Abstract
The Tudor domain comprises a family of motifs that mediate protein-protein interactions required for various DNA-templated biological processes. Emerging evidence demonstrates a versatility of the Tudor family domains by identifying their specific interactions to a wide variety of histone methylation marks. Here, we discuss novel functions of a number of Tudor-containing proteins (including JMJD2A, 53BP1, SGF29, Spindlin1, UHRF1, PHF1, PHF19 and SHH1) in ‘reading’ unique methylation events on histones in order to facilitate DNA damage repair or regulate transcription. This review covers our recent understanding of the molecular bases for histone-Tudor interactions and their biological outcomes. As deregulation of Tudor-containing proteins is associated with certain human disorders, pharmacological targeting of Tudor interactions could provide new avenues for therapeutic intervention.
Keywords: histone methylation, epigenetic ‘reader’, Tudor domain, JMJD2A, 53BP1, SGF29, Spindlin1, UHRF1, PHF1, PHF19, SHH1
Histone modification and its ‘reader’ proteins in gene regulation
In eukaryotic cells, DNA is packaged with core histones H2A, H2B, H3, and H4 to form the basic building unit of chromatin – nucleosomes. These histones possess many sites for post-translational modification (PTM) such as methylation, acetylation, ubiquitination, and phosphorylation, which constitute a hypothetical ‘histone code’ for chromatin organization and gene regulation [1]. It has been postulated that functional interpretation of histone PTMs is executed at least in part by so-called histone ‘reader’ proteins, which use structurally conserved domains to recognize and engage histone PTMs in a sequence- and modification-specific fashion. ‘Reader’-mediated chromatin interaction helps recruit and/or stabilize the associated multi-protein complexes to specific loci to further alter chromatin structure and regulate DNA-dependent processes in various biological contexts [2–4]. Deregulation in interpretation of histone PTMs has been causally linked to the development of various human diseases including cancer [5–7], immune dysfunction [8, 9], and neurological disorders [8, 10]. Therefore, dissecting the biochemical bases for histone-‘reader’ interactions could promote a deeper understanding of the fundamental mechanism that underlies gene regulation and pathogenesis.
A number of protein domain families have been identified that specifically recognize histone PTMs. For example, bromodomains bind histone lysine acetylation in a promiscuous manner [2, 4, 11], 14-3-3 and BRCT (BRCA1 C-terminus) domains bind to histone serine or threonine phosphorylation [2, 4], and different subsets of PHD (plant homeodomain) finger motifs are able to recognize different degrees of methylation status at histones H3 lysine 4 [2, 4, 12–17]. Furthermore, a large family of so-called Royal family domains including Tudor, chromo, MBT (malignant brain tumor), and PWWP (pro-trp-trp-pro) domains have been shown to interact with methylated histone tails [2, 4]. In this review, we focus on recent studies that reveal the multifaceted capacities of various Tudor domains in ‘reading’ different histone methylation marks on chromatin and discuss how these Tudor-containing ‘readers’ and associated protein complexes further direct chromatin state-dependent regulation of gene transcription and DNA damage repair.
Tudor domain as ‘readers’ of histone PTMs
The Tudor domain was named after the Drosophila tudor (tud) gene identified in a screen for maternal-effect recessive lethality or sterility [18]. Drosophila tud contains eleven repeats of a conserved motif, subsequently termed Tudor, which appears in many proteins throughout various species [19, 20]. The Tudor domain typically contains approximately sixty amino acids that comprise 4–5 antiparallel β-strands to form a barrel-like structure. A number of Tudor-containing proteins were found to interact with methylated arginine residues in non-histone proteins involved in the regulation of RNA metabolism, alternative splicing, small RNA pathways, or germ cell development [21, 22], whereas other Tudor domains were shown to form a chromodomain-like cage [23] at their surfaces to accommodate a methylated lysine [24–26]. Among the approximately 30 mammalian Tudor-containing proteins, 53BP1 and JMJD2A were the first ones that were shown to harbor histone methylation-binding capacities via Tudor [27–29]. Recent studies have identified novel functions of a number of other Tudor-containing proteins including SGF29 [30, 31], Spindlin1 [32, 33], UHRF1 [34–37], PCL family proteins (PHF1 [38–40] and PHF19 [38, 40–42]), and SHH1 [43], in ‘reading’ a variety of different histone methylations (summarized in Table 1); in addition, recent works also demonstrated that JMJD2A and 53BP1 binding to dimethylated histone H4 lysine 20 (H4K20me2) is critical for regulation of cellular response to DNA damage [44, 45]. In the following sections, we discuss our current understanding of the molecular basis and biological function of these new Tudor-histone interactions.
Table 1.
Mammalian Tudor domain-containing proteins as histone methylation ‘readers’, and their biological functions
| Protein | Domain architecture | Tudor ligand | Kd (μM) | Biological functions | Ref. |
|---|---|---|---|---|---|
| JMJD2A |
|
H3K4me3 | ~0.5 | H3K9me3 and H3K36me3- specific demethylase; transcriptional regulation and regulator of DNA damage response | 28, 45, 48 |
| H4K20me3 | ~0.4 | ||||
| H4K20me2 | ~2 | ||||
| 53BP1 |
|
H4K20me2 | 20~50 | substrate of ATM; promote non-homologous end joining DNA repair | 29, 44 |
| SGF29 |

|
H3K4me3 | 1~4 | component of SAGA complex; mediate transcriptional activation | 30, 31 |
| Spindlin1 |

|
H3K4me3 | ~0.8 | nucleolar protein; promote rRNA transcription | 32, 33 |
| UHRF1 |
|
H3K9me3 by Tudor | 1~3 | partner of DNMT1; maintain the level of DNA methylation during DNA replication | 35, 37, 66, 88, 89 |
| H3 N-terminus& K9me3 byTudor-PHD | ~0.4 | ||||
| PHF1 |
|
H3K36me3 | 5~50# | accessory component of PRC2 complex; promote transcriptional repression | 38, 39, 40 |
| PHF19 |
|
H3K36me3 | 6~35# | accessory component of PRC2 complex; promote transcriptional repression | 38, 40, 41, 42 |
| LBR |
|
H4K20me2 | N.D. | inner nuclear membrane protein; promote formation of nuclear peripheral heterochromatin | 84 |
| TDRD3 |
|
H4R3me2a; H3R17me2a; H3R2me2a | >500 | transcriptional coactivator and interacts with CARM1 and PRMT1 | 105, 109 |
Abbreviations: Kd, dissociation constant; N.D., not defined; ATM, Ataxia telangiectasia mutated; PRC2, polycomb repressive complex 2 Modifications: me1, monomethylation; me2, dimethylation; me3, trimethylation; me2a, asymmetric dimethylation.
Protein domains: UBA, ubiquitin-associated domain; JmjN, jumonji N domain; JmjC, jumonji C domain; PHD, plant homeodomain; BRCT, BRCA1 C terminus domain; UBL, ubiquitin-like domain; SRA, SET and RING finger associated domain; RING, really interesting new gene finger domain.
for PHF1 or PHF19 binding to H3K36me3 peptides, a Kd of 5~6 μM was obtained at 4 degree in a buffer of 100 mM NaCl and 20 mM Tris-HCl pH 7.5, and a higher Kd of 35–50 μM obtained at 25 degree in a buffer of 150 mM NaCl and 20 mM Tris, pH 6.8–7.5.
Hybrid tandem-Tudor domain as a histone PTM ‘reader’
JMJD2A, JMJD2B and JMJD2C, three members of the Jumonji domain – containing 2 (JMJD2) family of proteins, all contain a JmjN-JmjC domain that specifically removes tri-/di-methylation marks on histone H3 Lys9 and Ly36 (H3K9me3/2 and H3K36me3/2), two PHD fingers, and two Tudor domains in tandem (termed tandem-Tudor) near their C-termini (Table 1) [46]. Although the tandem-Tudor domain is not essential for demethylating activities [47], it harbors binding activities towards tri-methylation of histone H3 Lys4 and histone H4 Lys20 (H3K4me3 and H4K20me3), indicating a chromatin-targeting mechanism for these enzymes [28, 48].
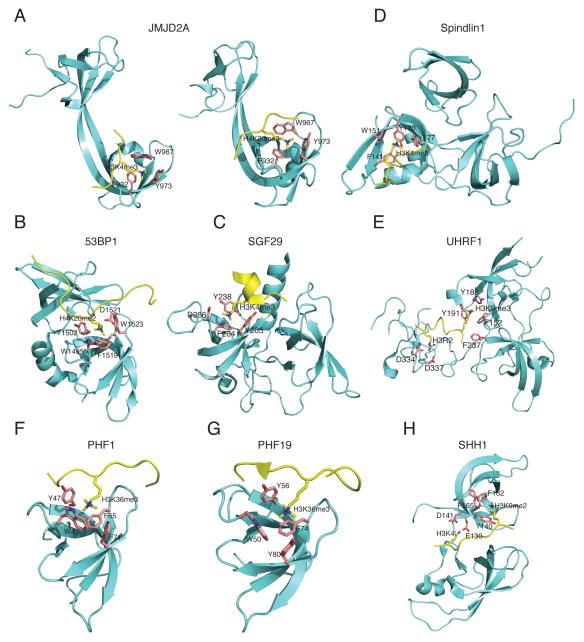
Structurally, the two Tudor domains in the JMJD2A tandem-Tudor inter-digitate with two shared β-strands to form a bilobal, saddle-shaped structure, with each hybrid lobe resembling a canonical Tudor fold (Figure 1A) [28]. The second lobe uses a cluster of aromatic residues, F932, W967 and Y973 to establish an open ‘cage’-like structure for binding the side chain of H3K4me3 or H4K20me3 (Figure 1A, left and right panels) [28]. The two complexes share high similarity in the overall hybrid lobe structure, the aromatic ‘cage’, and the binding affinities (Table 1). The H3 and H4 peptides, however, contact to the Tudor domains in opposite orientations and at different surfaces of the second hybrid Tudor domain (Figure 1A, left versus right) [48].
Figure 1. Structure of Tudor domains bound to their histone lysine methylation ligands.
Panels shown are the structure for a tandem-Tudor domain of JMJD2A in complex with H3K4me3 (panel A, left) or H4K20me3 (panel A, right); a tandem-Tudor domain of 53BP1 in complex with H4K20me2 (panel B); that of SGF29 (panel C) and Spindlin1 (panel D) in complex with H3K4me3; the linked Tudor-PHD modules of UHRF1 in complex with H3K9me3 and the unmodified N-terminus of histone H3 (panel E); a single Tudor motif of PHF1 (panel F) or PHF19 (panel G) in complex with H3K36me3; and a cryptic, tandem Tudor-like module of SHH1 in complex with H3K9me3 and the unmodified H3K4 (panel H). The Tudor sequences and histone peptides are colored in green and yellow, respectively. Identification of each of the aromatic residues involved in the formation of histone methylation-binding ‘cage’ or ‘pocket’ are labeled with their side chains colored in purple. The Protein Data Bank (PDB) accession numbers for structures presented in panels A to H are 2GFA, 2QQS, 2LVM, 3ME9, 4H75, 3ASK, 4HCZ, 4BD3, and 4IUT, respectively.
The JMJD2A tandem-Tudor domain also binds in relatively high affinity to H4K20 dimethylation (H4K20me2) (Table 1), a histone PTM known to mark the site of DNA damage and to recruit a critical DNA repair factor 53BP1 (which also uses a tandem-Tudor domain to bind H4K20me2; see the next section) [45]. A recent study showed that JMJD2A and JMJD2B proteins engage H4K20me2 and ‘mask’ the accessibility of this histone PTM to 53BP1 in non-damaged cells (Figure 2A) [45]. Upon DNA damage, the E3 ubiquitin ligases RNF8 and RNF168 degrade JMJD2 via an ubiquitination-dependent mechanism, thus allowing exposure of H4K20me2 and induction of 53BP1-mediated loci formation (Figure 2A) [45]. This highlights an elegant mechanism to specifically expose H4K20me2 at DNA damage sites where 53BP1 is recruited. Taken together, binding to histone methylation by the JMJD2A Tudor controls DNA damage-induced cellular response by antagonizing 53BP1.
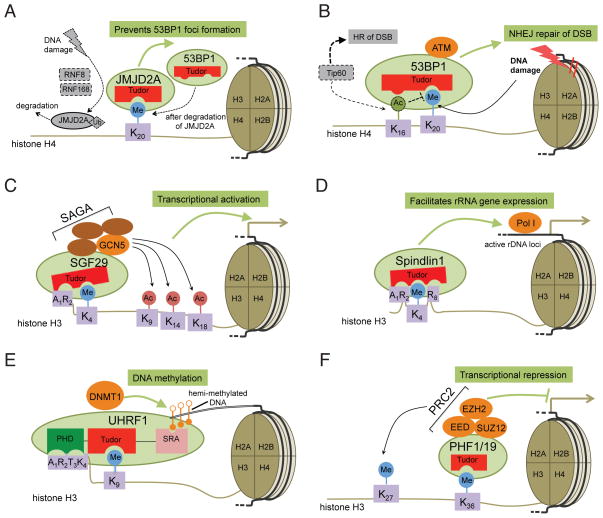
Figure 2. Functional ‘read-out’ of histone methylation by Tudor-containing proteins and their associated complexes.
(A) JMJD2A Tudor domain (in red) binds to H4K20me2 in undamaged cells and prevents the damage-associated formation of 53BP1 loci. Upon DNA damage, E3 ubiquitin ligases RNF8 and RNF168 promote JMJD2A ubiquitination and degradation at sites of DNA damage, which allow 53BP1 to bind to H4K20me2 and form loci at the damaged sites.
(B) During cellular response to DNA damage, recognition of H4K20me2 by the 53BP1 Tudor (in red) helps to efficiently recruit 53BP1 to sites of double-strand DNA breaks (DSB) where 53BP1 promotes the non-homologous end joining (NHEJ) repair pathway. TIP60-mediated H4K16 acetylation inhibits 53BP1 interaction to H4K20me2, blocks 53BP1 recruitment, and promotes BRCA1-mediated homologous repair (HR) pathway.
(C) ‘Reading’ H3K4me3 and the N-terminus of H3 (exampled by A1R2) by the SGF29 Tudor (in red) is critical for recruitment of GCN5 and other SAGA complex components to target gene promoters where GCN5-SAGA promotes histone acetylation and gene transcription.
(D) The nucleolar protein Spindlin1 utilizes a Tudor domain (in red) to recognize H3K4me3 and the surrounding H3 residues (exemplified by H3R2 and H3R8), which facilitates its recruitment to active rDNA genes and its promotion of rRNA expression.
(E) A multivalent engagement of the unmodified N-terminus of histone H3 (shown as A1R2T3K4), H3K9me3, and the hemi-methylated DNA by the linked UHRF1 PHD finger (in green), Tudor (in red) and SRA (in pink) motifs, respectively, helps to recruit UHRF1 and associated DNMT1 to heterochromatin. DNMT1 subsequently methylates the newly synthesized DNA and maintains a normal cellular level of DNA methylation.
(F) Binding to H3K36me3 by the PHF1 or PHF19 Tudor (in red) provides a novel mechanism for recruiting PRC2 to a subset of actively transcribed genes, which results in optimal H3K27me3 and repression of gene transcription.
Ac, acetylation; Me, methylation; Ph, phosphorylation; rDNA, ribosomal DNA; All Tudor-containing proteins are colored in light green.
Independent tandem-Tudor domain as a histone PTM ‘reader’
53BP1 Tudor, a ‘reader’ of methylated H4K20
The mammalian 53BP1 and its yeast homologue Crb2 are evolutionarily conserved checkpoint proteins involved in DNA damage response. ATM family kinases phosphorylate 53BP1/Crb2 upon insults such as DNA double-strand breaks (DSB). 53BP1/Crb2 subsequently re-localizes to DSB sites and promotes formation of the ionizing radiation-induced foci, an assembly of numerous DNA repair and checkpoint proteins [49]. The tandem Tudor domain of 53BP1 has been shown to bind damage-induced dimethylation of p53 and facilitate p53 accumulation during DNA repair [50, 51]. Radiation sensitivity and elevated tumor risk in 53BP1-deficient mice lend credence to the role of this protein in DNA damage response [52]. Recruitment of 53BP1 occurs, in part, through its association with methylated H4K20 (H4K20me) at DSB sites (Figure 2B) [27, 29, 53], a process negatively controlled by JMJD2A (Figure 2A). 53BP1 promotes the non-homologous end-joining (NHEJ) repair pathway whereas BRCA1 antagonizes 53BP1 to promote homologous recombination (HR) [54–56]. Biochemical and structural analysis indicate that a conserved tandem-Tudor domain of 53BP1 (Table 1) and Crb2 preferentially interacts with H4K20me2, though it also binds to H4K20me1 [29]. The 53BP1 tandem-Tudor domain forms two independently folded structures, which is different from the JMJD2A hybrid-tandem Tudor domain described above, despite their sequence similarity. The H4K20me2-binding cage comprises four aromatic residues, W1495, Y1502, F1519, and W1523, and an aspartic residue D1521, all of which reside in the first Tudor domain (Figure 1B). These aromatic residues interact with the dimethyllysine ammonium group of H4K20me2 through van der Waals and cation-π interactions, while a direct hydrogen bond formed between the amino proton of H4K20me2, and the carboxylate group of D1521 dictates selectivity for di- or mono-methylation over tri-methylation [29]. Mutations of these critical residues impaired binding to H4K20me2 and also compromised efficient 53BP1 targeting to DSB [29], which is in agreement with the genetic interaction between Crb2 and the H4K20 site observed in yeast [27, 53, 57].
The 53BP1 Tudor motif also forms extensive contacts with histone residues adjacent to H4K20me2 such as H4K16 [44] and H4H18 [29]. It is believed that association with adjacent histone sequences contributes to binding specificity, selectivity, and/or affinity, which is a common theme for almost all of the structurally defined histone methylation-‘readers’, including Tudor. Indeed, a recent study showed that recognition of H4K20me2 by 53BP1 is inhibited by TIP60-mediated acetylation of H4K16 (Figure 2B) [44]. Essentially, H4K16 acetylation disrupted a critical salt bridge formed between H4K16 and an acidic residue (E1551) of 53BP1 Tudor, and therefore destabilized 53BP1 binding to H4K20me2 [44]. As a result, 53BP1-mediated NHEJ is inhibited, and BRCA1-mediated HR takes over for repair (Figure 2B). Taken together, these studies demonstrate a direct role of 53BP1-mediated ‘read-out’ of H4K20me2 for promoting DNA repair, and the histone PTM contexts at sites of DSBs provide an elaborate regulatory mechanism for controlling 53BP1 association and dissociation, which fine-tunes the decision-making among different options available for repair.
SGF29 Tudor, a ‘reader’ of H3K4me3/2
SAGA (Spt–Ada–Gcn5 acetyltransferase) is an evolutionarily conserved multi-protein complex that facilitates gene transcription by mediating histone acetylation and deubiquitination [58]. Among SAGA subunits, SGF29 is the only one that contains a tandem-Tudor domain that is conserved across species from yeast to human [30]. The SGF29 tandem-Tudor domain (Table 1) has recently been identified as an H3K4me3/2-specific ‘reader’ by mass spectrometry-based screening [31]. H3K4me3 is its preferred ligand, with a Kd of about 1–4 μM [30, 31], which is consistent with ChIP-sequencing studies showing that SGF29 localizes to gene promoters and largely overlaps with H3K4me3 [31]. Structural analyses further demonstrate the SGF29 tandem-Tudor domains form independently but pack tightly against each other with interactions between the first two β-strands of each motif (Figure 1C) [30]. When bound by SGF29, H3K4me3 is anchored in a negatively charged ‘pocket’ that consists of three conserved aromatic residues, Y238, F264, and Y265, and an acid residue D266, at the surface of the second Tudor domain (Figure 1C). Multiple interactions including cation-π, van der Waals, and hydrophobic interactions, as well as a salt bridge between H3K4me3 and the aromatic ‘pocket’, establish the intermolecular binding [30]. Similar to H3K4me3/2-engaging PHD fingers [2, 8, 15], the SGF29 Tudor domain also interacts with the N-terminus of H3 including residues A1 and R2, which contributes to binding specificity towards H3K4me3 [30]. Knockdown of SGF29 or introduction of point mutations at its aromatic ‘pocket’ abolishes interaction between H3K4me3 and SAGA complexes, leading to loss of SAGA binding at target promoters and decreased acetylation of H3K9, H3K14 and H3K18 [30, 31]. Taken together, these observations demonstrate a critical role for the SGF29 tandem-Tudor domain in linking SAGA complexes to H3K4me3/2-marked promoters to mediate transcriptional regulation through subsequent chromatin modifications (Figure 2C).
Spindlin1 Tudor, a ‘reader’ of H3K4me3
The nucleolar protein Spindlin1 contains three Tudor-like domains in tandem as revealed by its crystal structure [59]. Initially, Spindlin1 was identified as an H3K4me3-interacting factor in a proteomics screen using protein affinity purification with pre-methylated nucleosomes [60]. A later measurement of Spindlin1 interaction with H3K4me3 indeed revealed a high affinity, with a Kd of ~0.3–0.8 μM (Table 1) [32, 33]. Structural analyses show the second Tudor domain is the sole contributor to H3K4me3 association, with a binding ‘cage’ consisting of four aromatic residues F141, W151, Y170, and Y177 that tap the H3K4-trimethylated side chain (Figure 1D) [32]. Other histone residues, including H3A1, H3R2, and H3R8, form hydrogen bonds with several negatively charged residues from the second Tudor domain [32]. These studies also show that all these interactions confer a tight binding of Spindlin1 to the H3K4me3-‘marked’ promoters among the ribosomal DNA gene repeats in the nucleolus, where Spindlin1 facilitates rRNA transcription (Figure 2D) [32, 33]. However, the mechanism by which Spindlin1 stimulates the rRNA gene expression is currently unknown.
UHRF1 Tudor, a ‘reader’ of H3K9me3
Ubiquitin-like with PHD and RING finger domains 1 (UHRF1) contributes to the maintenance of DNA methylation by recruiting DNA methyltransferase DNMT1 to replication forks [61, 62]. UHRF1 contains multiple conserved protein motifs, including an ubiquitin-like domain (UBL) at N-terminus, followed by a tandem-Tudor domain, a PHD finger, a SET- and RING-associated (SRA) domain, and a RING domain at C-terminus (Table 1). Previously, it was demonstrated that the UHRF1 SRA domain specifically recognizes replication-induced, hemi-methylated CpG dinucleotides (Figure 2E), providing a mechanism for targeting DNMT1 to the newly synthesized DNA fibers in order to restore the cellular level of DNA methylation [63–65].
Several recent studies further revealed an equally critical role of the UHRF1 tandem-Tudor domain for maintenance of DNA methylation [34, 36, 37, 66, 67]. These studies show the UHRF1 tandem-Tudor domain binds to H3K9me3 with high affinity (Table 1). Indeed, such binding is required for UHRF1-mediated recruitment of DNMT1 to heterochromatic regions to promote DNA methylation (Figure 2E) [35, 66, 67]. The crystal structure of UHRF1 tandem-Tudor domain plus PHD finger in association with H3K9me3-containing histone H3 peptides was solved [35]. Similar to SGF29, the two UHRF1 Tudor domains in tandem also packs tightly against each other using their first two β-strands, while the first Tudor accommodates the H3K9me3 side chain using an aromatic ‘cage’ formed by F152, Y188 and Y191 (Figure 1E) [35]. Genetic complementation assays performed among Uhrf1-null ES or UHRF1-knockdown cells demonstrate that the UHRF1 mutants, deficient in binding to either H3K9me3 or hemi-methylated CpG, only exhibit a partial or subtle defect in their association with heterochromatin and in their abilities to maintain DNA methylation, whereas those with deficiencies in both show a much more dramatic defect with a complete failure in rescuing loss of DNA methylation [34, 36]. Together, these studies demonstrate a multilayered, compensatory mechanism provided by various structural modules of UHRF1 in order to enforce an efficient chromatin targeting and to maintain the fidelity and level of DNA methylation (Figure 2E; also see section of ‘multivalent readout of histone PTMs’ below for further discussion of the linked Tudor and PHD modules of UHRF1).
Single Tudor domain as a histone PTM ‘reader’
PHF1 and PHF19 Tudor, ‘readers’ of H3K36me3/2
The polycomb-like (PCL) protein family acts as an accessory component of PRC2 (Polycomb Repressive Complex-2), the complex that catalyzes tri-methylation of histone H3 Lys27 (H3K27me3) to repress gene expression [68, 69]. Three mammalian PCL members, PHF1 (also known as PCL1), MTF2 (also known as PCL2), and PHF19 (also known as PCL3), all contain a single Tudor motif, two PHD fingers (Table 1), and a C-terminal chromo-like domain [42]. In vitro studies suggest that PCL proteins modulate PRC2 enzymatic activities and appear to help recruit PRC2 to a subset of target genes important for development and differentiation [70–76]. Recently, a flurry of reports further demonstrated that the Tudor domain of PHF1 and PHF19 specifically ‘reads’ H3K36me3/2, a histone PTM that marks the gene body of actively transcribed genes [38, 39, 41, 42]. Binding to H3K36me3 by the PHF1/PHF19 Tudor (Table 1)[38–40, 42] is much tighter than that by the previously reported H3K36me3 ‘readers’ such as the chromodomain of Eaf3 [77, 78] and PWWP domains[79, 80]. Structural analyses of the PHF1 and PHF19 Tudors reveal two highly similar β-barrel structures (Figure 1F–G) with each comprising five antiparallel β-strands [38, 39, 42]. The trimethylammonium side chain of H3K36me3 fits into an aromatic ‘cage’ at one end of the β-barrel (Figure 1F–G) [38, 39]; the histone H3 residue T32 to R40 make additional contacts to Tudor, which include a salt bridge formed between H3K37 and an acidic residue of Tudor (E66 of PHF1 or E75 of PHF19) [38, 39, 42]. Biochemically, extensive direct interactions between PHF1/PHF19 Tudors and the histone sequences surrounding H3K36me3 contribute to their binding specificity and affinity.
Using overexpression and knockdown of PHF1/PHF19, these recent studies collectively showed that ‘reading’ of H3K36me3/2 by PHF1/PHF19 Tudors mediates targeting and/or spreading of PRC2 complexes to a number of H3K36me3-containing loci among HeLa or pluripotent stem cells (Figure 2F) [38, 41, 42]. Mutations at the ‘cage’ residues of PHF1/PHF19 Tudors abolished the H3K36me3/2 binding and prevented PRC2-mediated repression of certain development genes such as Hox and Fgf [38, 41, 42]. Furthermore, PHF19 or the PRC2 complex was found to be associated with an H3K36me3 demethylase NO66 [41] or coexisting with an H3K36me2 demethylase KDM2B [42, 81] at a subset of PRC2 target genes, promoting a simultaneous H3K36 demethylation and H3K27 methylation in order to complete conversion from a state of active gene transcription to de novo silencing. Interestingly, a recent mass spectrometry-based study identified a form of asymmetrically methylated mono-nucleosomes that carry both H3K36me3/2 and H3K27me3/2 on two separate H3 tails in ES or HeLa cells [82], and such a bivalent mono-nucleosomes may represent the sites where PHF1/19-PRC2 complexes act [38]. However, the overall biological role of the PCL proteins is quite complex, as it has recently been shown that in vitro, binding of the PHF1 Tudor domain to H3K36me3-containing nucleosomes decreases the methyltransferase activity of PRC2 [39]; furthermore, Tudor-mediated binding to H3K36me3/2 was also found required for efficient recruitment of PHF1 to sites of DSBs during response to DNA damage [39], but the exact function of PHF1 in DNA repair remains to be studied. Taken together, these studies provide a novel mechanism for PRC2 complexes to gain access and target to the chromatin regions that harbor active genes, where PRC2 and associated factors establish de novo transcriptional silencing that is required for differentiation and development.
LBR Tudor, a ‘reader’ of heterochromatin and H4K20me2
The Lamin-B receptor (LBR) is an inner nuclear membrane protein that plays a crucial role in functional organization of nuclear architecture, particularly, in the formation and maintenance of nuclear peripheral heterochromatin [83]. In human, LBR mutations cause Pelger-Huët anomaly which is characterized by an aberrant neutrophil nuclear shape [84]; in murine models, deletions of LBR and Lamin-A/C lead to loss of peripheral heterochromatin, an inverted architecture with heterochromatin localizing to the nuclear interior, and perturbation in expression of genes associated with development [85]. The N-terminal part of LBR was known to be responsible for heterochromatin association [83], and contains a Tudor domain (Table 1). Deletion of Tudor renders LBR more mobile at nuclear envelope [84, 86]. A recent study reported the LBR Tudor domain binds H4K20me2, a heterochromatin-associated histone PTM [84], although another study indicates that the domain confers a ‘chaperone-like’ binding to histones [86]. These studies suggest a role of Tudor, possibly via interaction to heterochromatin PTMs, for LBR-mediated heterochromatin formation at the nuclear peripheral. Further examination of LBR Tudor and its binding partners needs to be performed.
Multivalent read-out of histone PTMs by Tudor and linked ‘reader’ modules
Tudor domains not only engage their preferred histone PTMs by a structurally defined ‘cage’ or ‘pocket’ (Figure 1), but also establish direct contacts to the surrounding histone sequences. The combination of these interactions contributes to the binding specificity, selectivity, and affinity of Tudor-domain proteins. In addition, Tudor domains often exist in proximity to other putative ‘reader’ domains (Table 1), indicating multivalent engagement of different histone PTMs by the linked ‘reader’ modules [3]. A prominent case of multivalency is the ‘PHD-linker-bromodomain’ cassette of BPTF, where the two separated ‘reader’ domains harbor capacities to bind to H3K4me3 and H4K16ac, respectively. The helical linker region in between dictates a precise relative orientation of two modules, ensuring a simultaneous, combinatorial read-out of the two PTMs located at separated histone tails within the same mono-nucleosome [3, 87].
Here, we discuss recent advances indicating a new mode of multivalent recognition utilized by the ‘tandem-Tudor-linker-PHD’ cassette of UHRF1 (Table 1). Initially, studies of individual domains within this cassette showed the UHRF1 tandem-Tudor and PHD finger motif engage H3K9me3 and the N-terminus of histone H3, respectively [66, 88]. However, examination of the whole domain cassette revealed that the inter-modular linker directly interacts with ‘reader’ modules and facilitates formation of a compact, ring-shaped architecture [35, 37, 89]. In this structure, the N-terminus of H3 (A1-R2-T3-K4) is engaged by the PHD finger, the residues 5–7 of H3 engaged by neither PHD nor Tudor, and only a rather short histone sequence (R8-K9me3) bound to Tudor (Figure 1E and 2E) [35, 37, 89]. As a result of such structural arrangement, binding to H3 primarily relies on the PHD finger, whereas Tudor appears only to confer additional selectivity for H3K9me3 [35, 37, 89]. Using a series of elegantly designed H3 peptides and UHRF1-mutants, a recent study provided supporting evidence for a mode of multivalent binding where the linked UHRF1 modules appear to scan from the extreme N-terminus of H3 towards PTMs located downstream [37], and this sequential ‘read-out’ of histone PTMs proposed for UHRF1 differs from a simultaneous, combinatorial mode of engagement [87].
Interestingly, the positioning of H3 in complex with the individual UHRF1 tandem-Tudor domain [66] is distinct from that observed in linked modules described above; in the former structure, residues 1–9 of H3 establish extensive contacts to a groove on the surface of Tudor[66], whereas this H3-binding groove is masked by the inter-modular linker and becomes non-accessible to H3 in the latter[35, 89]. These studies demonstrate an essential role of the linker in defining and reshaping the mode of binding to histones. In support, mutagenesis of two critical linker residues, R295 and R296, disconnect the coordinated action between ‘reader’ modules, leading to abrogation of combinatorial binding to H3/H3K9me3, reduction in chromatin localization of UHRF1 and loss of global DNA methylation [35]. Phosphorylation of S298, a conserved target site of PKA kinases within the linker, show a similar phenotype, indicating that modulation of the linker region might serve as a switching mechanism to regulate UHRF1 activities under a physiological condition [35]. Of note, unlike UHRF1, the PHD finger adjacent to PHF1/19 Tudors does not exhibit detectable histone-associating activities and does not alter binding to H3K36me3 by Tudor [40]. Taken together, these studies show adjacent ‘reader’ modules can evolve and form a high-order structure to establish various delicate mechanisms for multivalent ‘read-out’ of chromatin PTMs.
Targeting Tudor-histone interactions as potential therapeutic interventions
Many human diseases including cancer, possess mutations that deregulate chromatin PTM-specific ‘writers’, ‘erasers’ or ‘readers’[5, 8]. Pharmacological manipulation of these ‘writing’, ‘erasing’, and ‘reading’ processes has recently become an area of intense investigation [5, 90, 91]. Recently, small-molecule inhibitors for the BET bromodomain family of acetylation ‘readers’ have shown early promise in treatment of the genetically defined midline carcinoma [92] and hematopoietic malignancies [93–95]. Similar compounds could be developed to target other epigenetic ‘readers’ that are disease-associated [5, 6, 90, 91]. Notably, many of Tudor-containing ‘readers’ were found to be deregulated in cancer— all three Tudor-containing JMJD2 proteins are frequently overexpressed in various cancers [96], altered expression of UHRF1 is commonly found in cancer [97], and the recurrent chromosomal translocation of PHF1 and up-regulation of PHF19 were reported among endometrial sarcoma and solid tumors, respectively [98, 99]. Designing inhibitors that target the Tudor-histone binding interfaces may provide a unique tool not only for dissecting the role of these interactions in normal biological processes, but also for studying their relevance to pathogenesis. For instance, UHRF1 inhibitors could represent an alternative way to inhibit DNA methylation, in addition to the currently available DNA demethylating agents used for cancer therapies [100]. A recent study has developed the first-in-class inhibitor for histone methylation ‘readers’ [101]. Taken together, pioneering studies support druggability of histone methylation ‘readers’, and investigation is needed to develop potent, specific inhibitors that target Tudor-histone interactions.
Concluding remarks
Dissecting the fundamental mechanism by which chromatin modifications regulate various biological processes has become a major focus in chromatin biology. Studies aimed at understanding the interpretation of various chromatin modifications have focused on identifying novel epigenetic effectors. Mass spectrometry-based protein identification following pull-down with pre-modified histone peptides or nucleosomes [17, 31, 38, 60, 102, 103] has proven powerful in identifying novel, site-specific ‘readers’ for chromatin PTMs when combined with the high-throughput peptide or protein array technologies [38, 104, 105]. Subsequent structural and biological elucidation of these chromatin-‘reading’ modules allows a deeper understanding for the molecular bases that underlie processes regulated by histone-‘reader’ interactions. Recent identification of the Tudor family as versatile effectors of histone methylation re-enforces this theme and expands our current list of epigenetic ‘readers’. Future experiments should help characterize linked modules, and their role in combinatorial ‘read-out’ of multiple PTMs. For instance, a novel, dual histone-‘reading’ activity (binding of unmethylated H3K4 and H3K9 methylation) has recently been identified in a cryptic, tandem Tudor-like domain of SHH1 (SAWADEE homeodomain homolog-1) (Figure 1H) in Arabidopsis, and this activity is required for maintaining the level of siRNAs and RNA-directed DNA methylation in this organism [43]. In addition, evidence starts to emerge showing that Tudors also ‘read’ lysine methylation of non-histone partners, in addition to arginine methylation[21, 22]. For example, the 53BP1 tandem-Tudor domain binds to H4K20me3 and dimethylated p53 [50, 51], and that of PHF20 has been shown to bind to lysine dimethylation of histones [106] and p53 [107]; in the latter case, PHF20 binding to p53 stabilizes p53 and promotes its activation during DNA damage response [107]. Similarly, the Tudor domain of TDRD3 recognizes arginine dimethylation present in non-histone proteins such as the alternative splicing factor SmB [108] and in histones, such as asymmetric dimethylation of H4R3, H3R17 and H3R2 (Table 1) [105, 109], and studies have demonstrated a critical role of the TDRD3 Tudor domain in facilitating gene transcription[105]. It has become increasingly critical to dissect effects that are dependent on chromatin PTM and those that are directed through non-histone partners. Lastly, small-molecule inhibitors that specifically target epigenetic ‘readers’ hold promise for novel therapeutic means. Initial development of compounds could take advantage of the solved Tudor domain structures, while parallel efforts could be directed at examining the causality of Tudor-containing ‘readers’ in oncogenesis or other pathologies. These new pharmacological tools could allow a dynamic manipulation of histone-‘reader’ interactions and prove to be useful as therapeutic interventions.
Highlights.
Evidence reveals the versatility of Tudor in ‘reading’ various histone methylation.
Tudor domain uses an aromatic ‘cage’ to accommodate its histone methylation ligand.
Tudor and adjacent modules can combinatorially ‘read’ multiple histone modifications.
Tudor-histone interaction is critical for regulating various DNA-templated processes.
Acknowledgments
We graciously thank Jikui Song, David Allison, and Doan On for their help on structural illustration and critical reading. G.G.W. is a Martin D. Abeloff, M.D. V Scholar of the V Foundation for Cancer Research, and also supported by an NCI “Pathway to Independence” Award in Cancer Research (CA151683) and University Cancer Research Fund (UCRF) of the State of North Carolina. We apologize to researchers whose work could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Taverna SD, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruthenburg AJ, et al. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musselman CA, et al. Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chi P, et al. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang GG, et al. Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends in molecular medicine. 2007;13:363–372. doi: 10.1016/j.molmed.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Baker LA, et al. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008;647:3–12. doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwase S, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nature structural & molecular biology. 2011;18:769–776. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS letters. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 12.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pena PV, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruthenburg AJ, et al. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Molecular cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 16.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 18.Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 19.Callebaut I, Mornon JP. The human EBNA-2 coactivator p100: multidomain organization and relationship to the staphylococcal nuclease fold and to the tudor protein involved in Drosophila melanogaster development. The Biochemical journal. 1997;321 ( Pt 1):125–132. doi: 10.1042/bj3210125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ponting CP. Tudor domains in proteins that interact with RNA. Trends in biochemical sciences. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- 21.Chen C, et al. Deciphering arginine methylation: Tudor tells the tale. Nature reviews. Molecular cell biology. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 22.Pek JW, et al. Tudor domain proteins in development. Development. 2012;139:2255–2266. doi: 10.1242/dev.073304. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 24.Selenko P, et al. SMN tudor domain structure and its interaction with the Sm proteins. Nature structural biology. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 25.Friesen WJ, et al. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Molecular cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 26.Sprangers R, et al. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. Journal of molecular biology. 2003;327:507–520. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 27.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, et al. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science (New York, N Y. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 29.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bian C, et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. The EMBO journal. 2011;30:2829–2842. doi: 10.1038/emboj.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 32.Yang N, et al. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17954–17959. doi: 10.1073/pnas.1208517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, et al. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011;12:1160–1166. doi: 10.1038/embor.2011.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothbart SB, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nature structural & molecular biology. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arita K, et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, et al. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 37.Rothbart SB, et al. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes & development. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai L, et al. An H3K36 Methylation-Engaging Tudor Motif of Polycomb-like Proteins Mediates PRC2 Complex Targeting. Molecular cell. 2013;49:571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musselman CA, et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nature structural & molecular biology. 2012;19:1266–1272. doi: 10.1038/nsmb.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin S, et al. Tudor domains of the PRC2 components PHF1 and PHF19 selectively bind to histone H3K36me3. Biochem Biophys Res Commun. 2013;430:547–553. doi: 10.1016/j.bbrc.2012.11.116. [DOI] [PubMed] [Google Scholar]
- 41.Brien GL, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nature structural & molecular biology. 2012;19:1273–1281. doi: 10.1038/nsmb.2449. [DOI] [PubMed] [Google Scholar]
- 42.Ballare C, et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nature structural & molecular biology. 2012;19:1257–1265. doi: 10.1038/nsmb.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Law JA, et al. Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature. 2013 doi: 10.1038/nature12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang J, et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature structural & molecular biology. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallette FA, et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. The EMBO journal. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Molecular cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 48.Lee J, et al. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nature structural & molecular biology. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: insert bad pun here. DNA Repair (Amst) 2011;10:1071–1076. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Kachirskaia I, et al. Role for 53BP1 Tudor domain recognition of p53 dimethylated at lysine 382 in DNA damage signaling. The Journal of biological chemistry. 2008;283:34660–34666. doi: 10.1074/jbc.M806020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy S, et al. Structural insight into p53 recognition by the 53BP1 tandem Tudor domain. Journal of molecular biology. 2010;398:489–496. doi: 10.1016/j.jmb.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward IM, et al. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Molecular and cellular biology. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du LL, et al. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes & development. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aly A, Ganesan S. BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability. Journal of molecular cell biology. 2011;3:66–74. doi: 10.1093/jmcb/mjq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao L, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Molecular cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanders SL, et al. Requirement for the phospho-H2AX binding module of Crb2 in double-strand break targeting and checkpoint activation. Molecular and cellular biology. 2010;30:4722–4731. doi: 10.1128/MCB.00404-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samara NL, Wolberger C. A new chapter in the transcription SAGA. Current opinion in structural biology. 2011;21:767–774. doi: 10.1016/j.sbi.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Q, et al. Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation. The Journal of biological chemistry. 2007;282:647–656. doi: 10.1074/jbc.M604029200. [DOI] [PubMed] [Google Scholar]
- 60.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bostick M, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 62.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 63.Arita K, et al. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- 64.Avvakumov GV, et al. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 65.Hashimoto H, et al. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nady N, et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. The Journal of biological chemistry. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rottach A, et al. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic acids research. 2010;38:1796–1804. doi: 10.1093/nar/gkp1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 69.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao R, et al. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Molecular and cellular biology. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarma K, et al. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Molecular and cellular biology. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nekrasov M, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. The EMBO journal. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hunkapiller J, et al. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS genetics. 2012;8:e1002576. doi: 10.1371/journal.pgen.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Savla U, et al. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- 76.Casanova M, et al. Polycomblike 2 facilitates the recruitment of PRC2 Polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development. 2011;138:1471–1482. doi: 10.1242/dev.053652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun B, et al. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. The Journal of biological chemistry. 2008;283:36504–36512. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu C, et al. Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure. 2008;16:1740–1750. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu H, et al. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vezzoli A, et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nature structural & molecular biology. 2010;17:617–619. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- 81.Tzatsos A, et al. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. The Journal of clinical investigation. 2013;123:727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Voigt P, et al. Asymmetrically modified nucleosomes. Cell. 2012;151:181–193. doi: 10.1016/j.cell.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olins AL, et al. Lamin B receptor: multi-tasking at the nuclear envelope. Nucleus. 2010;1:53–70. doi: 10.4161/nucl.1.1.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hirano Y, et al. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. The Journal of biological chemistry. 2012;287:42654–42663. doi: 10.1074/jbc.M112.397950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 86.Liokatis S, et al. Solution structure and molecular interactions of lamin B receptor Tudor domain. The Journal of biological chemistry. 2012;287:1032–1042. doi: 10.1074/jbc.M111.281303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruthenburg AJ, et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rajakumara E, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Molecular cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng J, et al. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. The Journal of biological chemistry. 2013;288:1329–1339. doi: 10.1074/jbc.M112.415398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 91.Arrowsmith CH, et al. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 92.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berry WL, Janknecht R. KDM4/JMJD2 Histone Demethylases: Epigenetic Regulators in Cancer Cells. Cancer research. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bronner C, et al. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacology & therapeutics. 2007;115:419–434. doi: 10.1016/j.pharmthera.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 98.Wang S, et al. A novel human homologue of Drosophila polycomblike gene is up-regulated in multiple cancers. Gene. 2004;343:69–78. doi: 10.1016/j.gene.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 99.Panagopoulos I, et al. Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS ONE. 2012;7:e39354. doi: 10.1371/journal.pone.0039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Baylin SB. DNA methylation and gene silencing in cancer. Nature clinical practice. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 101.James LI, et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nat Chem Biol. 2013;9:184–191. doi: 10.1038/nchembio.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eberl HC, et al. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Molecular cell. 2013;49:368–378. doi: 10.1016/j.molcel.2012.10.026. [DOI] [PubMed] [Google Scholar]
- 103.Spruijt CG, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 104.Rothbart SB, et al. Peptide microarrays to interrogate the “histone code”. Methods Enzymol. 2012;512:107–135. doi: 10.1016/B978-0-12-391940-3.00006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang Y, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Molecular cell. 2010;40:1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Adams-Cioaba MA, et al. Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins. FEBS letters. 2012;586:859–865. doi: 10.1016/j.febslet.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 107.Cui G, et al. PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53. Nature structural & molecular biology. 2012;19:916–924. doi: 10.1038/nsmb.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. The Journal of biological chemistry. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 109.Liu K, et al. Crystal structure of TDRD3 and methyl-arginine binding characterization of TDRD3, SMN and SPF30. PLoS ONE. 2012;7:e30375. doi: 10.1371/journal.pone.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]