Abstract
The bacterial community of maple sap was characterized by analysis of samples obtained at the taphole of maple trees for the 2001 and 2002 seasons. Among the 190 bacterial isolates, 32 groups were formed according to the similarity of the banding patterns obtained by amplified ribosomal DNA restriction analysis (ARDRA). A subset of representative isolates for each ARDRA group was identified by 16S rRNA gene fragment sequencing. Results showed a wide variety of organisms, with 22 different genera encountered. Pseudomonas and Ralstonia, of the γ- and β-Proteobacteria, respectively, were the most frequently encountered genera. Gram-positive bacteria were also observed, and Staphylococcus, Plantibacter, and Bacillus were the most highly represented genera. The sampling period corresponding to 50% of the cumulative sap flow percentage presented the greatest bacterial diversity according to its Shannon diversity index value (1.1). γ-Proteobacteria were found to be dominant almost from the beginning of the season to the end. These results are providing interesting insights on maple sap microflora that will be useful for further investigation related to microbial contamination and quality of maple products and also for guiding new strategies on taphole contamination control.
Maple sap is collected from maple trees (Acer saccharum Marsh.) during the spring season for the production of maple syrup and other maple products. The sap is a solution containing between 1 and 5.4% solids composed of different organic compounds, such as sucrose, glucose, fructose, organic acids, and nitrogenous and phenolic compounds, as well as different minerals (19). This solution is known to be a good nutrient medium for microorganisms, and their growth in the sap will eventually affect the quality of maple syrup and other end products (15, 18, 21). These microorganisms can enter the sap, notably by unsanitary tapping, since the taphole constitutes their first point of entry into maple sap. They will then grow in the sap as it progresses in the tubing collection system and storage tanks. For a number of years, taphole microbial contamination has been a subject of interest for scientists and people involved in maple syrup production. Solutions to control this contamination have been proposed without any real success. Nevertheless, bacteria have already been found to be predominant in the sap compared to yeasts and moulds (15), and efforts have also been made in the past for their identification. The most significant work was reported by Sheneman and Costilow (25), who found several species of bacteria in the tapholes of maple trees, consisting mainly of gram-negative rod-shaped aerobic bacteria identified as Pseudomonas, Achromobacter, and Flavobacterium. Among them, Pseudomonas geniculata was the most frequently encountered species.
Since these pioneer works on maple sap microbiology, maple syrup production operations have significantly changed (10). Tapping procedures now use smaller spouts made of plastic materials instead of larger metallic spouts. Maple sap collection procedures also differ from historical ones by using plastic tubing and vacuum instead of metal buckets. Added to the evolution of the sugarbush operations is the fact that the microbiological identification methods have also considerably evolved over the years. Molecular tools for the identification of microorganisms are now in common use, and 16S rRNA gene analysis is intensively used in phylogeny studies. Among the 16S rRNA gene analysis methods is amplified ribosomal DNA restriction analysis (ARDRA). This molecular technique has been successfully used for bacterial community analysis in a great variety of environments, including food (2, 7, 22, 23).
All these changes are contributing to the need for updated information on the microbial ecology of maple sap. Therefore, the aim of this work was to analyze the cultivable bacterial community of maple sap coming out of the taphole by molecular techniques. Better knowledge of bacterial composition in sap will undoubtedly contribute to improvement of control strategies for prevention of taphole contamination.
MATERIALS AND METHODS
Maple sap sampling.
Microbial contamination of maple sap was assessed by performing bacterial counts on samples aseptically collected from the tapholes. Maple sap samples were collected during the 2001 and 2002 spring seasons at the Centre ACER experimental sugarbush located near the city of Victoriaville (Québec, Canada) in one of the world's biggest maple syrup production region. The samples were obtained from randomly selected tapholes at five different periods evenly distributed over the season in terms of sap volume percentage (0, 25, 50, 75, and 100%), determined by historical monitoring of seasonal sap yield. Tapping procedures were applied according to the current recommendations, and no chemical products were used during tapping to prevent microbial contamination. For each sampling period, 10 samples were collected for a total of 50 samples per season. In order to collect sap samples from the taphole, the spout was delicately removed and a sterile device made of a spout and a test tube was inserted into the taphole to aseptically obtain 20 ml of sap by applying vacuum to the device. Sap samples were kept frozen prior to analysis.
Bacterial counts and culture isolation.
Bacterial counts were performed on each maple sap sample. Total aerobic counts were obtained by diluting the samples in 0.1% peptone water (Difco Laboratories, Detroit, Mich.) and spread plating in duplicate on plate count agar (Difco Laboratories) supplemented with 0.5% of sucrose. Incubation was performed at 30°C for 48 h followed by 7°C for 10 days to increase the differences in colony appearance. Bacterial isolates were obtained from plate count agar plates presenting between 30 and 150 colonies (from one to eight colonies per sample). The isolates were picked to represent the widest variation of colony appearance (form, color, and texture) and then purified by three successive cultivations on tryptic soy agar (Difco) at 30°C for 24 to 48 h. Stock cultures were prepared in brain heart infusion medium (Difco) supplemented with glycerol and kept frozen at −80°C prior to use.
Total DNA extraction.
The DNA of bacterial isolates was prepared according to the procedures of Vincent et al. (27) with the exception that for gram-negative bacteria, no mutanolysin was used. The concentration of purified DNA was measured using Hoechst 33258 dye and a fluorometer (Hoefer DyNA Quant 200; Pharmacia Biotech, Piscataway, N.J.).
PCR amplification.
The amplified 16S rRNA gene was obtained from each isolate and reference strain by PCR with the universal primers F27 (5′-AGAGTTTGATCMTGGCTCAG-3′) (3) and R1492 (5′-TACGGYTACCTTGTTACGACTT-3′) (28), which are targeted to universally conserved regions and permit the amplification of an approximately 1,500-bp fragment. PCR amplification was carried out in a GeneAmp PCR System 9600 thermocycler (Perkin-Elmer Corporation, Norwalk, Conn.). Reaction tubes contained 25 ng (5 μl) of DNA extract, 1 U of Taq DNA polymerase (Pharmacia Biotech), 1× buffer (10 mM Tris-HCl [pH 9.0], 1.5 mM MgCl2, 500 mM KCl), 10 mM deoxynucleoside triphosphate, and 20 pmol of each primer/μl. Initial DNA denaturation and enzyme activation steps were performed at 95°C for 4 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 1 min and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. The presence and yield of specific PCR product (16S rRNA gene) was monitored by 1% agarose (wt/vol) (BioShop Canada) gel electrophoresis at 200 V for 1 h in 1× Tris-acetate-EDTA buffer and made visible by ethidium bromide staining and UV transillumination.
ARDRA procedure.
Three restriction enzymes were selected on the basis of simulated digests of the complete 16S rRNA gene sequences of chosen reference strains retrieved from GenBank. These strains are listed along with the accession numbers in Table 1. The selection of the reference strains was based on a preliminary study performed in our laboratory on bacterial population of maple sap from which different isolates were identified (data not shown) using the API 20E and 20NE strips (Biomerieux, Marcy-l'Etoile, France). The enzymatic reactions were simulated via the NEB cutter software, version 1.0, of the New England BioLabs website, using all commercially available enzymes. For digest experiments, cost-effective combinations of three enzymes were chosen, revealing clearly different (and thus by agarose gel electrophoresis clearly resolvable) restriction site polymorphisms in the reference strain sequences. Unpurified PCR products were each digested with the three selected restriction enzymes in separated reactions. The chosen enzymes were RsaI, HaeIII, and AluI. The digestions were performed for 2 h at 37°C in 20-μl reaction volumes containing 10 μl of the PCR product solution, 2 μl of the commercially supplied incubation buffer, 7.5 μl of water, and 0.5 μl (10 U/μl) of the restriction enzyme. Reaction products (10 μl) were run on a 2% agarose 1000 gel (GibcoBRL, Life Technologies Inc., Burlington, Ontario, Canada) in 1× Tris-borate-EDTA buffer for 4 h at 250 V under refrigeration. Gels were stained with ethidium bromide, made visible by UV transillumination, and digitalized with the gel print 2000i system (Bio/Can Scientific Inc., Mississauga, Ontario, Canada). The images were analyzed with the GelCompar software (Molecular Analyst Software Fingerprinting Plus; Bio-Rad Laboratories, Hercules, Calif.). The background was subtracted by the rolling disk method, and the normalized patterns obtained with each enzyme were combined to obtain a single pattern for each isolate or reference strain. The patterns were used to construct a dendrogram by using the unweighted pair group method using arithmetic averages (UPGMA) clustering algorithm using the Pearson correlation coefficient along with the fine optimization option. Bacterial isolates showing the same banding pattern were assembled to form a group. After distribution of the isolates according to their group and the sampling period (Table 2), the bacterial diversity was calculated for each sampling period by the Shannon diversity index (H) (17). For the diversity index calculation (see Fig. 5), the following equation was used: H = 1/N Σ − (logNi − logN) Ni, in which N represents the total number of isolates in a sampling period and Ni is the number of isolates for a single group.
TABLE 1.
List of reference strains and accession numbers used for ARDRA method development
| Name | ATCC no. | Accession no. |
|---|---|---|
| Pseudomonas fluorescens | 14150 | |
| Enterobacter aerogenes | 13048 | AJ251468 |
| Pseudomonas putida | 12633 | D84020 |
| Pseudomonas fluorescens | 13525 | D84013 |
| Pseudomonas chlororaphis | 9446 | AF094723 |
| Brevundimonas vesicularis | 11426 | AB021414 |
| Burkholderia thailandensis | 700388 | U91838 |
| Stenotrophomonas maltophilia | 19374 | AB021404 |
| Alcaligenes latus | 29712 | D88007 |
| Flavimonas oryzihabitans | 43272 | D84004 |
| Kocuria rosea | 186 | X87756 |
| Rhizobium rhizogenes | 11325 | D12788 |
| Burkholderia cepacia | 25416 | M22518 |
| Pseudomonas aeruginosa | 10145 | X06684 |
| Sphingomonas paucimobilis | 29837 | D16144 |
| Chryseobacterium indologenes | 29897 | M58773 |
| Chryseobacterium meningosepticum | 13253 | M58776 |
| Aeromonas hydrophila | 7966 | X60404 |
| Alcaligenes faecalis | 8750 | M22508 |
TABLE 2.
List of bacterial isolates obtained in this study representing each ARDRA group and their closest affiliation according to the V1, V2, and V6 hypervariable regions (8) partial sequencing (525 bp) of 16S rRNA gene.
| Isolate | ARDRA group | Sourcea | Accession no. | Closest EMBL library strain and accession no. | Similarity (%) |
|---|---|---|---|---|---|
| MSB2122 | 1 | 75%; 2001 | AY275474 | Pedobacter cryoconitis,AJ438170 | 97.8 |
| MSB2100 | 2 | 75%; 2001 | AY275473 | Pedobacter cryoconitis,AJ438170 | 97.4 |
| MSB3023 | 3 | 50%; 2002 | AY275498 | Pedobacter sp., AF385531 | 94.8 |
| MSB3020 | 4 | 50%; 2002 | AY275497 | Chryseobacterium sp., AY043370 | 97.1 |
| MSB2060 | 5 | 50%; 2001 | AY275475 | Flavobacterium indoltheticum,M58774 | 97.1 |
| MSB3005 | 6 | 0%; 2002 | AY275496 | Paenibacillus borealis,AJ011322 | 98.3 |
| MSB2004 | 7 | 0%; 2001 | AY275491 | Ralstonia eutropha,AF027407 | 97.9 |
| MSB2010 | 7 | 0%; 2001 | AY275489 | Ralstonia sp., AF500587 | 98.8 |
| MSB2080 | 7 | 50%; 2001 | AY275490 | Ralstonia taiwanensis,AF300324 | 98.5 |
| MSB2054 | 8 | 50%; 2001 | AY275495 | Bacillus cereus,AY138278 | 99.8 |
| MSB2047 | 8 | 50%; 2001 | AY275493 | Bacillus cereus,AY138271 | 99.6 |
| MSB2029 | 8 | 25%; 2001 | AY275494 | Bacillus cereus,AY138273 | 99.8 |
| MSB2042 | 9 | 50%; 2001 | AY275501 | Staphylococcus warneri,Z26903 | 100.0 |
| MSB2065 | 9 | 50%; 2001 | AY275502 | Staphylococcus warneri,Z26903 | 100.0 |
| MSB2016 | 9 | 0%; 2001 | AY275500 | Staphylococcus warneri,Z26903 | 100.0 |
| MSB2005 | 10 | 0%; 2001 | AY275504 | Staphylococcus epidermidis,AF269927 | 99.8 |
| MSB2032 | 10 | 25%; 2001 | AY275505 | Staphylococcus caprae,AB009935 | 99.6 |
| MSB2024 | 11 | 25%; 2001 | AY275503 | Staphylococcus sp., X86640 | 98.9 |
| MSB2146 | 12 | 100%; 2001 | AY275492 | Brenneria alni,AJ223468 | 96.8 |
| MSB2071 | 13 | 50%; 2001 | AY275482 | Pseudomonas graminis,Y11150 | 99.8 |
| MSB2083 | 13 | 75%; 2001 | AY275480 | Pseudomonas sp., AY131214 | 99.6 |
| MSB2084 | 13 | 75%; 2001 | AY275481 | Pseudomonas graminis,Y11150 | 99.8 |
| MSB2019 | 13 | 25%; 2001 | AY275479 | Pseudomonas sp., AB013843 | 99.8 |
| MSB2074 | 14 | 50%; 2001 | AY275484 | Pseudomonas sp., AF177916 | 99.8 |
| MSB2111 | 14 | 75%; 2001 | AY275485 | Pseudomonas fulgida,AJ492830 | 100.0 |
| MSB2037 | 15 | 50%; 2001 | AY275476 | Pseudomonas syringae,AB001446 | 100.0 |
| MSB2015 | 15 | 0%; 2001 | AY275477 | Pseudomonas syringae,AB001450 | 99.8 |
| MSB2018 | 15 | 0%; 2001 | AY275478 | Pseudomonas syringae,AB001445 | 99.8 |
| MSB2097 | 16 | 75%; 2001 | AY275483 | Pseudomonas sp., AY131221 | 99.0 |
| MSB2082 | 17 | 75%; 2001 | AY275488 | Pseudomonas sp., AF511509 | 98.7 |
| MSB2046 | 18 | 50%; 2001 | AY275486 | Pseudomonas sp., AF408942 | 99.8 |
| MSB2138 | 19 | 100%; 2001 | AY275487 | Pseudomonas sp., AF4088867 | 97.7 |
| MSB3007 | 20 | 0%; 2002 | AY275499 | Uncultured γ-proteobacteria, AF424755 | 99.8 |
| MSB2139 | 21 | 100%; 2001 | AY275509 | Plantibacter flavus,AJ310417 | 100.0 |
| MSB2110 | 21 | 75%; 2001 | AY275508 | Plantibacter flavus,AJ310417 | 99.8 |
| MSB3012 | 22 | 50%; 2002 | AY275519 | Agreia sp., AF513393 | 97.4 |
| MSB2127 | 23 | 75%; 2001 | AY275511 | Frigoribacterium sp., AF157479 | 99.8 |
| MSB2131 | 23 | 75%; 2001 | AY275510 | Frigoribacterium sp., AF157479 | 99.8 |
| MSB2040 | 24 | 50%; 2001 | AY275514 | Arthrobacter rhombi,Y15885 | 96.2 |
| MSB2038 | 25 | 50%; 2001 | AY275512 | Micrococcus luteus,AF057289 | 99.6 |
| MSB3029 | 25 | 50%; 2002 | AY275513 | Micrococcus luteus,AF057289 | 99.8 |
| MSB2098 | 26 | 75%; 2001 | AY275515 | Microbacterium sp., AJ391205 | 100.0 |
| MSB2114 | 27 | 75%; 2001 | AY275516 | Sanguibacter suarezii,X79451 | 100.0 |
| MSB3037 | 28 | 50%; 2002 | AY275520 | Cellulomonas flavigena,AF140036 | 96.8 |
| MSB3004 | 29 | 0%; 2002 | AY275506 | Curtobacterium flaccumfaciens,AJ312209 | 99.8 |
| MSB2033 | 30 | 25%; 2001 | AY275507 | Brevibacterium casei,X76564 | 99.2 |
| MSB3008 | 31 | 0%; 2002 | AY275518 | Rothia dentocariosa,AF543276 | 99.6 |
| MSB3003 | 32 | 0%; 2002 | AY275517 | Rhodococcus erythropolis,AY147846 | 99.8 |
Source, sap flow cumulative percentage and season.
FIG. 5.
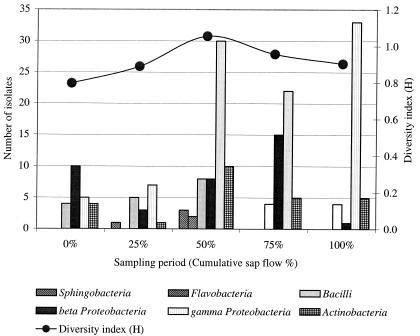
Change in the maple taphole bacterial composition over the sap flow season. Composition of the taphole bacterial community is described by the different classes of bacteria and the Shannon diversity index (H) for each sampling period (cumulative sap flow percentage).
Cloning, plasmid extraction, and digestion.
Amplified PCR products of representatives of each ARDRA group were randomly selected and ligated into the pCR2.1-TOPO cloning vector (Invitrogen, Burlington, Ontario, Canada). Ligated DNA was transformed into competent Escherichia coli TOP10 cells (Invitrogen) by electroporation (Gene Pulser, 25 μF, 2.5 kV; Bio-Rad). To determine positive clones, plasmid inserts were extracted by the method of Holmes and Quigley (11) except that isopropanol precipitation of DNA was replaced by two successive precipitations with ammonium acetate (7.5 M) and ethanol (100%). The extracts were digested with the restriction endonuclease EcoRI. Two microliters of the extracts were digested with 1 U of the restriction endonuclease in a total volume of 20 μl. The digestions were carried out as recommended by the manufacturer, with the appropriate restriction buffer at 37°C for 2 h. DNA fragments were resolved in 1% (wt/vol) agarose gels (BioShop Canada) with 1× Tris-acetate-EDTA buffer. DNA was visualized by transillumination with UV light after the gels were stained with ethidium bromide. Plasmid DNA of positive clones was then purified with the QIAprep Spin Miniprep kit (Qiagen, Mississauga, Ontario, Canada) according to the recommendations of the manufacturer.
16S rRNA gene partial sequencing and sequence analysis.
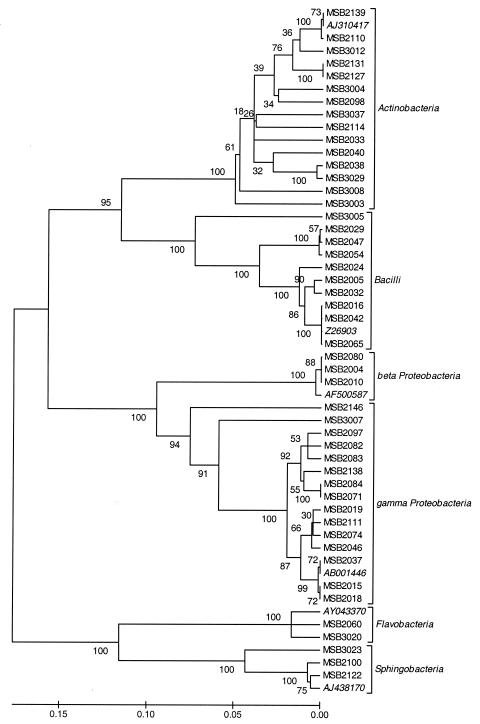
PCR products obtained from 48 cloned bacterial isolates were sequenced using an Applied Biosystems 310 sequencer (ABI 310 DNA sequencer, Big Dye Terminator cycle sequencing ready kit; Perkin-Elmer). Sequences of the PCR products (each 1,500 bp) obtained with the F27 and R1492 primers were partially sequenced using primers M13F (Invitrogen), 342R (16), and 800R (1). The partial sequences used to determine similarities were analyzed from the F27 primer to the conserved region corresponding to the sequence 5′-GTGCCAGCMGCCGCGGTAATAC-3′, thus giving an approximately 525-bp fragment. The 16S rRNA gene sequences which have been determined in the present study were deposited in the National Center for Biotechnology Information database and are available under the accession numbers shown in Table 2. The FASTA database of the European Bioinformatics Institute (EMBL), accessible on the internet (http://www.ebi.ac.uk/fasta/) was used to find nearly identical sequences for the 16S rRNA gene sequences determined. The ClustalW program, from the European Bioinformat-ics Institute (EMBL), accessible on the net (http://www.ebi.ac.uk/clustalw/), was used to align the sequences. Sequence dissimilarities were converted to evolutionary distances according to the method of Jukes and Cantor (13). The construction of neighbor-joining tree (see Fig. 4) (24) and bootstrap analysis of 500 resamplings (6) were performed using Mega 2 software (molecular evolutionary genetics analysis, version 2.1).
FIG. 4.
Phylogenetic tree of 48 maple sap bacterial isolates based on 16S rRNA gene partial sequences. The bootstrap consensus tree (50% cutoff value) was constructed by using the UPGMA.
RESULTS
Taphole bacterial contamination.
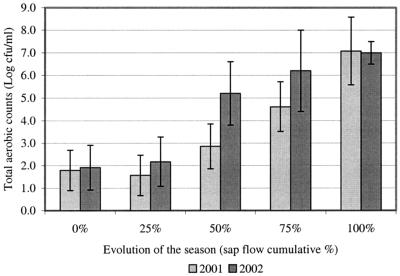
The log number of CFU per milliliter of sap directly obtained from maple tree tapholes was measured at five sampling periods over the 2001 and 2002 seasons to assess the change in the contaminating bacterial population (Fig. 1). Results show that bacterial contamination of the taphole was quite low at the beginning of the season but that it progressively reached a higher concentration of bacteria from the middle of the season to the end. Contamination of the 2001 samples contamination tended to be lower than but not significantly different from that of the 2002 samples. The mean daily air temperatures during the 2001 and 2002 seasons were 6.2 and 5.1°C, respectively, and the average sap volumes per taphole obtained for 2001 and 2002 were relatively similar at 88 and 83 liters/taphole, respectively. The extent of the 2001 season was 42 days with 34 days of sap flow, while the extent of the 2002 season was 56 days with 35 days of sap flow.
FIG. 1.
Change in the contaminating bacterial population of the sap obtained from maple tree tapholes during the 2001 and 2002 seasons. Average results (n = 10) are presented with standard deviations (bars) for each sap flow cumulative percentage.
ARDRA analysis on reference strains.
The ARDRA method was first applied on reference strains obtained from American Type Culture Collection (Table 1) to verify whether the method was able to provide enough discrimination between the banding patterns of different genera and species of bacteria. The results of ARDRA analysis on reference strains (Fig. 2) show that the method was able to differentiate between reference strains at the genus level, with a similarity lower than 50% between strains of different genera. The method was also able to differentiate between species of the same genus, like Pseudomonas, where similarity was 70% or less. Species of the genera Burkholderia and Chryseobacterium seemed to be harder to differentiate, with similarities ranging between 70 and 80%.
FIG. 2.
Agarose gel-generated ARDRA dendrogram illustrating the relationship (percent similarity) between different reference bacterial strains obtained from American Type Culture Collection that were used for ARDRA method development. ARDRA banding patterns were obtained by independent digestion of the amplified 16S rRNA gene with RsaI, HaeIII, and AluI endonucleases. The dendrogram was constructed with Molecular Analysis Software Fingerprinting Plus and grouped with the UPGMA.
ARDRA analysis and 16S rRNA gene partial sequencing on maple sap bacterial isolates.
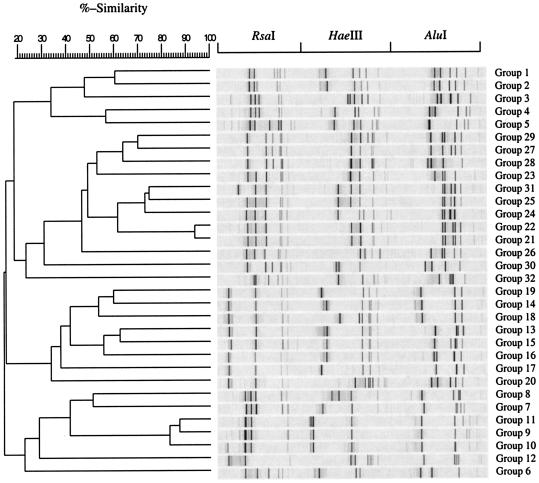
As described for the reference strains, 190 maple sap isolates obtained from each of the sampling periods of the 2001 and 2002 seasons were subjected to ARDRA analysis by digestion of the amplified 16S rRNA gene with RsaI, HaeIII, and AluI. Similar banding patterns obtained after combination of the three independent digestions were grouped (Fig. 3) to finally obtain a total of 32 groups. Representatives of each group were selected for cloning and partial 16S rRNA gene sequencing so as to retrieve sequence similarity and bacterial identity from sequence databases (Table 2). As shown in Fig. 3, each group presented a specific banding pattern, and groups further known to be of different genera (Table 2) were clearly differentiated, with a similarity lower than 70%. However, exceptions were found for groups 24, 25, and 31, where the similarity was between 70 and 80%, and for groups 21 and 22, where similarity was higher than 90%. These last groups were further known to be of different genera after 16S rRNA gene partial sequencing (Table 2) and nevertheless gave relatively similar banding patterns. Some groups formed according to ARDRA analysis were more easily distinguishable and have been differentiated at the species level. For instance, groups among the Pseudomonas genus (groups 13 to 19) had a similarity lower than 65% and were clearly differentiated. After partial sequencing of the 16S rRNA gene corresponding to a 525-bp fragment coding for the hypervariable V1, V2, and V6 regions (8) of ARDRA group representatives, closest identities of isolates were retrieved from sequence databases. Identity results showed a strong similarity with library sequences, close to 100% for each isolate (Table 2). Some isolates, like those of group 9, were strongly related to library sequences, with 100% similarity. Relatedness between maple sap isolate sequences is demonstrated in Fig. 4, where isolates were clustered according to the organism class. Isolates were therefore clustered in six different classes: Actinobacteria, bacilli, β- and γ-Proteobacteria, Flavobacteria, and Sphingobacteria. Actinobacteria and γ-Proteobacteria encountered the highest number of different ARDRA groups, with 12 and 9, respectively.
FIG. 3.
Dendrogram of maple sap bacterial isolates representing each ARDRA group. ARDRA banding patterns were obtained after independent restriction digestion of the amplified 16S rRNA gene with three different enzymes (RsaI, HaeIII, and AluI). The dendrogram was constructed with Molecular Analysis Software Fingerprinting Plus and grouped with the UPGMA.
Distribution and diversity of maple sap isolates.
Results in Table 2 demonstrated a wide variety of organisms, with 22 different genera encountered. Gram-negative isolates were more frequently encountered, with 140 isolates, compared to 50 gram-positive isolates. Distribution of maple sap isolates according to groups and the evolution of the sap flow season (Table 3) revealed that groups 7, 13, 14, and 15 had the most numerous isolates and that these groups belonged to gram-negative organisms corresponding to the Ralstonia and Pseudomonas genera. For gram-positive organisms, groups 9, 21, 10, and 8 were the most highly represented, with less than 7% total frequency. These last groups corresponded to the Staphylococcus, Plantibacter, and Bacillus genera. Sampling periods corresponding to sap flow cumulative percentages of 50 and 75 had the highest numbers of isolates, with 59 and 49, respectively. According to the data presented in Fig. 5, all six classes of bacteria were encountered only at midseason, and the γ-Proteobacteria class dominated from 25% of cumulative sap flow to the end of the season. Diversity, as indicated by the Shannon diversity index (H), was highest at 1.1 at midseason and was less at the beginning and at the end, according to data in Fig. 5.
TABLE 3.
Maple taphole bacterial isolates distribution according to ARDRA groups, restriction patterns, and the evolution of the season as determined by the cumulative sap flow percentage
| Group no. | ARDRA banding pattern according to restriction enzyme used
|
No. of isolates for each sampling period (cumulative sap flow %)
|
Group total frequency (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| RsaI | HaeIII | AluI | 0% | 25% | 50% | 75% | 100% | ||
| 1 | A | A | A | 2 | 1.1 | ||||
| 2 | B | A | A | 1 | 0.5 | ||||
| 3 | C | B | B | 1 | 0.5 | ||||
| 4 | C | D | D | 1 | 0.5 | ||||
| 5 | D | C | C | 1 | 0.5 | ||||
| 6 | E | E | E | 1 | 0.5 | ||||
| 7 | F | F | F | 10 | 3 | 8 | 15 | 1 | 19.5 |
| 8 | F | G | G | 1 | 1 | 3 | 2.6 | ||
| 9 | G | H | H | 1 | 1 | 4 | 3 | 3 | 6.3 |
| 10 | G | H | I | 1 | 2 | 1 | 1 | 1 | 3.2 |
| 11 | G | H | J | 1 | 0.5 | ||||
| 12 | H | I | K | 4 | 2.1 | ||||
| 13 | I | J | L | 1 | 4 | 6 | 10 | 11.1 | |
| 14 | I | J | M | 4 | 17 | 9 | 7 | 19.5 | |
| 15 | I | J | N | 4 | 2 | 6 | 4 | 9 | 13.2 |
| 16 | I | J | O | 1 | 0.5 | ||||
| 17 | I | K | L | 1 | 0.5 | ||||
| 18 | I | L | M | 3 | 1 | 2 | 3.2 | ||
| 19 | I | M | M | 1 | 0.5 | ||||
| 20 | I | N | P | 1 | 0.5 | ||||
| 21 | J | O | Q | 2 | 1 | 4 | 3.7 | ||
| 22 | J | P | Q | 1 | 0.5 | ||||
| 23 | J | Q | R | 2 | 1.1 | ||||
| 24 | J | S | V | 2 | 1.1 | ||||
| 25 | J | T | W | 2 | 1.1 | ||||
| 26 | J | U | U | 1 | 0.5 | ||||
| 27 | K | O | S | 1 | 1 | 1 | 1.6 | ||
| 28 | K | O | T | 1 | 0.5 | ||||
| 29 | K | R | U | 2 | 1 | 1.6 | |||
| 30 | K | V | X | 1 | 0.5 | ||||
| 31 | L | T | W | 1 | 0.5 | ||||
| 32 | M | W | Y | 1 | 0.5 | ||||
| Total | 23 | 16 | 59 | 49 | 43 | 100.0 | |||
DISCUSSION
Bacterial contamination of maple sap obtained from the taphole was assessed in this study. Results obtained emphasize the need to elaborate strategies to control microbial contamination of the taphole, since this contamination will serve as inoculum for the sap collection and holding system. Research in the past has demonstrated that microorganisms can adversely affect the quality of maple syrup (5, 15, 18, 21). This quality is primarily based on the color, flavor, and texture of maple syrup. Microorganisms of maple sap have the ability to break down the sucrose molecule, the main organic component of the sap, into glucose and fructose subunits. These sugar molecules will then react during the heat evaporation process to produce darker syrup and intense caramel flavor through nonenzymatic browning reactions (caramelization and Maillard reaction). Microorganisms contaminating the maple tree taphole will also decrease, at least in part, the flow of sap by blocking the wood vessels, resulting in a shortage of productivity (26). Better knowledge of taphole contamination would bring interesting insight that could help determine contamination control strategies. For instance, control could be oriented against microorganisms having a negative impact on the quality of maple sap and/or to promote the presence of microorganisms having a positive impact. The data obtained on maple sap microflora will permit further pursuit of this type of investigation.
In this way, results obtained in this study have demonstrated the great bacterial diversity of the maple taphole. Gram-negative organisms were among the prevailing bacteria, and γ-Proteobacteria, including the Pseudomonas genus, were most frequently encountered. This observation was also made by Sheneman and Costilow (25), who have found that P. geniculata, now known as Pseudomonas fluorescens biotype G, was dominant in the maple taphole when old techniques of production were applied. The presence of P. geniculata in maple sap has already been found to enhance the characteristic flavor of maple syrup (20, 29). Our results showed that the ARDRA method was able to differentiate the Pseudomonas genus from others and that it was also useful for the discrimination of other genera and species of bacteria. The presence of the genus Pseudomonas was expected due to its importance in natural ecosystems, in relation notably with the degradation of xenobiotic compounds, its effect as a plant pathogen, and its plant growth-promoting activities (9, 12). Our study also reports the important presence at the maple tree taphole of β-proteobacteria from the genus Ralstonia (group 7), observed throughout the season. This genus was not reported in previous publications. The only reported genus closed to Ralstonia was Achromobacter, which is also part of the β-Proteobacteria class (25).
A total of 32 patterns were identified among the 190 isolates from maple taphole. The 32 ARDRA patterns represented organisms belonging to six bacterial divisions, namely Actinobacteria, bacilli, β- and γ-Proteobacteria, Flavobacteria, and Sphingobacteria. Actinobacteria and γ-Proteobacteria encountered the highest number of ARDRA groups. The community diversity in maple sap was observed by determining phylotype richness, distribution, and similarity for 32 16S rRNA gene clones from each ARDRA group. Our results are in agreement with those of previous studies using conventional microbiological methods. Looking at our results, some analogies could be made with the bacterial community of maple sap and those observed for the forest soils. For instance, Morselli and Whalen (18) have demonstrated that the wood tissue of a healthy maple tree is sterile or practically sterile, suggesting that contamination of the taphole would necessarily come from the surrounding environment of the tree. Thus, microorganisms encountered in the forest soil could possibly contaminate the tree bark and find their way into the taphole and consequently into maple sap. The rhizosphere is a dynamic niche containing complex microbial communities, and microbial members may participate in a variety of beneficial and detrimental interactions with plants. Bacterial community members of rhizosphere from British Columbia (Canada) forest soils were characterized by DNA sequence analysis of 16S rRNA gene fragments following direct DNA isolation from soil, PCR amplification, and cloning. Phylogenetic analyses revealed that 85% of 709 16S rRNA gene clones were classified as α-, β-, γ-, and δ-Proteobacteria, Actinobacteria, Cytophaga-Flexibacter-Bacteroides group, Acidobacterium, and Verrucomicrobia. Members of the Proteobacteria had an important contribution, representing 55% of the clone library (4).
In addition, the predominant presence of proteobacteria and gram-positive bacteria, such as bacilli and staphylococci, isolated from the maple taphole may be due in part to the use of aerobic-growth media. Kasahara and Ahttori (14) observed that in a study of two soils in Japan, 71% of the isolates that formed visible colonies within 18 h of incubation were members of the gram-positive species. As the length of the incubation period increased, the relative abundance of gram-positive organisms in the total collection of isolates decreased. This observation might explain in part our results on dominant members of maple sap microflora and on its diversity.
This study examined the cultivable bacterial community of maple sap by using the ARDRA method and 16S rRNA gene fragment sequencing. Results showed the great diversity of this bacterial community by the large number of ARDRA patterns observed in relation to the evolution of the sap flow season. After partial 16S rRNA gene sequencing, identities of ARDRA group representatives were revealed. Although sugaring operations have significantly changed since earlier studies on maple taphole microflora, the members of the genus Pseudomonas are still dominant in this environment. Dominance of the genus Ralstonia was also observed, which has not been reported before. Our results also show a greater bacterial diversity than previous work. For instance, an analogy was made between the bacterial community profile of the maple taphole and the bacterial community of the forest soil rhizosphere. These results provide interesting insights into maple taphole microflora that will be useful for further investigations of phenomena related to microbial contamination and quality of maple products and also of microbial control strategies.
Acknowledgments
This work was supported by the Centre ACER Research Fund (St-Hyacinthe, Québec, Canada) and Agriculture and Agri-food Canada (Ottawa, Ontario).
We thank Carmen Charron, Mélissa Cournoyer, and René Desruisseaux for their technical work on sample collection and microbial counts.
REFERENCES
- 1.Anzai, Y., K. Hongik, J. Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50:1563-1589. [DOI] [PubMed] [Google Scholar]
- 2.Cambon-Bonavita, M. A., F. Lesongeur, S. Menoux, A. Lebourg, and G. Barbier. 2001. Microbial diversity in smoked salmon examined by a culture-independent molecular approach—a preliminary study. Int. J. Food Microbiol. 70:179-187. [DOI] [PubMed] [Google Scholar]
- 3.Chèneby, D., L. Philippot, A. Hartmann, C. Hénalut, and J. C. Germon. 2000. 16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microb. Ecol. 24:121-128. [DOI] [PubMed] [Google Scholar]
- 4.Chow, M. L., C. C. Radomski, J. M. McDermott, J. Davies, and P. E. Axelrood. 2002. Molecular characterization of bacterial diversity in lodgepole pine (Pinus contorta) rhizosphere soils from British Columbia forest soils differing in disturbance and geographic source. FEMS Microbiol. Ecol. 42:347-357. [DOI] [PubMed] [Google Scholar]
- 5.Fabian, F. W., and H. H. Buskirk. 1935. Aerobacter aerogenes as a cause of ropiness in maple syrup. Ind. Eng. Chem. 27:349-350. [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Giraffa, G., P. De Vecchi, and L. Rossetti. 1998. Note: identification of Lactobacillus delbrueckii subspecies bulgaricus and subspecies lactis dairy isolates by amplified rDNA restriction analysis. J. Appl. Microbiol. 85:918-924. [DOI] [PubMed] [Google Scholar]
- 8.Gray, M. W., D. Sankoff, and R. J. Cedergren. 1984. On the evolutionary descent of organisms and organelles: a global phylogeny based on a highly conserved structural core in small subunit ribosomal RNA. Nucleic Acids Res. 12:5837-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harayama, S., and K. N. Timmis. 1989. Catabolism of aromatic hydrocarbons by Pseudomonas, p. 151-174. In D. A. Hopwood and K. F. Chater (ed.), Genetics of bacterial diversity. Academic Press, London, United Kingdom.
- 10.Heiligmann, R. B., M. R. Koelling, R. R. Morrow, L. Staats, L. Myott, G. Cook, G. Buzzell, and S. Williams. 1996. Maple sap production—tapping, collection and storage, p. 49-71. In M. R. Koelling and R. B. Heiligmann (ed.), North American maple syrup producers manual. The Ohio State University, Columbus, Ohio.
- 11.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 12.Johnsen, K., and P. Nielsen. 1999. Diversity of Pseudomonas strains isolated with King's B and Gould's S1 agar determined by repetitive extragenic palindromic-polymerase chain reaction, 16S rDNA sequencing and Fourier transform infrared spectroscopy characterization. FEMS Microbiol. Lett. 173:155-162. [DOI] [PubMed] [Google Scholar]
- 13.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-123. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 14.Kasahara, Y., and T. Ahttori. 1991. Analysis of bacterial populations in a grassland soil according to rates of development on solid media. FEMS Microbiol. Ecol. 86:95-102. [Google Scholar]
- 15.Lagacé, L., C. Girouard, J. Dumont, J. Fortin, and D. Roy. 2002. Rapid prediction of maple syrup grade and sensory quality by estimation of microbial quality of maple sap using ATP bioluminescence. J. Food Sci. 67:1851-1854. [Google Scholar]
- 16.Lane, D. J. 1991. 16S/23S rDNA sequencing, p. 115-176. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, New York, N.Y.
- 17.Legendre, L., and P. Legendre. 1984. Les données qualitatives multidimensionnelles, p. 187-203. In Écologie numérique, le traitement multiple des données écologiques. Masson, Paris, France, and les Presses de l'Université du Québec, Sainte-Foy, Québec, Canada.
- 18.Morselli, M. F., and M. L. Whalen. 1991. Aseptic tapping of sugar maple (Acer saccharum) results in light color grade syrup. Can. J. For. Res. 21:999-1005. [Google Scholar]
- 19.Morselli, M. F., and M. L. Whalen. 1996. Maple chemistry and quality, p. 162-171. In M. R. Koelling and R. B. Heiligmann (ed.), North American maple syrup producers manual, The Ohio State University, Columbus, Ohio.
- 20.Naghski, J., L. L. Reed, and C. O. Willits. 1957. Maple sirup. X. Effect of controlled fermentation of maple sap on the color and flavor of maple sirup. Food Res. 22:176-181. [Google Scholar]
- 21.Naghski, J., and C. O. Wilits. 1957. Maple sirup. XI. Relationship between the type and origin of reducing sugars in sap and the color and flavor of maple sirup. Food Res. 22:567-571. [Google Scholar]
- 22.Ross, I. L., Y. Alami, P. R. Harvey, W. Achouak, and M. H. Ryder. 2000. Genetic diversity and biological control activity of novel species of closely related pseudomonads isolated from wheat field soils in South Australia. Appl. Environ. Microbiol. 66:1609-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy, D., S. Sirois, and D. Vincent. 2001. Molecular discrimination of lactobacilli used as starter and probiotic cultures by amplified ribosomal DNA restriction analysis. Curr. Microbiol. 42:282-289. [DOI] [PubMed] [Google Scholar]
- 24.Saitou, N., and M. Nei. 1987. The neighbour-joining method: a new method for reconstruction of phylogenetic trees. Mol. Biol. E vol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 25.Sheneman, J. M., and R. N. Costilow. 1958. Identification of microorganisms from maple tree tapholes. Food Res. 24:146-151. [Google Scholar]
- 26.Sheneman, J. M., R. N. Costilow, P. W. Robbins, and J. E. Douglas. 1958. Correlation between microbial populations and sap yields from maple trees. Food Res. 24:152-159. [Google Scholar]
- 27.Vincent, D., D. Roy, F. Mondou, and C. Déry. 1998. Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 43:185-193. [DOI] [PubMed] [Google Scholar]
- 28.Wang, G. C. Y., and Y. Wang. 1996. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142:1107-1114. [DOI] [PubMed] [Google Scholar]
- 29.Willits, C. O., H. A. Frank, and R. A. Bell. 1957. Maple sirup. XIX. Flavor and color through controlled fermentation of maple sap. Food Technol. 15:473-474. [Google Scholar]