Abstract
Purpose
MicroRNAs are small noncoding RNAs and microRNA-214 (miR-214) has been associated with the inhibition of cancer cell growth, migration, and invasion. The aim of this study was to investigate whether cell-free miR-214 isolated from urine could be used as a biomarker for non-muscle-invasive bladder cancer (NMIBC).
Materials and Methods
A total of 138 patients with primary NMIBC and 144 healthy normal controls were enrolled in this study. By use of quantitative polymerase chain reaction (PCR), the urinary levels of cell-free miR-214 were measured and the clinicopathological parameters of patients with NMIBC were compared with those of the controls.
Results
The urinary levels of cell-free miR-214 were significantly higher in the NMIBC patients than in the controls (20.08±3.21 vs. 18.96±2.68, p=0.002). However, the urinary levels of cell-free miR-214 were neither graded nor staged for the NMIBC patients (p>0.05, each). When we compared the urinary levels of cell-free miR-214 according to clinical outcomes, patients with recurrence had lower levels of miR-214 than did those with no recurrence (19.24±2.67 vs. 20.41±3.41, p=0.023). By contrast, there were no significant differences in the urinary level of cell-free miR-214 between the NMIBC patients showing progression and those showing no progression (p=0.919). Multivariate Cox regression analysis revealed that urinary levels of cell-free miR-214 were an independent predictor of NMIBC recurrence (hazard ratio, 2.011; 95% confidence interval, 1.027 to 3.937; p=0.041).
Conclusions
Urinary levels of cell-free miR-214 could be an independent prognostic parameter for NMIBC recurrence. Thus, urinary cell-free microRNA-214 might be a useful prognostic marker for NMI BC.
Keywords: MicroRNAs, Neoplasms, Recurrence, Urinary bladder
INTRODUCTION
Bladder cancer is the second most common urological malignancy in humans, and it was estimated in 2012 in the United States that 73,510 new cases of cancer of the urinary bladder were diagnosed along with 14,680 deaths [1]. In Korea also, bladder cancer is the second most common genitourinary tumor and is about five times as common in men as in women [2,3]. Bladder cancer is classified into two large groups of non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC) according to pathology and clinical features [4]. Although only 20% of bladder cancer is confirmed as MIBC at first diagnosis, MIBC accounts for the majority of cancer-specific deaths [5,6]. More than three-quarters of all bladder cancer cases are NMIBC that can be treated by transurethral resection (TUR). Unfortunately, about 70% of patients with NMIBC who have undergone TUR experience a tumor recurrence within 2 years. Furthermore, 20% to 30% of patients who have had a complete TUR and intravesical therapy performed with bacillus Calmette-Guérin (BCG) have experienced progression to MIBC [7]. Thus, many diagnostic tools and biomarkers have been proposed and developed to predict the recurrence and progression of NMIBC [8-10], but most of these have proven to be inadequate in terms of efficacy and accuracy because of the heterogeneous behavior of bladder cancer.
MicroRNAs (miRNAs) are small, noncoding RNAs that are 18 to 22 nucleotides in length in their mature form [11,12]. MicroRNAs act as posttranscriptional gene modulators and play an important role in cell proliferation, differentiation, survival, programmed cell death, and oncogenesis of cells and organisms, usually by inhibiting translation [13]. MicroRNAs are aberrantly expressed in human cancer and may function as a novel class of oncogenes or tumor suppressor genes. Actually, numerous studies have detected miRNA deregulation in human malignancies, including chronic lymphocytic leukemia, breast cancer, primary glioblastoma, lung cancer, thyroid cancer, colon cancer, and pancreatic cancer [14]. Also, when histologically matched with normal urothelium, various types of mi-RNAs are aberrantly expressed in bladder cancer. Accordingly, it has been suggested that miRNAs play roles as oncogenes or tumor suppressors in the tumorigenesis and progression of bladder cancer [15]. However, although many miRNAs, their targets, and their mechanism of action as well as their clinical value have recently been described, few studies have focused on cell-free miRNAs isolated from urine, particularly in bladder cancer.
In the present study, we measured the expression levels of urinary cell-free miR-214 in NMIMC patients and then investigated whether urinary cell-free miR-214 could be a prognostic biomarker for NMIBC.
MATERIALS AND METHODS
1. Study population and samples
A total of 138 patients with primary urothelial carcinoma of the urinary bladder and 144 healthy controls were enrolled in the study. Controls were selected to match the age and gender proportions of the bladder cancer patients, and subjects were screened to ensure that they were within the normal range of all laboratory results with no history of malignant tumors. Urine samples were collected in the morning and stored at 4℃, then centrifuged at 25,000 rpm for 15 minutes. Each supernatant and sediment was portioned into Eppendorf tubes and stored at -20℃ until use. All primary tumor samples were obtained from patients who underwent TUR and were histologically verified to have urothelial carcinoma. To reduce confounding factors affecting the analyses, and to delineate a more homogeneous study population, any patients diagnosed with a concomitant carcinoma in situ or for whom data collection was incomplete were excluded. The collection and analysis of all samples was approved by the Institutional Review Board of our institute, and written informed consent was obtained from each subject (IRB approval number 2006-01-001).
Patients who had a T1 tumor, multiple tumors, large tumors (>3 cm in diameter), or high-grade Ta NMIBC received one cycle of intravesical treatment (BCG or mitomycin-C). Response to treatment was assessed by cystoscopy and urinary cytology. Patients who were free of disease within 3 months after treatment were assessed every 3 months for the first 2 years, and then every 6 months thereafter. Patients who refused or did not complete an imaging work-up such as a computed tomographic scan or an magnetic resonance imaging at least once every 3 months to evaluate their response were excluded from the study. Tumors were staged and graded according to the 2002 TNM classification and the 1973 World Health Organization grading system, respectively. Recurrence was defined as a recurrence of primary NMIBC at either a lower or the same pathological stage, and progression was defined as disease with a higher TNM stage upon relapse of NMIBC.
2. Purification of miRNA
A Genolution urine miRNA purification kit (Genolution Pharmaceuticals Inc., Seoul, Korea) was used to purify the urine samples. Urine sample supernatant (500 µL) was added to each tube containing Genolution proprietary miRNA separation solution, which was then vortexed for 20 seconds. Next, 200 µL of chloroform was added and the samples were vortexed for 10 seconds, after which they were centrifuged at 13,000 rpm for 10 minutes at 4℃. A 600-µL fraction from the top aqueous phase was taken and transferred into a new 1.5 mL tube, and 0.8 mL of isopropanol was added, followed by centrifugation for 5 minutes at 13,000 rpm and 4℃. After removing the aqueous solution, 1 mL of 70% ethanol was added and the sample was again centrifuged for 2 minutes at 13,000 rpm and 4℃. After removing the ethanol, the pellet was dissolved in 30 µL RNase-free water and stored at -80℃ until use. A fixed concentration of microRNA (5 ng/µL) from a given volume of starting urine was used as the input into the reverse transcription reaction.
3. Reverse transcription of miRNA
The miScript reverse transcription kit (Qiagen Korea, Seoul, South Korea) was used for reverse transcription of the miRNAs. After mixing with template RNA, 5X miScript buffer, miScript reverse transcriptase mix, and RNase-free water in a final volume of 20 µL, the mixture was centrifuged briefly and incubated for 60 minutes at 37℃. To inactivate the miScript reverse transcriptase mix, the samples were incubated for 5 minutes at 95℃ and placed on ice.
4. Real-time PCR detection of miRNA
For quantification of the miRNA expression, real-time amplification was performed with a Rotor-Gene 6000. Real-time PCR assays were performed in microreaction tubes (Corbett Research, Mortlake, Australia) using the miScript PCR starter kit (Qiagen Korea, Seoul, Korea). To amplify target miRNAs, forward primers were designed for the miR-214 (5'-ACA GCA GGC ACA GAC AGG CA GT-3'). After thawing the reagents and template cDNA, the PCR reaction was carried out in a final volume of 20 µL containing 10 µL 2X QuantiTect SYBR Green PCR master mix, 2 µL 10X miScript universal primer, 2 µL 10 pmol forward primer, 2 µL template cDNA, and RNase-free water. Real-time PCR conditions were as follows: one cycle of initial activation for 15 minutes at 95℃, followed by 50 cycles of 15 seconds at 94℃ for denaturation, annealing for 30 seconds at 55℃, and extension for 30 seconds at 70℃. The melting program was performed at 70℃ to 99℃ at a heating rate of 1℃ per 5 seconds. Spectral data were captured and analyzed by using Rotor-Gene real-time analysis software 6.0, Build 14. All samples were run in triplicate. For accurate and reproducible results by real-time PCR, it was necessary to normalize the amount of target miRNA by using a suitable endogenous reference RNA. This approach is known as relative quantification. In the present experiment, U6 was used as a control for the normalization of real-time PCR results in miRNA quantification studies using the miScript PCR system. U6 was analyzed in parallel as an endogenous RNA reference gene and all data were normalized to U6 expression.
5. Statistical analysis
A Student t-test was applied to assess the association of the miRNA expression level with the clinical variables. To compare the levels of miRNA-214 in enrolled patients and controls, an analysis of variance was applied. The prognostic value of the levels of miR-214 in NMIBC was studied by using multivariate Cox proportional hazards regression models. Statistical analysis was performed by using IBM SPSS ver. 19.0 (IBM Co., Armonk, NY, USA), and a p-value of <0.05 was considered statistically significant.
RESULTS
1. Baseline characteristics
The baseline characteristics of the enrolled patients and controls are listed in Table 1. The mean age of the patients was 62.08±13.38 years for 110 males and 28 females; the mean age of the controls was 63.79±12.65 years for 114 males and 30 females. The number of patients in grades G1, G2, and G3 was 53, 64, and 21, respectively. The number of patients in stages Ta and T1 was 64 and 74, respectively. The median follow-up period was 49.22±33.04 months. Other clinical and pathological features of the enrolled patients are presented in Table 1.
TABLE 1.
Baseline clinical and pathological features of patients with bladder cancer
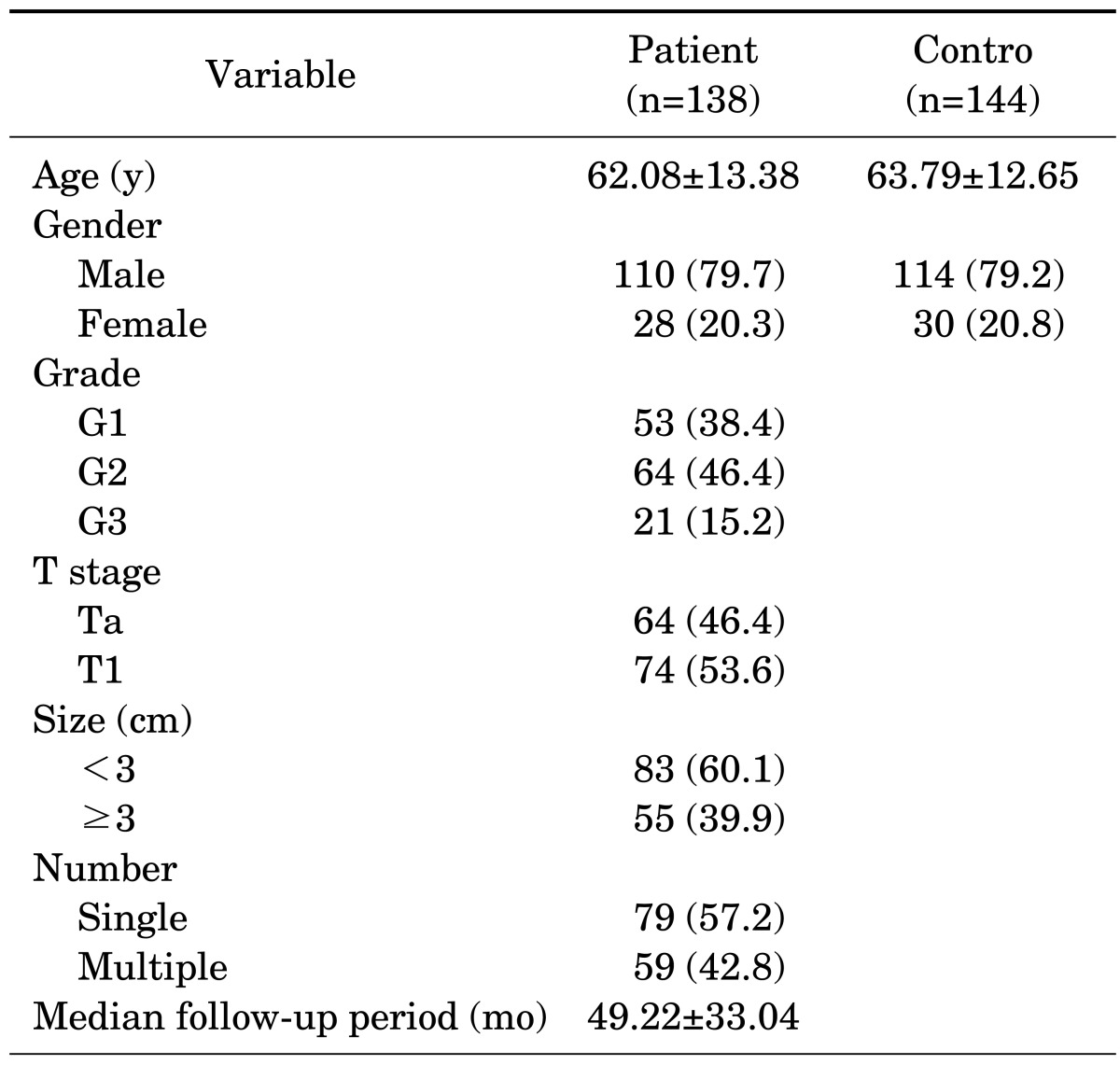
Values are presented as mean±standard deviation or number (%).
2. Comparison of miR-214 in controls and patients
Table 2 summarizes the association between urinary levels of miRNA-214 and the clinicopathological parameters of bladder cancer patients. The levels of miR-214 were significantly higher in the NMIBC patients than in the controls (20.08±3.21 vs. 18.96±2.68, p=0.002). However, the urinary levels of miR-214 were not significantly associated with either the grade or the stage of bladder cancer (p>0.05, each).
TABLE 2.
Urinary levels of microRNA-214 according to the clinicopathologic features of bladder cancer
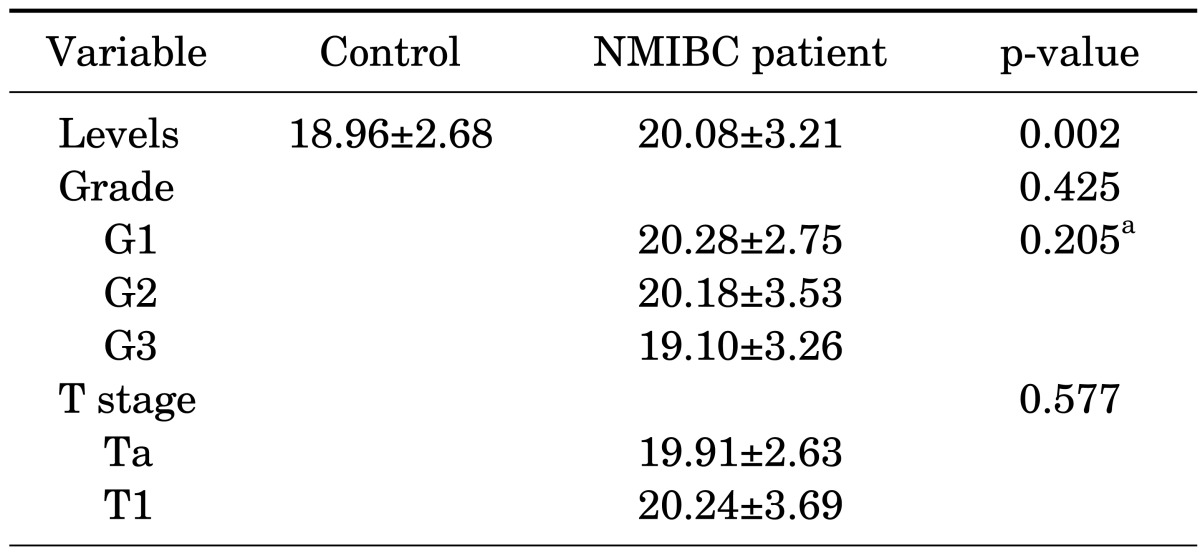
Values are presented as mean±standard deviation.
NMIBC, non-muscle-invasive bladder cancer.
a:p for trend was analyzed by analysis of variance.
3. Comparing the levels of miR-214 in predicting the clinical outcomes of bladder cancer
When we compared the urinary levels of miR-214 according to clinical outcomes, we found that the urinary levels of miR-214 were significantly different between the recurrence and the nonrecurrence groups (Table 3). Patients with recurrence of NMIBC had lower levels of miR-214 than did those who had no recurrence (19.24±2.67 vs. 20.41±3.41, p=0.023). There was no significant difference in the levels of miR-214 between the progression and the nonprogression groups (p=0.919).
TABLE 3.
Comparison of microRNA-214 in predicting recurrence and progression
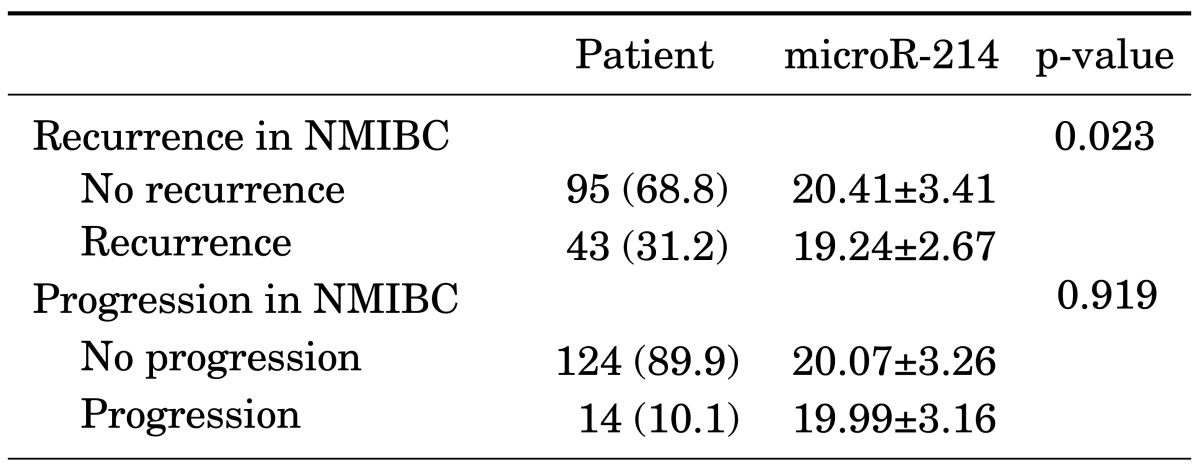
Values are presented as number (%) or mean±standard deviation.
NMIBC, non-muscle-invasive bladder cancer.
4. Prediction of recurrence in patients with NMIBC
In the univariate Cox regression analysis, the number of tumors (especially ≥8) and the levels of miR-214 were influential factors in the recurrence of NMIBC (hazard ratio [HR], 2.636; 95% confidence interval [CI], 1.220 to 5.699; p=0.014; HR, 2.231; 95% CI, 1.175 to 4.238; p=0.014, respectively). Multivariate Cox regression analysis revealed that a low level of urinary miR-214 was the only independent predictor of NMIBC recurrence (HR, 2.011; 95% CI, 1.027 to 3.937; p=0.041) (Table 4). The patients with low levels of miR-214 had a significantly longer recurrence-free survival time than did the patients with high level of miR-214 (p=0.012) (Fig. 1).
TABLE 4.
Univariate and multivariate cox regression analysis for the prediction of recurrence in non-muscle-invasive bladder cancer according to microRNA-214 Level
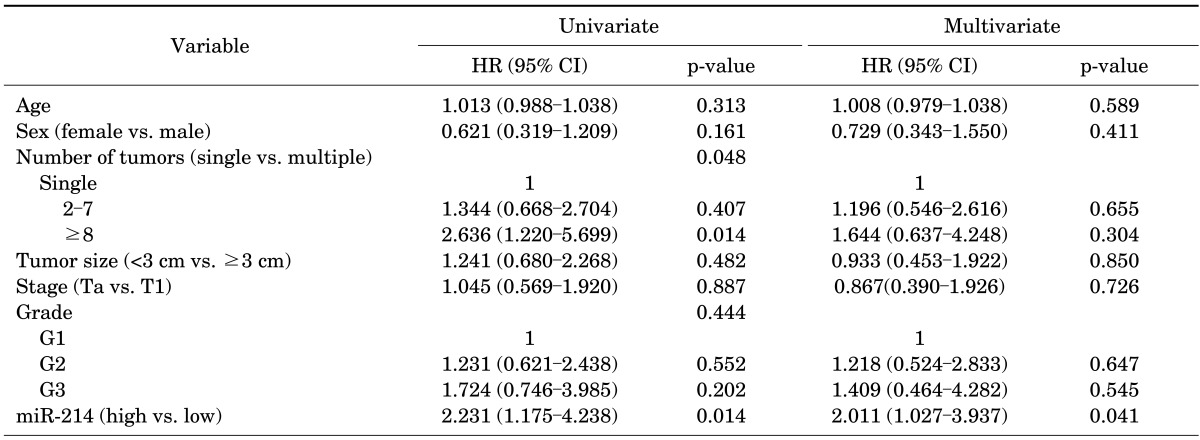
HR, hazard ratio; CI, confidence interval.
FIG. 1.
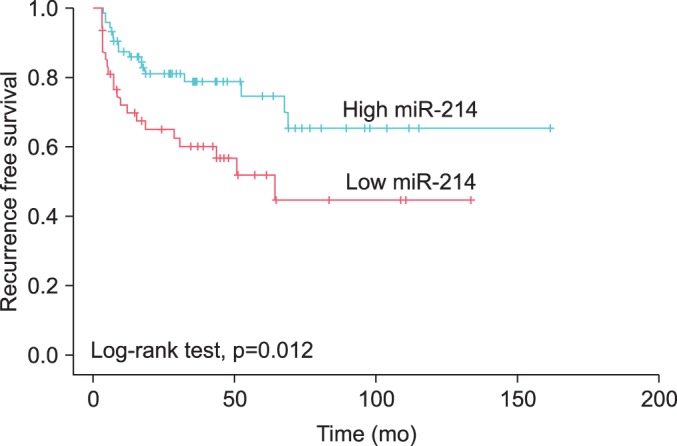
Recurrence-free survival in patients with non-muscle-invasive bladder cancer according to urinary cell-free microRNA-214 (miR-214).
DISCUSSION
The current study showed that urinary cell-free miRNAs can serve as noninvasive prognostic markers for the recurrence of NMIBC. MicroRNAs have been identified in all types of bodily fluids, including cerebrospinal fluid, pleural fluid, peritoneal fluid, and even tears, saliva, and urine [16]. Numerous studies have recently shown that miRNAs are emerging as a new class of cancer biomarkers [17]. In the case of urothelial carcinoma, urine is a particularly desirable source of such biomarkers. An ideal biomarker must be accessible by noninvasive protocols, be inexpensive to quantify, be specific to the disease of interest, and be a reliable indicator of disease before the appearance of clinical symptoms [16]. Urine is more convenient and less invasive to collect than blood; in addition, miRNAs exist in a stable form in urine even after seven cycles of freezing and thawing or 72 hours at room temperature [18]. Also, miRNA levels in the urine rarely change. Cell-free miRNAs in urine could be direct indicators of urological conditions including injury, malignancy, and so on. The exact mechanisms of release and the roles of cell free circulating miRNAs are unclear. Recent studies have suggested, however, that the interaction between cells via mRNA and miRNA is accomplished by microvesicle transfer [19]. Circulating miRNAs might be released from cancer cells and communicate with recipient cells in the surrounding microenvironment by microvesicles [20]. For this reason, it is certain that urine would have the highest exposure to microvesicles from urothelial cancer tissue.
A number of studies have established either the over-expression or under-expression of miRNAs in different types of human malignancy [21-27]. Accumulated evidence has shown that miRNA expression signatures correlate well with the specific characteristics of each malignancy and can be used to classify normal and malignant tissues as well as the subtype of malignancy [25,26]. These observations suggest that miRNAs can function either as tumor suppressors or oncogenes, regulating different cellular processes by targeting hundreds of genes and conferring a predictive diagnostic value to miRNA expression [27].
In solid tumors, such as stomach, pancreatic, and prostate cancer, alteration of the levels of a small number of miRNAs including miR-214 has been identified as a signature. In breast cancer, miR-214 expression is reduced [28,29], and in pancreatic and ovarian cancers, miR-214 has a relationship with chemoresistance [21,30]. These studies have proposed that miR-214 negatively modulates angiogenic activity and inhibits cell proliferation. However, studies that have evaluated the relationship between the level of miR-214 and bladder cancer are rare, which, of course, includes urinary miR-214.
In the present study, we confirmed that urinary miR-214 can serve as a noninvasive prognostic biomarker of bladder cancer. The levels of miR-214 capably distinguished NMIBC patients from the noncancerous controls, but could show no correlation with either the grade or the stage of NMIBC. We suggest that the NMIBC patients had relatively high levels of miR-214 compared with the control patients owing to the tumor-suppressive effects of miR-214.
In NMIBC patients, low levels of urinary miR-214 could be associated with a recurrence of NMIBC owing to relatively low tumor-suppressive effects. On the basis of these results, we inferred that miR-214 could be related to the inhibition of angiogenesis, to cell proliferation, and to tumor recurrence.
As far as we could ascertain, only one study has evaluated urinary cell-free miRNAs as diagnostic and prognostic biomarkers of bladder cancer. The current study is the first to link urinary miRNA-214 with bladder cancer. The results of this study indicated that miR-214 in urine might serve as a noninvasive biomarker for predicting the prognosis of bladder cancer.
CONCLUSIONS
Low levels of urinary miR-214 can be used as an independent prognostic parameter for the recurrence of NMIBC. Thus, cell-free urinary microRNA-214 might be a useful noninvasive biomarker for the recurrence of NMIBC.
ACKNOWLEDGEMENT
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012R1A1A4A01008753).
Footnotes
The authors have nothing to disclose.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Jung KW, Park S, Kong HJ, Won YJ, Lee JY, Seo HG, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2009. Cancer Res Treat. 2012;44:11–24. doi: 10.4143/crt.2012.44.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha YS, Kim MJ, Yoon HY, Kang HW, Kim YJ, Yun SJ, et al. mRNA expression of S100A8 as a prognostic marker for progression of non-muscle-invasive bladder cancer. Korean J Urol. 2010;51:15–20. doi: 10.4111/kju.2010.51.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Bohle A, Palou-Redorta J, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008;54:303–314. doi: 10.1016/j.eururo.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 5.Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. 2009;374:239–249. doi: 10.1016/S0140-6736(09)60491-8. [DOI] [PubMed] [Google Scholar]
- 6.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 7.Messing EM. Urothelial tumors of the bladder. In: Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA, editors. Campbell-Walsh urology. 9th ed. Philadelphia: Saunders; 2007. pp. 2426–2427. [Google Scholar]
- 8.Wu TT, Chen JH, Lee YH, Huang JK. The role of bcl-2, p53, and ki-67 index in predicting tumor recurrence for low grade superficial transitional cell bladder carcinoma. J Urol. 2000;163:758–760. doi: 10.1016/s0022-5347(05)67798-1. [DOI] [PubMed] [Google Scholar]
- 9.Quan C, Park MS, Jo SW, Lee SC, Kim WJ. Effects of transforming growth factor-beta1 and its receptor on the development, recurrence and progression of human bladder cancer. Korean J Urol. 2006;47:426–435. [Google Scholar]
- 10.Ha YS, Yun SJ, Kim YJ, Lee SC, Kim WJ. Utility of Smo as a prognostic marker for human bladder tumors. Korean J Urol. 2007;48:997–1003. [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Chen S, Luan X, Li Y, Liu M, Li X, et al. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life. 2009;61:1075–1082. doi: 10.1002/iub.252. [DOI] [PubMed] [Google Scholar]
- 15.Han Y, Chen J, Zhao X, Liang C, Wang Y, Sun L, et al. MicroRNA expression signatures of bladder cancer revealed by deep sequencing. PLoS One. 2011;6:e18286. doi: 10.1371/journal.pone.0018286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffrey SS. Cancer biomarker profiling with microRNAs. Nat Biotechnol. 2008;26:400–401. doi: 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- 18.Yun SJ, Jeong P, Kim WT, Kim TH, Lee YS, Song PH, et al. Cell-free microRNAs in urine as diagnostic and prognostic biomarkers of bladder cancer. Int J Oncol. 2012;41:1871–1878. doi: 10.3892/ijo.2012.1622. [DOI] [PubMed] [Google Scholar]
- 19.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 20.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 22.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 24.Yamada Y, Enokida H, Kojima S, Kawakami K, Chiyomaru T, Tatarano S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 25.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 26.Tricoli JV, Jacobson JW. MicroRNA: potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 27.Hagan JP, Croce CM. MicroRNAs in carcinogenesis. Cytogenet Genome Res. 2007;118:252–259. doi: 10.1159/000108308. [DOI] [PubMed] [Google Scholar]
- 28.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang XJ, Ye H, Zeng CW, He B, Zhang H, Chen YQ. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J Hematol Oncol. 2010;3:46. doi: 10.1186/1756-8722-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]


