Abstract
Seven mammalian sirtuins are nicotinamide adenine dinucleotide (NAD)+-dependent deacetylases and are important modulators of energy metabolism and stress resistance. Two new studies by Du et al. (2011) and Peng et al. (2011) identify a new enzymatic activity for SIRT5, expanding the cellular repertoire of posttranslational modifications targeted by the sirtuins.
The sirtuins are a family of NAD+-dependent protein deacetylases named after the yeast silent information regulator 2 (Sir2) that regulate important biological pathways in eubacteria, archaea, and eukaryotes (see Imai and Guarente, 2010 for review). Bacteria and archaea express one or two sirtuins, but mice and humans have seven, named SIRT1–7. The seven mammalian sirtuins occupy different subcellular compartments, including the cytoplasm (SIRT1 and 2), nucleus (SIRT1, 2, 3, 6, and 7), and mitochondria (SIRT3, 4, and 5). The sirtuins catalyzea deacetylation reaction that uses NAD+ as a cofactor, yielding O-acetyl-ADP-ribose, the deacetylated substrate, and nicotinamide. Given the large number of members of the sirtuin family and their varied cellular distribution, it has long been suspected that some family members may have primary activities other than deacetylation. Now two reports describe novel enzymatic activities for SIRT5 (Du et al., 2011; Peng et al., 2011)—desuccinylase and demalonylase activity—which seem to predominate its previously reported weak deacetylase activity.
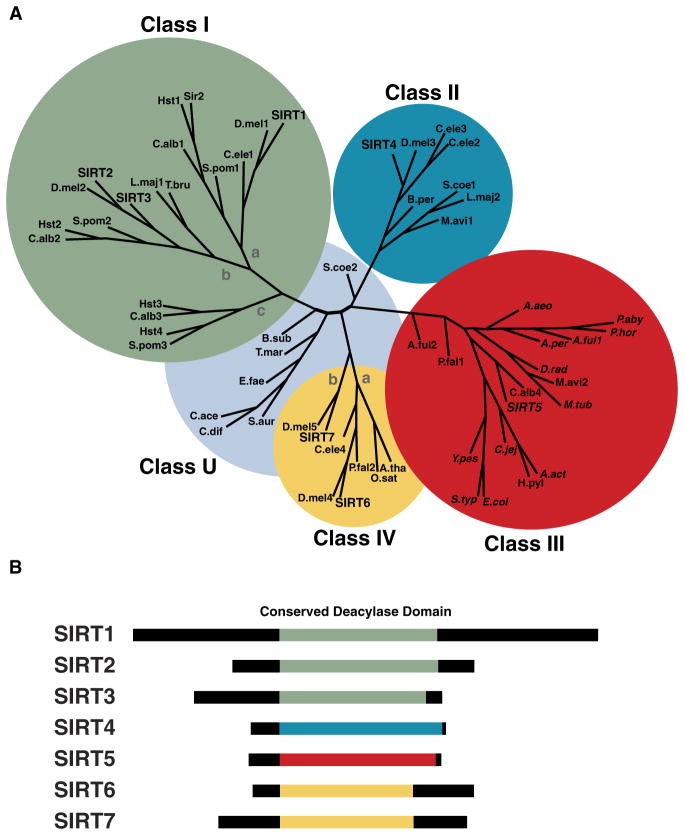
The sirtuins are assigned to five classes (I–IV and U; Figure 1A) based on the conservation of an ~250 amino acid core domain (Frye, 2000). Among mammalian sirtuins, SIRT1, 2, and 3 are class I sirtuins, have high homology to the yeast sirtuins Sir2, Hst1, and Hst2, and exhibit robust deacetylase activity (North et al., 2003). Class II sirtuins, including mammalian SIRT4, have no detectable deacetylase activity and instead show weak ADP-ribosyltransferase activity (Haigis et al., 2006). Class III sirtuins, including mammalian SIRT5, were heretofore described as having weak deacetylase activity. Class IV sirtuins have ADP ribosyltransferase and deacetylase activity (SIRT6) or unknown activity (SIRT7). Finally, class U sirtuins are intermediate between class I and IV, and have thus far only been observed in bacteria. Collectively, the sirtuins play important roles in regulating genomic stability, energy metabolism, and stress resistance in a variety of organisms (reviewed in Finkel et al., 2009).
Figure 1. Molecular Phylogeny of the Sirtuins.
(A) An unrooted tree diagram of a phylogenetic analysis of the conserved sirtuin core deacylase domain sequences, divided into class I, II, III, IV, and U groups; classes I and IV are further divided into subclasses indicated by lowercase letters. Adapted from Frye (2000).
(B) Schematic of the conserved sirtuin core deacylase domains in human SIRT1–7, color coded to match the phylogenetic class from (A): green, class I; blue, class II; red, class III; yellow, class IV. Italic print indicates conservation of tyrosine (Y102) and arginine (R105) residues (numbering based on hSIRT5) required for the demalonylase and desuccinylase activities of hSIRT5.
SIRT5 interacts with carbamoyl phosphate synthetase 1 (CPS1), which catalyzes the first step of ammonia detoxification and disposal via the urea cycle (Nakagawa et al., 2009). Incubation of CPS1 with SIRT5 was shown to reduce the amount of acetyllysine detectable with an antibody specific for this posttranslational modification, and to increase CPS1 activity. Mice lacking SIRT5 fail to upregulate CPS1 activity and show elevated blood ammonia during fasting, long-term calorie restriction, and high-protein diet feeding. However, Du et al. (2011) show that while CPS1 is both acetylated and succinylated at three lysine residues (K44, K287, and K1291), in the absence of SIRT5 succinylation of K1291 increases, whereas succinylation of K44 and K287 or acetylation of K44, K287, and K1291 remain unchanged. These new succinylation data contradict the earlier acetylation data and suggests that SIRT5 regulates the enzymatic activity of CPS1 by desuccinylating K1291. Because acetylation and succinylation can occur on the same CPS1 lysine residues, further studies are needed to resolve this discrepancy.
Posttranslational protein succinylation was first observed in rat liver mitochondrial extracts and hypothesized to play a role in regulating enzymatic activity of 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HGMCS2) (Quant et al., 1989). Recently, a proteomic survey verified the presence of succinylated lysines in proteins from E. coli extracts, further characterized this modification, and reported several new succinylated proteins (Zhang et al., 2011). However, the existence or identity of an enzyme to remove this modification remained unknown. Together, the reports by Du et al. (2011) and Peng et al. (2011) conclusively show that lysine succinylation is a new posttranslational modification and demonstrated that SIRT5 serves as a key enzyme for removing succinyl groups from lysine residues of multiple cellular proteins.
Furthermore, lysine malonylation, which is structurally homologous to succinylation, is a posttranslational modification also targeted by SIRT5. Using a combination of chemical and biological approaches, Peng et al. (2011) show that several proteins are malonylated on lysine residues in the cytoplasm and in the mitochondria, and that loss of SIRT5 increases the malonylation signal (Peng et al., 2011). The biological significance of protein malonylation remains to be determined, but the clear demonstration of malonylation and succinylation as new posttranslational modifications regulated by SIRT5, and at least one example of effects of succinylation to modify a key metabolic enzyme activity (CPS1), implies that a potentially broader impact of SIRT5 on other key enzymes will be uncovered as this research field moves forward.
These findings also imply that novel enzymatic activities could exist for the other sirtuins. Of the seven mammalian sirtuins, SIRT1–3 have strong deacetylase activity, whereas SIRT4–7 have weak or non-detectable deacetylase activity (North et al., 2003). Because SIRT5 demonstrated weak deacetylase activity and seems now to be primarily a demalonylase and desuccinylase, other sirtuins with weak deacetylase activities (SIRT4, SIRT6, SIRT7) could also have unique roles in targeting posttranslational modifications that remain to be discovered.
The deacetylating sirtuins SIRT1–3 are all class I sirtuins (Figure 1A). While the phylogenetic organization of the sirtuin family has been known for over ten years (Frye, 2000), the new enzymatic activities of SIRT5 discovered by Du et al. (2011) and Peng et al. (2011) suggest that the phylogenetic classification of the sirtuins could be key to their enzymatic activity. With the conserved sirtuin core domain aligned and coded for phylogenetic classification (Figure 1B), a pattern of enzymatic activity emerges. Class I sirtuins (SIRT1, 2, and 3) are strong deacetylases and weak demalonylases and desuccinylases. Class III sirtuins (SIRT5) are strong demalonylases and desuccinylases and weak deacetylases. Furthermore, Du et al. (2011) showed the critical tyrosine (Y102) and arginine (R105) residues required for the demalonylase and desuccinylase activities of SIRT5 are conserved across 14 species of class III sirtuins (Figure 1A; italics), suggesting these class III sirtuins from other organisms might also have demalonylase and desuccinylase activities. Future work on class II, IV, and U sirtuins could uncover novel enzymatic activities.
Together, the reports by Du et al. (2011) and Peng et al. (2011) broaden the cellular repertoire of posttranslational modifications that are targeted by the sirtuins, which now extend beyond acetylation and encompass other acyl-modifications, including succinylation and malonylation. These findings suggest that the sirtuins are not simply NAD+-dependent deacetylases but are more accurately described as NAD+-dependent deacylases. Further studies in this rapidly expanding field will shed light on the physiological significance of the succinyl- and malonyl-lysine modifications, and perhaps identify new acyl modifications targeted by the sirtuin family.
References
- Du J, Zhou Y, Su X, Yu J, Kahn S, Jiang H, Kim J, Woo J, Kim J, Choi B, et al. Science. 2011 in press. [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, et al. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Imai S-i, Guarente L. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, et al. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.012658. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quant PA, Tubbs PK, Brand MD. Biochem J. 1989;262:159–164. doi: 10.1042/bj2620159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]