanized mice provided evidence of heteroactivation of CYP3A that was consistent with in vitro data (5, 6). In contrast, a limited number of clinical studies in humans showed a lack of correlation of CYP allosteric effects (including that for CYP3A4 and CYP2C9) between in vitro and in vivo observations (1, 7, 8). Indirect evidence of CYP3A4 allosterism in humans consistent with in vitro observations was reported in a retrospective meta-analysis (9). However, direct clinical evidence of CYP allosterism remains absent from the literature. The lack of correlation between in vitro and in vivo CYP allosterism is due mainly to the fact that non-Michaelis-Menten kinetics can be discerned only from a large range of concentrations of effector or substrate that usually are not achieved or maintained in vivo (10). Other factors, such as multiple metabolic pathways and extensive protein binding, also may contribute to such discrepancies, especially when a single product formation or total drug clearance is measured (1, 11).
Midazolam (substrate) and fluconazole (effector) were selected in the present study to examine CYP3A4 allosterism in vivo and in vitro. Midazolam is metabolized almost exclusively by CYP3A enzymes to two primary metabolites, 1’-hydroxymidazolam (1’-OH-MDZ) and 4-hydroxymidazolam (4-OH-MDZ), which are converted subsequently to N- and O-glucuronides by UDP-glucuronosyltransferases (UGTs) (12). In vitro, regioselective formation of 1’-OH-MDZ and 4-OH-MDZ depends on enzyme isoform, midazolam concentration and the absence or presence of other allosteric molecules (2, 13–17). Fluconazole is a moderate inhibitor of CYP3A4 and a relatively less potent inhibitor of CYP3A5 both in vitro and in vivo (18). In vitro, fluconazole is 12 times more potent an inhibitor of 1’-OH-MDZ formation than 4-OH-MDZ formation (19). Such regioselective inhibition is incompatible with a simple competitive inhibition mechanism and suggests partial inhibition, which has been recognized as a contributor to the atypical kinetics of CYP3A enzymes. The mechanism of partial inhibition is attributed to interactions between inhibitor and substrate with multiple or overlapping binding domains of the enzymes (2, 20–22). Accordingly, we selected the regioselectivity of midazolam metabolism as a reporter of the allosteric effects of fluconazole with CYP3A4 and CYP3A5 and examined allosterism with both enzymes in vivo and in vitro.
RESULTS
Effect of fluconazole on 1’-OH-MDZ and 4-OH-MDZ formation in healthy volunteers
Concentrations of 1’-OH-MDZ and 4-OH-MDZ were determined in plasma obtained from healthy volunteers genotyped as CYP3A5*1/*1 (n=5), CYP3A5*1/*X (n=7) or CYP3A5*X/*X (n=6) (X represents a CYP3A5*3, *6 or *7 allele). Midazolam (1 mg) was administered intravenously alone or 2 hours after oral fluconazole (400 mg), when fluconazole had reached peak concentration. Our previous analysis showed that during the midazolam sampling period (5 to 720 min), average (± S.D.) fluconazole concentrations were 17.4 ± 2.3 µM (CYP3A5*1/*1), 19.5 ± 1.7 µM (CYP3A5*1/*X) or 22.0 ± 6 µM (CYP3A5*X/*X) (18). Concentration-time profiles of midazolam and primary metabolites for the three CYP3A5 genotype groups were similar. The elimination of 1’-OH-MDZ and 4-OH-MDZ conformed to formation rate-limited metabolite kinetics (Figure 1A–C). Consistent with our previous analysis (18), control AUCMDZ had no significant differences among the three CYP3A5 genotype groups, albeit a large inter-individual variability. In addition, fluconazole significantly increased AUCMDZ in all three genotype groups (Table 1).
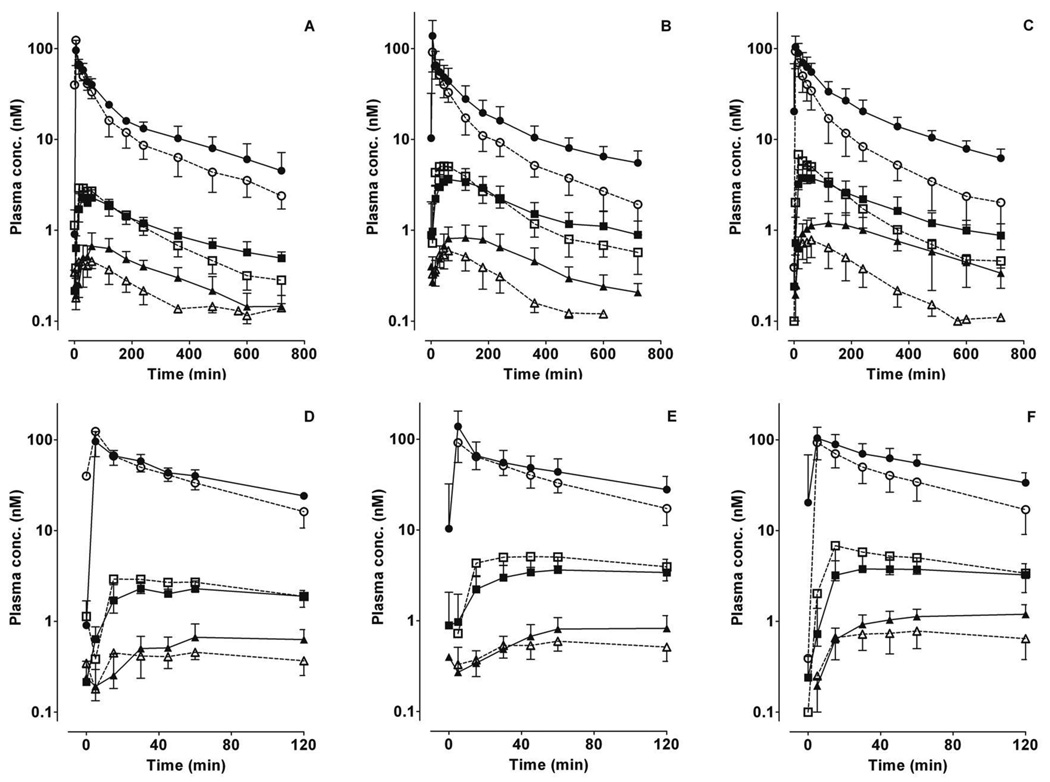
Figure 1. Plasma concentration-time profiles for midazolam and primary metabolites following intravenous administration of midazolam.
The mean ± S.D. data points for midazolam (circles), 1’-hydroxymidazolam (squares) and 4-hydroxymidazolam (triangles) in the absence (open symbols) or presence (solid symbols) of fluconazole for the three CYP3A5 genotype groups (A, D: CYP3A5*1/*1; B, E: CYP3A5*1/*X; C, F: CYP3A5*X/*X) are shown. Fluconazole (400 mg) was administered orally 120 min before midazolam. Panels D, E, F depict the same data from time 0 to 120 min.
Table 1. Pharmacokinetics of midazolam, 1’-hydroxymidazolam and 4-hydroxymidazolam following a single intravenous dose (1 mg) of midazolam alone (control) or with a single dose of oral fluconazole (400 mg) according to CYP3A5 genotype.
Data represent means ± S.Ds. Values in parentheses indicate the percentage compared with control.
| CYP3A5 genotype | *1/*1 (n=5) | *1/*X (n=7) | *X/*X (n=6) | |||
|---|---|---|---|---|---|---|
| Control | Fluconazole | Control | Fluconazole | Control | Fluconazole | |
| AUCMDZ (nM· hr)a | 160 ± 35 | 227.0 ± 60.0* (140 % ± 39%) | 150 ± 31 | 261.9 ± 69.0* (173.6 ± 18.6%) | 147.4 ± 39.5 | 308.1 ± 57.2* (214 ± 29.3%) |
| AUC1'-OH (nM·hr)b | 13.5 ± 3.1 | 16.6 ± 2.4* (130% ± 23%) | 25.6 ± 4.6 | 30.9 ± 9.5* (118.9% ± 19.7%) | 23.8 ± 8.7 | 30.8 ± 9.1* (133.8% ± 20.4%) |
| AUC4-OH (nM·hr)c | 2.2 ± 0.6 | 4.8 ± 1.4* (230% ± 67%) | 3.0 ±0.8 | 7.0 ± 2.2* (243.5% ± 95.7%) | 3.8 ± 1.3 | 10.6 ± 2.4* (290.3% ± 51.2%) |
| AUC1'-OH/AUCMDZd | 0.08 ± 0.02 | 0.07 ± 0.01 (95% ± 42%) | 0.18 ± 0.06 | 0.12 ± 0.05* (68.6% ± 10.6%) | 0.16 ± 0.05 | 0.10 ± 0.03* (62.6% ± 5.9%) |
| AUC4-OH/AUCMDZe | 0.01 ± 0.004 | 0.02 ± 0.006* (162.4 % ± 55.4%) | 0.020 ± 0.005 | 0.026 ± 0.004* (140.8% ± 56.9%) | 0.027 ± 0.006 | 0.035 ± 0.005* (135.3% ± 12.3%) |
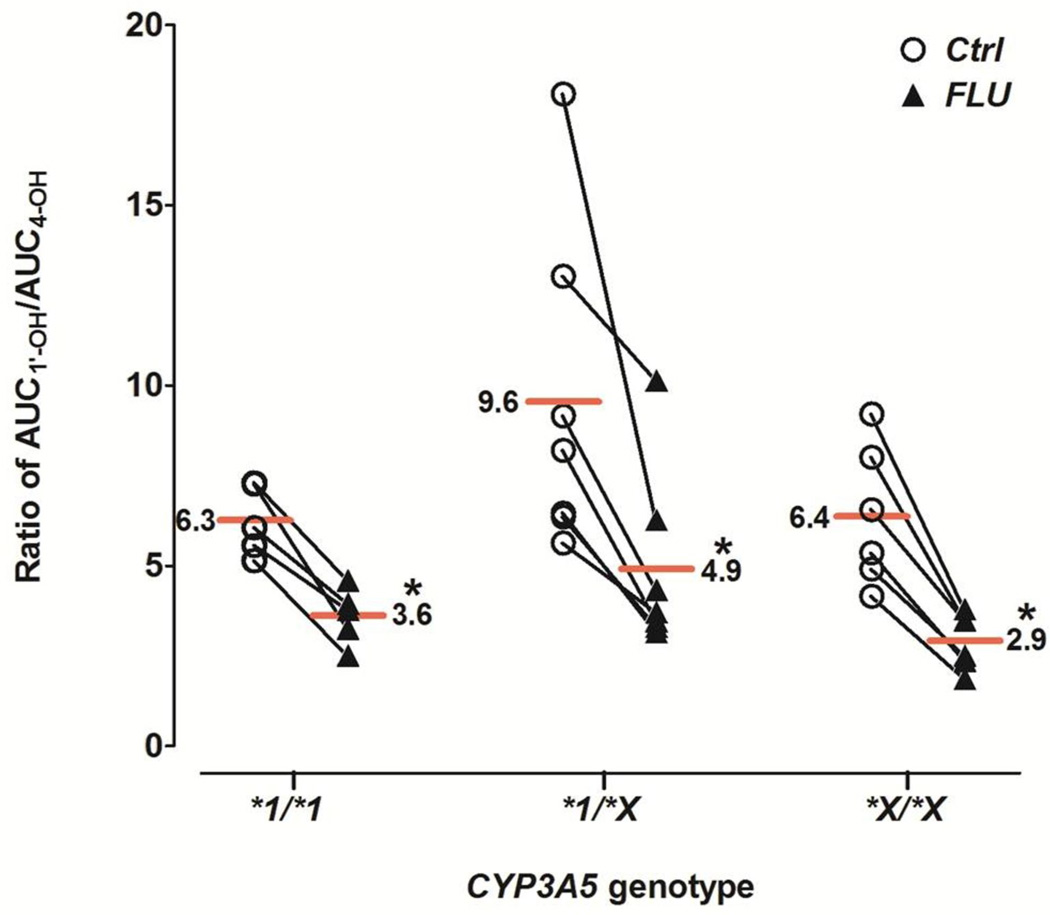
| AUC1'-OH/ AUC4-OHf | 6.3 ± 1.0 | 3.6 ± 0.8* (58.0 % ± 10.5%) | 9.6 ± 4.5 | 4.9 ± 2.5* (52.7% ± 14.6%) | 6.4 ± 2.0 | 2.9 ± 0.8* (46.4% ± 4.7%) |
p<0.05 compared with control within the genotype group.
a, c, d, interaction between treatments and genotypes. a, no difference in basal AUCMDZ across three genotype groups, and *X/*X is significant ly higher than the *1/*1 group (p=0.01) with fluconazole; c, no difference in basal AUC4-OH across three genotype groups, *1/*X, and*X/*X are significantly higher than the *1/*1 group (p=0.05 and p<0.0001) with fluconazole and AUC4-OH of *X/*X is significantly higher than *1/*X with fluconazole (p=0.0003); d, the basal ratio of AUC1’-OH/AUCMDZ of *1/*1 is significantly lower than *1/*X (p<0.0001) and *X/*X (p<0.0001), and *1/*1 is significantly lower than *1/*X with fluconazole (p=0.004) but not different from *X/*X (p=0.61).
b, e, f, no interaction between treatments and genotypes. b, *1/*X and *X/*X are significantly greater than *1/*1 (p=0.01 and 0.03); e, *X/*X is significantly greater than *1/*1 (p=0.0003) and *1/*X (p=0.009).
1’-OH, 1’-hydroxymidazolam; 4-OH, 4-hydroxymidazolam; AUC, area under the curve.
During the first 120 min after midazolam administration, 4-OH-MDZ concentrations were increased by fluconazole, whereas 1’-OH-MDZ concentrations decreased, an effect observed for all three genotype groups (Figure 1D–F). This observation suggested a metabolic switching (altered regioselective metabolism) induced by fluconazole. Considering the entire concentration-time profile (Table 1), fluconazole significantly increased total AUC4-OH 2.2- to 2.9-fold for all three CYP3A5 genotype groups, while total AUC1’-OH increased slightly (1.2- to 1.3-fold). The AUC4-OH/AUCMDZ ratio was increased 35–60%, while AUC1’-OH/AUCMDZ ratio was decreased 5–37%, by fluconazole. As a consequence, the AUC1’-OH/AUC4-OH ratio was significantly decreased 46–58% by fluconazole for all three genotype groups, from 6.3–9.6 in the control state to 2.9–4.9 with fluconazole. No significant difference in the AUC1’-OH/AUC4-OH ratio was observed among the genotype groups in the absence or presence fluconazole (Figure 2 and Table 1).
Figure 2. Effect of fluconazole on the AUC ratio of 1’-OH-MDZ/4-OH-MDZ.
Ratios for individual subjects in the absence (open circles) and presence (solid triangles) of fluconazole are shown for the three CYP3A5 genotype groups. Horizontal lines depict group means. Asterisk (*) indicates p<0.0001 vs. control. Ctrl, control; FLU, fluconazole.
Effect of fluconazole on midazolam metabolism in vitro
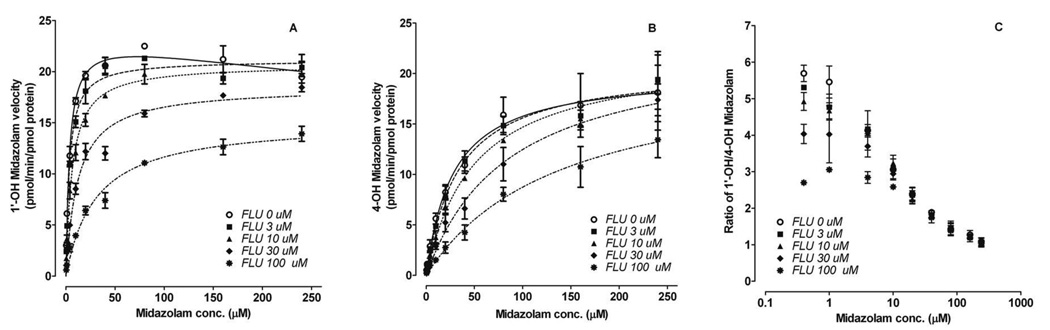
1’-OH-MDZ formation exhibited substrate inhibition kinetics with recombinantly expressed CYP3A4 (rCYP3A4) (Figure 3). In the presence of fluconazole, this profile was transformed into classic Michaelis-Menten kinetics, accompanied by a markedly increased Km and modestly decreased Vmax. In contrast, 4-OH-MDZ formation was well-described by a Michaelis-Menten model with or without fluconazole. In the presence of fluconazole, 4-OH-MDZ had markedly increased Km and unchanged Vmax. CLint of 1’-OH-MDZ and 4-OH-MDZ decreased from 6.5 to 1.5 and from 0.7 to 0.3 (pmol/min/pmol CYP3A4), respectively, at 30 µM fluconazole. Accordingly, the CLint,1’-OH/CLint, 4-OH ratio decreased from 9.1 in the absence of fluconazole to 5.7 (60%) in the presence 30 µM fluconazole (Table 2), which is similar to the peak in vivo concentrations observed after a single oral dose of fluconazole to humans (23).
Figure 3. Effect of fluconazole on midazolam metabolic kinetics.
Representative kinetic profiles of 1’-hydroxymidazolam (A) and 4-hydroxymidazolam (B) formation by rCYP3A4 in the absence and presence of fluconazole are shown. Also shown is the altered 1’-OH-MDZ/4-OH-MDZ ratio as a function of both midazolam concentration and fluconazole concentration incubated with rCYP3A4 (C). Symbols and error bars denote means and S.E., respectively, of triplicate incubations. FLU, fluconazole.
Table 2. Fluconazole effects on metabolic kinetic parameters of 1’-hydroxymidazolam and 4-hydroxymidazolam formation for recombinant CYP3A4.
Each value represents the mean ± S.E. of the parameter estimate.
| Fluconazole (µM) | 0 | 3 | 10 | 30 | 100 | |
|---|---|---|---|---|---|---|
| 1’-OH-MDZ (0.4–240 µM) | kcat (pmol/min/pmol CYP3A4) | 23.6 ± 0.9a | 21.1 ± 0.4* | 20.7 ± 0.4* | 18.5 ± 0.5* | 15.2 ± 0.6* |
| Km (µM) | 3.6 ± 0.5a | 3.5 ± 0.4 | 6.3 ± 0.6* | 12.0 ± 1.4* | 30.4 ± 4.1* | |
| Ki (µM) | 1480 ± 615a | |||||
| CLint (µl/min/pmol CYP3A4) | 6.55 | 6.03 | 3.29 | 1.54 | 0.50 | |
| 4-OH-MDZ (0.4–240 µM) | kcat (min−1) | 20.2 ± 1.0 | 20.8 ± 0.7 | 21.9 ± 1.0 | 23.1 ± 1.2 | 20.9 ± 2.0 |
| Km (µM) | 28.1 ± 4.7 | 33.4 ± 3.6 | 49.5 ± 6.2* | 85.3 ± 10.9* | 139.0 ± 26.9* | |
| CLint (µl/min/pmol CYP3A4) | 0.72 | 0.62 | 0.44 | 0.27 | 0.15 | |
| CLint, 1’-OH/CLint, 4-OHb | 9.1 | 9.7 | 7.5 | 5.7 | 3.3 | |
substrate inhibition kinetics model was fit to the data.
CLint is calculated as the ratio of mean values of kcat and Km (CLint = kcat/Km).
p<0.05 compared with control (absence of fluconazole). 1’-OH MDZ, 1’-hydroxymidazolam; 4-OH MDZ, 4-hydroxymidazolam
Unlike the observed increased formation of 4-OH-MDZ by fluconazole in vivo, both 1’-OH-MDZ and 4-OH-MDZ formation were inhibited in the presence of fluconazole in vitro. The IC50 of fluconazole was determined at a low concentration of midazolam (0.4 µM, << Km) was 15.3 µM for 1’-OH-MDZ, which was 50% lower than that for 4-OH-MDZ (33.8 µM), indicating that fluconazole preferentially inhibited 1’-OH-MDZ formation.
Comparison of allosteric interactions of midazolam and fluconazole with CYP3A4 and CYP3A5
Previous investigators (14, 24, 25) have reported that recombinantly expressed CYP3A5 (rCYP3A5) catalyzes 1’-OH-MDZ formation more efficiently than rCYP3A4, and rCYP3A5 catalyzes 4-OH-MDZ formation slightly less efficiently than rCYP3A4. In contrast to these in vitro observations, both the AUC1’-OH/AUCMDZ and AUC1’-OH/AUC4-OH ratios in the current study were similar for CYP3A5 expressors and CYP3A5 non-expressors, regardless of the presence of fluconazole.
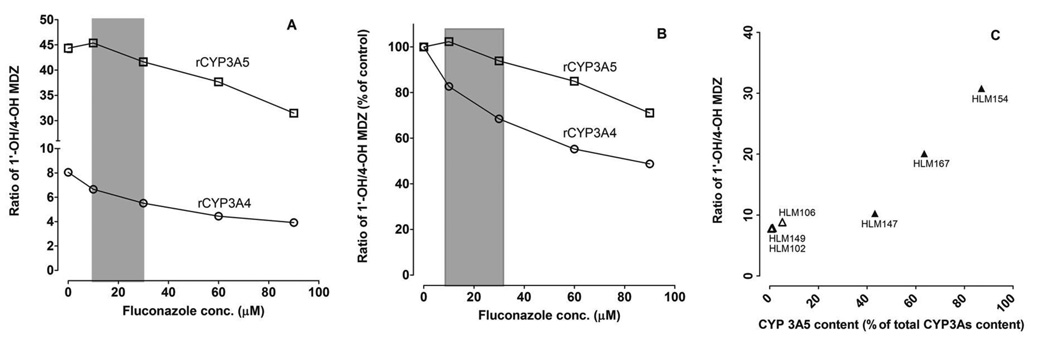
To investigate the effect of CYP3A5 genotype on the formation of midazolam metabolites under conditions predicted to occur in vivo, a sub-saturating concentration of midazolam (0.4 µM) was co-incubated with fluconazole (concentration range: 0 to 100 µM) and rCYP3A4, rCYP3A5 or HLMs from donors with different CYP3A5 genotypes. The ratio of 1’-OH-MDZ/4-OH-MDZ for rCYP3A5 (~45) was significantly greater than that for rCYP3A4 (~8) (Figure 4A). The ratio of 1’-OH-MDZ/4-OH-MDZ for rCYP3A4 decreased 20–30% by fluconazole at clinically relevant concentrations (10–30 µM) but no change was observed for CYP3A5 (Figure 4B), indicating that CYP3A5 is less susceptible than CYP3A4 to the allosteric effects of fluconazole. The ratio of 1’-OH-MDZ/4-OH-MDZ similarly was high for HLMs with a high CYP3A5 protein content, as observed for rCYP3A5, but the ratio was not significantly different for HLMs with low CYP3A5 content (Figure 4C).
Figure 4. Effect of fluconazole on the 1’-OH-MDZ/4-OH-MDZ ratio produced by rCYP3A4 and rCYP3A5.
A low concentration of midazolam (0.4 µM) was co-incubated with a range of fluconazole concentrations and rCYP3A4 or rCYP3A5 (A and B) or with different HLM preparations (C). Y-axis values are depicted as either the absolute ratio of 1’-OH-MDZ/4-OH-MDZ from the incubation (A and C) or the percentage of the control (without fluconazole) reaction (B). The grey area indicates the range of in vivo concentrations of fluconazole expected during the period of midazolam elimination after a single 400 mg oral dose of fluconazole (A and B). Open triangles (n=3) and solid triangles (n=3) represent microsomes isolated from CYP3A5 non-expressing and CYP3A5 expressing human livers, respectively (C). Symbols denote observed data and are means of duplicate incubations.
Effect of fluconazole on glucuronidation of hydroxymidazolam metabolites
After intravenous administration of midazolam, the metabolite-to-parent AUC ratio (AUCm/AUCp) is the ratio of formation clearance (CLf) to elimination clearance (CLm) of the metabolite (26). Since the determination of plasma midazolam metabolites in this study was limited solely to the primary metabolites, interpretation of the in vivo data assumed that fluconazole had no effect on CLm (i.e., glucuronidation) of hydroxymidazolam metabolites. That is, the changes in ratios of AUC4-OH/AUCMDZ, AUC1’-OH/AUCMDZ or AUC1’-OH/AUC4-OH by fluconazole reflected altered CLf, but not CLm, by fluconazole. To determine if CLm confounds any of these ratios, the effect of fluconazole on the glucuronidation of 1’-OH-MDZ and 4-OH-MDZ was examined in vitro using HLMs.
Glucuronide conjugates of hydroxymidazolam metabolites were identified as previously reported (27) (Supplementary Figure S1A). 1’-OH-MDZ forms 1’-OH-O-glucuronide-MDZ (mainly by UGT2B4 and UGT2B7) and 1’-OH-N-glucuronide-MDZ (mainly by UGT1A4). 4-OH-MDZ forms two peaks: peak 1 (by multiple UGTs) and peak 2 (mainly by UGT1A4) (27–29). At low concentrations of 1’-OH-MDZ or 4-OH-MDZ (1 µM), hecogenin (UGT1A4 inhibitor) and diclofenac (UGT2B7 substrate) decreased glucuronide formation significantly (Supplementary Figure S1B and S1C). In contrast, fluconazole had no effect on the glucuronidation of either hydroxymidazolam metabolite, even at 300 µM, a concentration 10-fold higher than that observed in vivo (Supplementary Figure S1D).
DISCUSSION
Following intravenous administration of midazolam to healthy volunteers, the major metabolic pathway (1’-hydroxylation) was inhibited, whereas the minor metabolic pathway (4-hydroxylation) was activated, by oral fluconazole, resulting in a decreased AUC1’-OH/AUC4-OH ratio. Metabolic switching of midazolam metabolism also was observed in vitro as a change in the 1’-OH-MDZ/4-OH-MDZ ratio, which decreased as a function of increasing concentrations of both fluconazole and midazolam. Because the same enzymes (CYP3A4/5) catalyze the two hydroxylation reactions, in vitro and in vivo results demonstrated the homotropic allosterism of CYP3As with midazolam alone, as well as the heterotropic allosterism of CYP3As with midazolam plus fluconazole. In vitro, 4-OH-MDZ formation from a low concentration of midazolam was less susceptible to inhibition by fluconazole than 1’-OH-MDZ formation. In vivo, the AUC1’-OH/AUC4-OH ratio was decreased significantly (46–58%) by fluconazole in a manner quantitatively similar to the decreased ratio of CLint,1’-OH/CLint,4-OH determined in vitro (60%) at clinically relevant concentrations of fluconazole. However, formation of 1’-OH-MDZ and 4-OH-MDZ were both inhibited by fluconazole in vitro, while only 1’-OH-MDZ was inhibited in vivo. This in vitro-in vivo discrepancy may reflect fundamental differences between the two systems, e.g., different substrate concentrations and temporal factors (30). Because fluconazole had no effect on the in vitro glucuronidation of either 1’-OH-MDZ or 4-OH-MDZ at clinically relevant concentrations, results of this study provide strong evidence that the metabolic shift of midazolam metabolites in vivo most likely was due to a heterotropic allosteric interaction between CYP3A4, midazolam and fluconazole. Alternatively, selective and significant contribution by a different enzyme to 4-OH-MDZ formation, such as a peroxidase, could account for the differential effect of fluconazole on the midazolam product ratio; however, to the authors’ knowledge, such a parallel metabolic process is not known to occur in vitro or in vivo.
Despite difficulties in elucidating mechanisms of CYP3A4 allosterism, prevailing opinion, based on extensive in vitro kinetic data and some crystallographic data, holds that the large substrate-binding pocket of CYP3A4 allows more than one ligand to bind simultaneously, leading to atypical kinetic behavior (3, 4, 31, 32). With midazolam, the mechanism could involve two substrate molecules or one substrate molecule and one effector molecule, such as fluconazole. 1’-OH-MDZ and 4-OH-MDZ formation by rCYP3A4 have different Km, Ki or IC50 values in the presence of the same inhibitors (15, 19, 33, 34). Moreover, the effect of fluconazole on 4-OH-MDZ formation appears competitive, whereas a mixed mechanism was observed for inhibition of 1’-OH-MDZ (Table 2). These observations support the hypothesis proposed by Khan et al (33) that two midazolam binding sites exist in the CYP3A4 active site. This hypothesis also is supported by the observation that the relative formation rates of 1’-OH-MDZ and 4-OH-MDZ changed as a function of the concentration of midazolam alone and of fluconazole (Figure 3), and is consistent with previous in vitro studies with other CYP3A4/5 effectors (13, 14, 16, 24). Crystal structures of the CYP3A4-effector complex have shown that the binding of ketoconazole to CYP3A4 increases the volume of the CYP3A4 active site significantly and that two molecules of ketoconazole bind to the active site (32). Fluconazole, like ketoconazole, is a more potent inhibitor of 1’-OH-MDZ than of 4-OH-MDZ formation (19). Thus, it is reasonable to speculate that fluconazole and midazolam can bind to CYP3A4 simultaneously and shift midazolam metabolism through an allosteric interaction.
Non-Michaelis-Menten kinetics resulting from enzyme allosterism can be discerned only when a large range of effector or substrate concentrations are employed. However, drug concentrations in most in vivo studies were too low to elicit a detectable allosteric effect (1, 10). The concentrations of substrate and effector in vivo usually were below the substrate Km and/or effector Ki (5–8, 10, 11), thus yielding largely inconclusive evidence for CYP allosterism (10). To enhance our ability to characterize CYP3A allosterism in vivo, both the major metabolite (1’-OH-MDZ) and minor metabolite (4-OH-MDZ) after intravenous midazolam administration were quantified in the absence and presence of an inhibitory effector, fluconazole. The rapid and near complete oral absorption of fluconazole, along with a slow elimination (t1/2 ~32 hrs) and low plasma protein binding (11–12%) (35), provided a stable and potent effector concentration over multiple half-lives of midazolam. This experimental design maximized the likelihood of detecting heterotropic allosterism and minimized confounding from midazolam homotropic interaction, which might best be explored in future dose-ranging studies with oral midazolam alone.
There are relatively few in vivo studies in which the plasma concentration of 4-OH-MDZ was quantified in addition to midazolam and 1’-OH-MDZ. To the authors’ knowledge, a metabolic switch between AUC1’-OH and AUC4-OH in the presence of an allosteric effector has not been reported. Although midazolam product ratios have been reported (36, 37), the current study, for the first time, revealed a good in vitro-in vivo correlation for the 1’-OH-MDZ/4-OH-MDZ ratio in the absence and presence of an allosteric effector. Importantly, unconjugated metabolites in plasma were measured in this study to focus on the initial CYP3A-dependent reactions. Previous studies showed that, depending on the CYP3A5 genotype, the ratio of total (unconjugated + conjugated) AUC1’-OH/AUC4-OH varied from 12 to 19 (36) or 24 to 36 (37). When total plasma metabolites (conjugated + unconjugated) are measured through β-glucuronidase-mediated hydrolysis, as with the two studies cited above, the product ratios represent a hybrid of the rates of primary and secondary metabolite formation. 4-OH-MDZ is known to be unstable under acidic conditions, which are optimal for β-glucuronidase activity (pH 5) (38), and N-glucuronides usually are more resistant to hydrolysis mediated by β-glucuronidase (39). Thus, it is possible that β-glucuronidase-mediated hydrolysis could cause degradation of 4-OH-MDZ and/or incomplete hydrolysis of hydroxymidazolam N-glucuronides. In addition, unlike the intravenous midazolam administration used in the present study, midazolam was given orally in one of the two previous studies (36). Based on the observation that significant first-pass metabolism of midazolam occurs in the small intestine after oral dosing (40), and that the ratio of 1’-OH-MDZ/4-OH-MDZ can be changed by the concentration of midazolam through a homotropic allosteric interaction with CYP3A4, it is probable that the in vivo 1’-OH-MDZ/4-OH-MDZ ratio is dependent on the route of midazolam administration. Therefore, different assay measurements and midazolam administration routes may contribute to the observed differences in the AUC1’-OH/AUC4-OH ratio between the present study and values reported by other investigators.
At clinically-relevant midazolam concentration (<0.5 µM), rCYP3A5 catalyzed 1’-OH-MDZ formation more efficiently and yielded a much higher basal ratio of 1’-OH-MDZ/4-OH-MDZ than did rCYP3A4 (16, 17), which was confirmed in the current study (Figure 4). However, the AUC1’-OH/AUCMDZ ratio in CYP3A5 expressors (*1/*1 and *1/*X) was similar to that in CYP3A5 nonexpressors (*X/*X) (Figure 2, Table 1). CYP3A5 genotype appeared have no effect on AUC1’-OH/AUC4-OH ratio under control and fluconazole-treated conditions. Yu et al. reported similar results (37), whereas Eap et al. reported a higher basal ratio in CYP3A5 expressors (36). This in vitro–in vivo discrepancy is unclear. It is possible that, despite the stratification of subjects based on CYP3A5 genotype, the contribution of CYP3A5 to total midazolam metabolism was too low (because of high CYP3A4 content) to detect the expected relationship between genotype and product ratio. A significant association between the in vivo Ki for total midazolam intrinsic clearance and CYP3A5 genotype has reported previously (18), but the magnitude of the Ki differences was small (compared to in vitro data) and also might reflect low CYP3A5 relative to CYP3A4 content in the CYP3A5*1 carrier groups.
Although the in vivo 1’-OH-MDZ/4-OH-MDZ ratio was altered significantly by fluconazole, and is attributed to an allosteric interaction with CYP3A4, this observation probably is not of major importance regarding the clinical use of midazolam. However, metabolic switching by allosteric effectors of other CYP3A substrates may have a greater pharmacological or toxicological significance if one or more of the metabolites have biological effects. For example, biotransformation is required for the carcinogenicity of aflatoxin B1 (AFB1) (41). CYP3A4 contributes significantly to the biotransformation of AFB1 to the carcinogenic metabolite (AFBO), as well as to the primary detoxification metabolite (AFQ1) (42). Albeit speculative, the AFBO/AFQ1 product ratio could be modulated by endogenous or exogenous substances through an allosteric interaction, altering an individual’s risk to developing cancer. Another example involves the anti-estrogen agent tamoxifen (TAM) used for the treatment of breast cancer. CYP3A4 plays a pivotal role in the formation of the major primary metabolite N-demethyl-TAM, which is converted subsequently by CYP2D6 into a much more potent secondary metabolite endoxifen (43). CYP3A4 also catalyzes the formation of a minor but putatively genotoxic primary metabolite, α-hydroxy-TAM (44). Again, although speculative, a metabolic shift due to allosteric interactions with CYP3A4 could contribute to inter-individual differences in both the therapeutic and toxicological response to TAM.
METHODS
Chemicals
Midazolam, 1’-OH-MDZ, 4-OH-MDZ, fluconazole, alamethicin, diclofenac, NADPH, UDPGA, and β-estradiol-17-glucuronide were purchased from Sigma-Aldrich (St. Louis, MO). Hecogenin was from Science Lab. Com. (Kingwood, TX). 15N3-midazolam was a gift from Roche Laboratories (Nutley, NJ). Internal standards (15N3-midazolam, 15N3-1’-OH-MDZ and 15N3-4-OH-MDZ) were prepared as previously described (45). rCYP3A4 and rCYP3A5 (with human P450 reductase and cytochrome b5; Supersomes™) were from BD Gentest (Woburn, MA). N-methyl-N-(t-butyl-dimethylsilyl)trifluoroacetamide (MTBSTFA) was from Pierce Chemical (Rockford, IL). All other chemicals were of analytical grade.
Human subject study
General issues: A midazolam-fluconazole interaction study involving healthy volunteers was approved by the Institutional Review Board and the Clinical Research Advisory Committee at the University of North Carolina at Chapel Hill. A complete description of the study and initial pharmacokinetic analysis of plasma midazolam and fluconazole was published previously (18). Some details are repeated here for clarity. Subjects: Subjects were recruited and grouped based on their CYP3A5 genotype (18). Homozygous CYP3A5 carriers (CYP3A5*1/*1, n=5; one subject less than the previous analysis because of a limited sample volume), heterozygous CYP3A5 carriers (CYP3A5*1/*X, n=7) and homozygous non-carriers (CYP3A5*X/*X, n=6) were examined, where X represents a CYP3A5*3, *6 or *7 allele. None of the subjects were taking prescription or over-the-counter medications known to alter CYP3A activity. Subjects were instructed to abstain from consuming grapefruit-containing products at least seven days before and during the study, as well as to abstain from caffeinated and alcoholic beverages during the study phases. Study design: The study consisted of two phases scheduled on two consecutive days. All subjects underwent an overnight fast before each phase. On the morning of phase 1, midazolam (1 mg; Bedford Laboratories, Bedford, OH) was administered intravenously. Serial blood (5 mL) was collected just before and from 5 to 720 min after midazolam administration. On the morning of phase 2, a single dose (400 mg) of fluconazole was given orally 120 minutes before midazolam administration. Serial blood collection was repeated. Plasma was isolated by centrifugation (3500g, 15 min, 4°C) and stored at −20°C prior to analysis. Quantification of midazolam and primary metabolite in plasma: Midazolam and metabolites were measured by GC/MS, as described previously (18, 45). Calibration standards (0.3 to 320 nM) for midazolam and metabolites were prepared using human plasma. Quality controls contained 6 and 160 nM of midazolam and metabolites. Inter-day coefficients of variation for midazolam and metabolites in quality controls were <8.0% at 6 nM and <3.6 % at 160 nM.
Midazolam metabolic studies
A mixture containing 20 pmol/ml rCYP3A or 0.1 mg/ml human liver microsomal protein, midazolam (0.4 to 240 µM), fluconazole (0 to 100 µM) and potassium phosphate buffer (0.1 M, pH 7.4) was pre-warmed at 37°C for 5 min. Reactions were initiated with NADPH (1 mM) and then terminated after 2 min by an equal volume of ice-cold Na2CO3 (0.1 M, pH 11). 1’-OH-MDZ and 4-OH-MDZ were quantified by GC-MS as described previously (18, 45).
Glucuronidation of hydroxymidazolams in HLMs
Pooled HLMs (0.75 mg/ml) were pre-treated with alamethicin (50 µg/mg protein) in Tris-HCl (50 mM, pH7.4), on ice, for 15 min. MgCl2 (50 mM) and substrate (1’-OH-MDZ or 4-OH-MDZ, 1 µM) with or without inhibitors were mixed with the pre-treated HLM and pre-warmed at 37°C for 5 min. Reactions were initiated with UDPGA (50 mM) and quenched with ice-cold acetonitrile (40% of aqueous mixture, v/v) after 20 min. The mixtures were centrifuged for 5 min (10,000g, 4°C). The supernatants were analyzed by LC/MS/MS.
Glucuronide conjugates of hydroxymidazolam metabolites were identified as described previously(27) with some modifications. Briefly, an API-4000 LC/MS/MS system (Applied Biosystems, Foster City, CA) coupled with a Shimadzu SCL-10AVP liquid chromatography system was used. Separation was accomplished with a Polar-RP column (2 mm i.d. × 250 mm, 4-µM particle size, Phenomenex, Torrance, CA). Analytes were eluted at 0.2 ml/min with a mobile phase consisting of 0.1% formic acid (A) and acetonitrile (B) using a linear gradient. Solvent B began at 25%, then increased to 40% (10 min), 50% (12 min) and 25% (15 min). The Turbo Spray interface was operated in positive ion mode at 4500 V and 500°C. Declustering potential and collision energy was 70V and 35eV. Selected ions were: m/z 514→342 (glucuronides of 1’-OH-MDZ and 4-OH-MDZ) and m/z 447→271 (β-estradiol-17-glucuronide, internal standard). Standard solutions with known amount of 1’-OH-MDZ-O-glucuronide (Cayman Chemical Company, Ann Arbor, MI) were used to quantify 1’-OH-MDZ-O-glucuronide. Analysis of 1’-OH-MDZ-N-glucuronide and 4-OH-MDZ–glucuronides was considered semi-quantitative due to the lack of authentic standards.
Data analysis
Pharmacokinetic analysis
AUC values were determined by the log-linear trapezoidal method, with extrapolation to infinity by dividing the last measurable plasma concentration by the terminal slope (WinNonlin version 5.2, Pharsight). The percent of extrapolated AUC was 13.0 ± 9.5%, 18.4 ± 10.6% and 19.0 ± 8.3 (mean ± S.D.) for midazolam, 1’-OH-MDZ and 4-OH-MDZ, respectively.
Determination of in vitro midazolam kinetic parameters
Kinetic parameters for 1’-OH-MDZ and 4-OH-MDZ formation were estimated by non-linear regression using GraphPad Prism5 (La Jolla, CA). Kinetic models (substrate inhibition and hyperbolic kinetics) were evaluated from visual inspection, extra sum-of-squares F test and randomness of the residuals. IC50 values were obtained by fitting the one-site competitive inhibition binding model to the data using GraphPad Prism5.
Statistical analysis
Statistical analysis of the in vivo data was performed using SAS version 9.2 (SAS Institute, Cary, NC). Repeated measures two-way analysis of variance was used for comparisons across the CYP3A5 genotype groups and fluconazole treatment, followed by the Bonferroni test for multiple comparisons. An unpaired t-test was used to compare the best-fit values (Vmax, Km) between control and fluconazole treatment. One way analysis of variance followed by Dunnett’s test was used for comparisons of glucuronidation of hydroxymidazolams between control and with inhibitors using GraphPad Prism5. p<0.05 was considered significant for all tests.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants P01 GM32165, R01 63666 and M01 RR00046. The authors thank Dr. David Blough for his assistance in statistical analysis, Dr. Yoshihisa Shitara for his help with in vitro kinetics study and the GCRC staff at the University of Carolina at Chapel Hill for their excellent assistance with the clinical study.
Footnotes
CONFLICT OF INTEREST/DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1.Tracy TS. Atypical cytochrome P450 kinetics: implications for drug discovery. Drugs in R&D. 2006;7:349. doi: 10.2165/00126839-200607060-00004. [DOI] [PubMed] [Google Scholar]
- 2.Atkins WM. Implications of the allosteric kinetics of cytochrome P450s. Drug Discov Today. 2004;9:478–484. doi: 10.1016/S1359-6446(04)03072-7. [DOI] [PubMed] [Google Scholar]
- 3.Wei T, Stearns RA. Heterotropic Cooperativity of Cytochrome P450 3A4 and Potential Drug-Drug Interactions. Current Drug Metabolism. 2001;2:185. doi: 10.2174/1389200013338658. [DOI] [PubMed] [Google Scholar]
- 4.Davydov DR, Halpert JR. Allosteric P450 mechanisms: multiple binding sites, multiple conformers or both? Expert Opinion on Drug Metabolism and Toxicology. 2008;4:1523–1535. doi: 10.1517/17425250802500028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang W, Stearns RA, Kwei GY, Iliff SA, Miller RR, Egan MA, et al. Interaction of Diclofenac and Quinidine in Monkeys: Stimulation of Diclofenac Metabolism. J Pharmacol Exp Ther. 1999;291:1068–1074. [PubMed] [Google Scholar]
- 6.van Waterschoot RAB, Rooswinkel RW, Sparidans RW, van Herwaarden AE, Beijnen JH, Schinkel AH. Inhibition and Stimulation of Intestinal and Hepatic CYP3A Activity: Studies in Humanized CYP3A4 Transgenic Mice Using Triazolam. Drug Metabolism and Disposition. 2009;37:2305–2313. doi: 10.1124/dmd.109.029397. [DOI] [PubMed] [Google Scholar]
- 7.Backman JT, Maenpaa J, Belle DJ, Wrighton SA, Kivisto KT, Neuvonen PJ. Lack of correlation between in vitro and in vivo studies on the effects of tangeretin and tangerine juice on midazolam hydroxylation. Clin Pharmacol Ther. 2000;67:382–390. doi: 10.1067/mcp.2000.105756. [DOI] [PubMed] [Google Scholar]
- 8.Hutzler JM, Frye RF, Korzekwa KR, Branch RA, Huang SM, Tracy TS. Minimal in vivo activation of CYP2C9-mediated flurbiprofen metabolism by dapsone. European Journal of Pharmaceutical Sciences. 2001;14:47–52. doi: 10.1016/s0928-0987(01)00144-0. [DOI] [PubMed] [Google Scholar]
- 9.Egnell AC, Houston B, Boyer S. In vivo CYP3A4 heteroactivation is a possible mechanism for the drug interaction between felbamate and carbamazepine. J Pharmacol Exp Ther. 2003;305:1251–1262. doi: 10.1124/jpet.102.047530. [DOI] [PubMed] [Google Scholar]
- 10.Jones JP. Metabolic ménages à trois: what does it mean for drug design? Drug Discovery Today. 2004;9:592. doi: 10.1016/S1359-6446(04)03109-5. [DOI] [PubMed] [Google Scholar]
- 11.Hutzler JM, Tracy TS. Atypical Kinetic Profiles in Drug Metabolism Reactions. Drug Metab Dispos. 2002;30:355–362. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 12.Reves JG, Fragen RJ, Vinik HR, Greenblatt DJ. Midazolam: pharmacology and uses. Anesthesiology. 1985;62:310–324. [PubMed] [Google Scholar]
- 13.Cameron MD, Wen B, Allen KE, Roberts AG, Schuman JT, Campbell AP, et al. Cooperative Binding of Midazolam with Testosterone and α-Naphthoflavone within the CYP3A4 Active Site: A NMR T1 Paramagnetic Relaxation Study. Biochemistry. 2005;44:14143–14151. doi: 10.1021/bi051689t. [DOI] [PubMed] [Google Scholar]
- 14.Williams JA, Ring BJ, Cantrell VE, Jones DR, Eckstein J, Ruterbories K, et al. Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos. 2002;30:883–891. doi: 10.1124/dmd.30.8.883. [DOI] [PubMed] [Google Scholar]
- 15.Ghosal A, Satoh H, Thomas PE, Bush E, Moore D. Inhibition and kinetics of cytochrome P4503A activity in microsomes from rat, human, and cdna-expressed human cytochrome P450. Drug Metabolism and Disposition. 1996;24:940–947. [PubMed] [Google Scholar]
- 16.Wandel C, Bocker R, Bohrer H, Browne A, Rugheimer E, Martin E. Midazolam is metabolized by at least three different cytochrome P450 enzymes. Br J Anaesth. 1994;73:658–661. doi: 10.1093/bja/73.5.658. [DOI] [PubMed] [Google Scholar]
- 17.Gorski JC, Hall SD, Jones DR, VandenBranden M, Wrighton SA. Regioselective biotransformation of midazolam by members of the human cytochrome P450 3A (CYP3A) subfamily. Biochem Pharmacol. 1994;47:1643–1653. doi: 10.1016/0006-2952(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 18.Isoherranen N, Ludington SR, Givens RC, Lamba JK, Pusek SN, Dees EC, et al. The Influence of CYP3A5 Expression on the Extent of Hepatic CYP3A Inhibition Is Substrate-Dependent: An in Vitro-in Vivo Evaluation. Drug Metabolism and Disposition. 2008;36:146–154. doi: 10.1124/dmd.107.018382. [DOI] [PubMed] [Google Scholar]
- 19.von Moltke LL, Greenblatt DJ, Schmider J, Duan SX, Wright CE, Harmatz JS, et al. Midazolam hydroxylation by human liver microsomes in vitro: inhibition by fluoxetine, norfluoxetine, and by azole antifungal agents. J Clin Pharmacol. 1996;36:783–791. doi: 10.1002/j.1552-4604.1996.tb04251.x. [DOI] [PubMed] [Google Scholar]
- 20.Wang RW, Newton DJ, Liu N, Atkins WM, Lu AYH. Human Cytochrome P-450 3A4: In Vitro Drug-Drug Interaction Patterns Are Substrate-Dependent. Drug Metab Dispos. 2000;28:360–366. [PubMed] [Google Scholar]
- 21.Wang RW, Newton DJ, Scheri TD, Lu AYH. Human Cytochrome P450 3A4-Catalyzed Testosterone 6β-Hydroxylation and Erythromycin N-Demethylation. Drug Metab and Dispos. 1997;25:502–507. [PubMed] [Google Scholar]
- 22.Houston JB, Galetin A. Progress towards prediction of human pharmacokinetic parameters from in vitro technologies. Drug Metab Rev. 2003;35:393–415. doi: 10.1081/dmr-120026870. [DOI] [PubMed] [Google Scholar]
- 23.Grant SM, Clissold SP. Fluconazole. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in superficial and systemic mycoses. Drugs. 1990;39:877–916. doi: 10.2165/00003495-199039060-00006. [DOI] [PubMed] [Google Scholar]
- 24.Huang W, Lin YS, McConn DJ, II, Calamia JC, Totah RA, Isoherranen N, et al. Evidence of Significant Contribution from CYP3A5 to Hepatic Drug Metabolism. Drug Metab Dispos. 2004;32:1434–1445. doi: 10.1124/dmd.104.001313. [DOI] [PubMed] [Google Scholar]
- 25.Wandel C, Bocker R, Bohrer H, Browne A, Rugheimer E, Martin E. Midazolam is metabolized by at least three different cytochrome P450 enzymes. Br J Anaesth. 1994;73:658–661. doi: 10.1093/bja/73.5.658. [DOI] [PubMed] [Google Scholar]
- 26.Houston JB. Drug metabolite kinetics. Pharmacology & Therapeutics. 1981;15:521–552. doi: 10.1016/0163-7258(81)90056-5. [DOI] [PubMed] [Google Scholar]
- 27.Seo K-A, Bae SK, Choi Y-K, Choi CS, Liu K-H, Shin J-G. Metabolism of 1'- and 4-Hydroxymidazolam by Glucuronide Conjugation Is Largely Mediated by UDP-Glucuronosyltransferases 1A4, 2B4, and 2B7. Drug Metabolism and Disposition. 2010;38:2007–2013. doi: 10.1124/dmd.110.035295. [DOI] [PubMed] [Google Scholar]
- 28.Hyland R, Osborne T, Payne A, Kempshall S, Logan YR, Ezzeddine K, et al. In vitro and in vivo glucuronidation of midazolam in humans. British Journal of Clinical Pharmacology. 2009;67:445–454. doi: 10.1111/j.1365-2125.2009.03386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu B, Bush D, Doss GA, Vincent S, Franklin RB, Xu S. Characterization of 1'-Hydroxymidazolam Glucuronidation in Human Liver Microsomes. Drug Metab Dispos. 2008;36:331–338. doi: 10.1124/dmd.107.017962. [DOI] [PubMed] [Google Scholar]
- 30.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 31.Shou M, Grogan J, Mancewicz JA, Krausz KW, Gonzalez FJ, Gelboin HV, et al. Activation of CYP3A4: Evidence for the Simultaneous Binding of Two Substrates in a Cytochrome P450 Active Site. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 32.Ekroos M, Sjögren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proceedings of the National Academy of Sciences. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan KK, He YQ, Domanski TL, Halpert JR. Midazolam Oxidation by Cytochrome P450 3A4 and Active-Site Mutants: an Evaluation of Multiple Binding Sites and of the Metabolic Pathway That Leads to Enzyme Inactivation. Mol Pharmacol. 2002;61:495–506. doi: 10.1124/mol.61.3.495. [DOI] [PubMed] [Google Scholar]
- 34.Miller GP, Guengerich FP. Elucidation of Distinct Ligand Binding Sites for Cytochrome P450 3A4. Biochemistry. 2000;39:5929–5939. doi: 10.1021/bi992765t. [DOI] [PubMed] [Google Scholar]
- 35.Dudley MN. Clinical pharmacology of fluconazole. Pharmacotherapy. 1990;10:141S–145S. [PubMed] [Google Scholar]
- 36.Eap CB, Buclin T, Hustert E, Bleiber G, Golay KP, Aubert A-C, et al. Pharmacokinetics of midazolam in CYP3A4- and CYP3A5-genotyped subjects. European Journal of Clinical Pharmacology. 2004;60:231–236. doi: 10.1007/s00228-004-0767-7. [DOI] [PubMed] [Google Scholar]
- 37.Yu K-S, Cho J-Y, Jang I-J, Hong K-S, Chung J-Y, Kim J-R, et al. Effect of the CYP3A5 genotype on the pharmacokinetics of intravenous midazolam during inhibited and induced metabolic states[ast] Clin Pharmacol Ther. 2004;76:104–112. doi: 10.1016/j.clpt.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 38.de Loor H, de Jonge H, Verbeke K, Vanrenterghem Y, Kuypers DR. A highly sensitive liquid chromatography tandem mass spectromety method for simultaneous quantification of midazolam, 1′-hydroxymidazolam and 4-hydroxymidazolam in human plasma. Biomedical Chromatography. doi: 10.1002/bmc.1576. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 39.Hawes EM. N +-Glucuronidation, a Common Pathway in Human Metabolism of Drugs With a Tertiary Amine Group. Drug Metabolism and Disposition. 1998;26:830–837. [PubMed] [Google Scholar]
- 40.Paine MF, Shen DD, Kunze KL, Perkins JD, Marsh CL, McVicar JP, et al. First-pass metabolism of midazolam by the human intestine[ast] Clin Pharmacol Ther. 1996;60:14–24. doi: 10.1016/S0009-9236(96)90162-9. [DOI] [PubMed] [Google Scholar]
- 41.Wild CP, Turner PC. The toxicology of aflatoxins as a basis for public health decisions. Mutagenesis. 2002;17:471–481. doi: 10.1093/mutage/17.6.471. [DOI] [PubMed] [Google Scholar]
- 42.Kamdem Landry K, Meineke Ingolf, Gödtel-Armbrust Ute, Brockmöller Jürgen, Wojnowski L. Dominant Contribution of P450 3A4 to the Hepatic Carcinogenic Activation of Aflatoxin B1. Chemical Research in Toxicology. 2006;19:577–586. doi: 10.1021/tx050358e. [DOI] [PubMed] [Google Scholar]
- 43.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 44.Rochat B. Role of cytochrome P450 activity in the fate of anticancer agents and in drug resistance: focus on tamoxifen, paclitaxel and imatinib metabolism. Clin Pharmacokinet. 2005;44:349–366. doi: 10.2165/00003088-200544040-00002. [DOI] [PubMed] [Google Scholar]
- 45.Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, et al. Characterization of Interintestinal and Intraintestinal Variations in Human CYP3A-Dependent Metabolism. J Pharmacol Exp Ther. 1997;283:1552–1562. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.