Abstract
High throughput sequencing technologies have become essential in studies on genomics, epigenomics, and transcriptomics. While sequencing information has traditionally been elucidated using a low throughput technique called Sanger sequencing, high throughput sequencing (HTS) technologies are capable of sequencing multiple DNA molecules in parallel, enabling hundreds of millions of DNA molecules to be sequenced at a time. This advantage allows HTS to be used to create large data sets, generating more comprehensive insights into the cellular genomic and transcriptomic signatures of various diseases and developmental stages. Within HTS technologies, whole exome sequencing can be used to identify novel variants and other mutations that may underlie many genetic cardiac disorders, whereas RNA sequencing (RNA-seq) can be used to analyze how the transcriptome changes. Chromatin immunoprecipitation sequencing (ChIP-seq) and methylation sequencing (Methyl-seq) can be used to identify epigenetic changes whereas ribosome sequencing (Ribo-seq) can be used to determine which mRNA transcripts are actively being translated. In this review, we will outline the differences in various sequencing modalities and examine the main sequencing platforms on the market in terms of their relative read depths, speeds, and costs. Lastly, we will discuss the development of future sequencing platforms and how these new technologies may improve upon current sequencing platforms. Ultimately, these sequencing technologies will be instrumental in further delineating how the cardiovascular system develops and how perturbations in DNA and RNA can lead to cardiovascular disease.
Keywords: DNA sequencing, RNA sequencing, chiP sequencing, cardiac disease, transcriptome, genomics, genetics
Introduction
Until the discovery of retroviruses, the central dogma of molecular biology stated that genes are transcribed to make RNA and in turn RNA is translated into protein 1, 2. This dogma outlines how the variable expression of genes can dynamically control cellular functionality and identity from a single genome. Gene expression is dynamically controlled and variations in transcription and translation can result in major functional changes within the cell. If the underlying DNA sequence is mutated, or if the downstream message is changed during transcription and translation, cellular function may be compromised, leading to various disease pathologies. While our environment affects the manifestation of disease, many diseases also have a strong underlying genetic component. Diseases that have a stronger genetic component rather than environmental component may include those that surface at birth (congenital diseases) and those which run in families (familial inheritance diseases). To determine how one’s genetic background contributes to disease, a large collection of genomic and transcriptomic data sets will be required. By sequencing multiple genomes, it is therefore possible to evaluate human genomic diversity, as demonstrated by the 1000 Genomes Project 3, 4. In addition, the ENCODE 5 and HapMap project 6 have employed many high throughput sequencing (HTS) applications outlined below to understand the functional attributes of each region of the genome. With advancements in HTS technologies, sequencing costs have now dramatically decreased and it may soon be possible to sequence the entire human genome for $1,000 or less 7. As the price of sequencing decreases, sequencing may become commonplace, which will vastly contribute our understanding on genomic variability and how this variability may increase one’s susceptibility to develop cardiovascular diseases. Ultimately, by lowering sequencing cost and in turn, making sequencing technologies mainstream, implementation of HTS technologies will be invaluable in determining the molecular pathways involved in cardiovascular development and disease.
First Generation Sanger Sequencing
DNA sequencing information has traditionally been elucidated using Sanger sequencing 8. This technique was developed by Dr. Frederick Sanger, who was subsequently awarded the 1980 Nobel prize in Chemistry 9. In this method, a complementary strand of DNA is made from the input template DNA from a mixture of 2′-deoxynucleotides (dNTPs) including 2′,3′-dideoxynucleotides (ddNTPs), which are labeled with fluorescent dyes 8. ddNTPs are nucleotides that lack a 3'-OH group required for cDNA elongation and when a ddNTP is incorporated into the elongating DNA strand, elongation is terminated, resulting in the generation of multiple DNA fragment sizes. The sizes of these fragments are separated using single base-pair resolution capillary electrophoresis, resulting in an electropheragram that appears to be a direct read-out of the nucleotide sequence from the original template molecule 10. Sanger sequencing can have an average read length of 800 base pairs, but it is limited by the amount of DNA that could be processed at a given time. To address the low throughput, newer sequencing technologies were developed that could read the sequence of multiple DNA molecules in parallel. Parallel capillary systems greatly increased the throughput of the number of DNA strands that could be analyzed 11, 12, because 1–6 megabases of DNA sequence could be acquired per day in a standard 96-capillary instrument 13. However, parallel capillary based systems are still limited by the amount of capillary columns which could be processed at a given time. Because the human genome consists of approximately 3 gigabases, containing approximately 20,000 genes that span 45 megabases (1.5% of the whole genome) 14–16, it took the Human Genome Project over a decade and billions of dollars to complete using Sanger sequencing 17, 18.
Key HTS Platforms
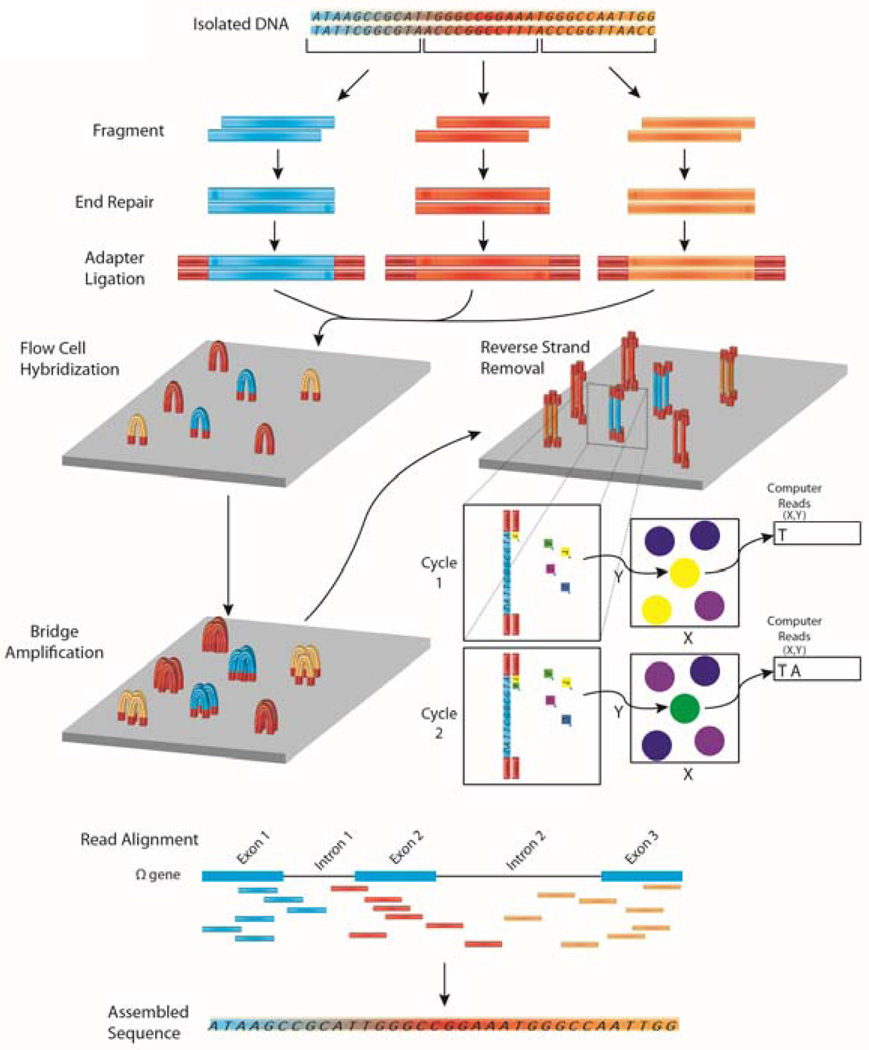
Commercially available sequencing platforms are expanding the potential of sequencing by exponentially increasing the throughput of their technologies. While many sequencing platforms are available, Illumina’s platforms (http://www.illumina.com/) have dominated much of the sequencing industry (Figure 1) 19. Illumina’s bridge amplification method allows for generation of small “clusters” with an identical sequence to be analyzed. Clusters formed on an Illumina flow cell create multiple primer hybridization steps allowing multiple sequencing start points. This allows the sequencing of both ends of the original template molecule known as paired-end sequencing. Sequencing information from paired-end reads play an important role in Illumina’s technology by increasing the output from a sequencing run, identifying splice variants in RNA-seq, and to deduplicate (remove duplicate copy) reads originating from the same original template molecule. Paired end reads are also important for identifying large structural variants such as inversions from whole genome sequencing, which are unnoticed with short sequencing techniques. A third read may also be used to separate out samples as long as each sample in the sequencing library had a unique barcode read engineered into the adapter construction as a third separate read. In Illumina sequencing however, all four nucleotides are available during incorporation which can lead to an overall substitution error rate of 0.11% 20.
Figure 1. Overview of DNA-sequencing using the Illumina platform.
In next generation DNA sequencing, DNA is first fragmented into smaller input-sized fragments by enzymes or by sonication. The ends of these fragments are repaired and specific adapters are ligated to the ends of the fragments, allowing hybridization to a flow cell to occur. A bridge amplification step is performed to create a “cluster” of fragments with the same sequence. One stand of DNA is removed and fluorescently labeled nucleotides are passed by each cluster. An image of the flow cells is recorded for the first cycle and a computer processes which nucleotide was incorporated at each cluster’s co-ordinates. The fluorescent label is cleaved and a second round of fluorescently labeled nucleotides is passed by each cluster. Again (cycle 2) the nucleotide is recorded and each cycle leads to the sequence of each fragment (a “read”). These reads are then aligned to a reference genome. By assembling reads (merging short reads together), it is therefore possible to reconstruct the unfragmented original sequence.
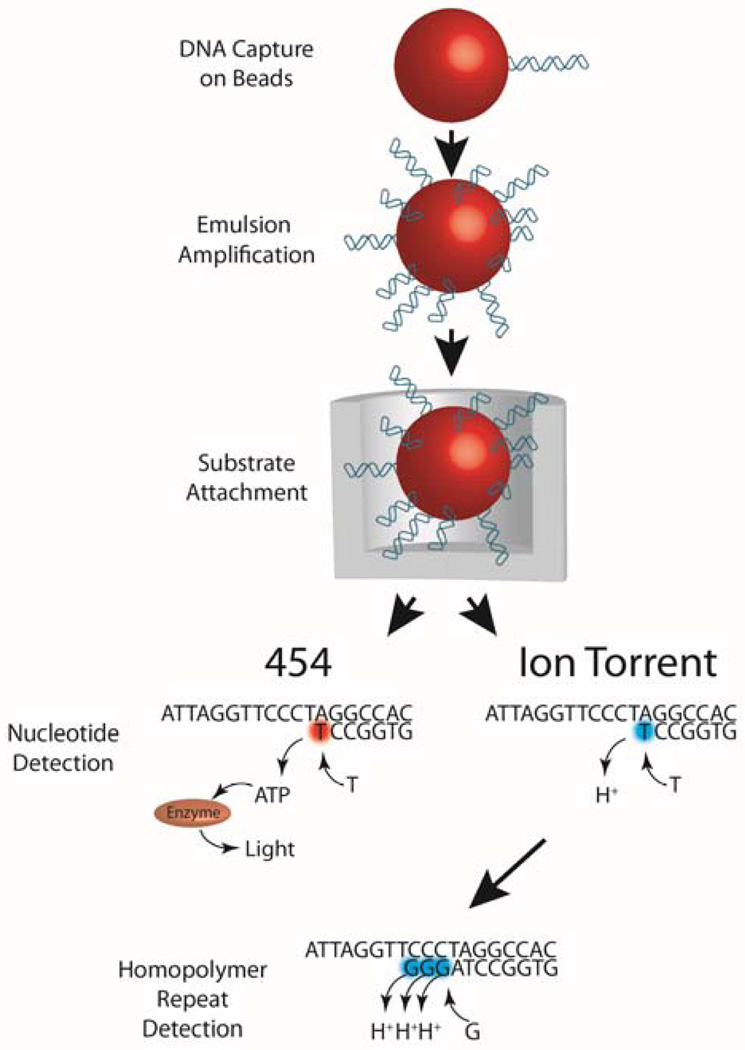
Both Ion Torrent (http://www.iontorrent.com/) and 454 (http://www.454.com/) employ the use of polymerase chain reactions to amplify DNA within an emulsified droplet (Figure 2). Sequencing information is correlated with either light (in 454) or hydrogen ions (in Ion Torrent) detection during each nucleotide incorporation event. If multiple nucleotide incorporation events occur, this is interpreted as stretches in the sequence of a particular nucleotide (homopolymers). All high throughput sequencing technologies have difficulties sequencing homopolymers, but sequencing homopolymers in Ion Torrent and 454 are more problematic because the nucleotides used lack a blocking moiety, resulting in entirely incorporating homopolymers during one cycle. Difficulties in interpreting homopolymers arise because these homopolymer signals are non-linear and have a Poisson distribution. This results in homopolymers of just two or three nucleotides to sometimes be contracted or expanded.
Figure 2. Ion Torrent and 454 sequencing.
Both Ion Torrent and 454 immobilize DNA fragments onto beads. In both platforms, template molecules are first immobilized on a bead which is emulsified so that subsequent amplification can occur clonally within the droplet. After clonal amplification, enrichment for DNA positive beads is performed using additional beads that can bind and isolate the available end of the library molecule, thus removing DNA negative beads. Enriched beads are deposited at the bottom of a well and sequencing is performed by flowing one base at a time over the templates. In Ion Torrent, an incorporation event is measured by a pH change from the release of protons resulting from the incorporation, whereas 454 uses a cascade of reactions resulting from pyrophosphate being released from each incorporation reaction. This leads to a photon being released by the enzyme luciferase. Therefore, the detection of light (in 454) or hydrogen ions (in Ion Torrent) when adenine is passed over each chamber is interpreted as thymine being the next nucleotide in the DNA sequence. Amplification of DNA fragments occurs in an emulsion and each bead is placed into a well large enough for each bead. Nucleotides are sequentially passed by each well where nucleotide incorporation occurs. If nucleotide incorporation occurs, a series of enzymatic reactions occurs that results in light being detected in the 454 platform. In Ion Torrent platforms, nucleotide incorporation results in the release of hydrogen ions and these ions are detected by each well. In Ion Torrent, if homopolymer repeats of the same nucleotide are present (GGG), multiple hydrogen ions will be released, generating a higher electrical signal. This is subsequently interpreted as multiple identical nucleotides being present in the sequence.
The Pacific Biosciences Real Time Sequencer (PacBio RS) (www.pacificbiosciences.com/) requires that each circular library molecule be bound to a polymerase enzyme as the input for sequencing on their single-molecule real-time sequencing (SMRT) cells. The library/polymerase complex is diffused over the SMRT cell’s Zero Mode Waveguides (ZMW’s), allowing the template to occupy the lumen of the ZMW sometimes. The RS uses video imaging of fluorescent nucleotides pausing at the bottom of the ZMW to record an incorporation event 21. Because the “pausing” can range from 1–3 seconds per incorporation event and nucleotides can diffuse freely into the ZMW, insertions are more common. It is possible to get multiple passes of sequencing around the same circular library molecule to generate what is called circular consensus sequencing (CCS) because of the long read length 22. The RS has seemingly random errors, whereas the other sequencing technologies tend to be less random with mistakes. These random errors, combined with multiple passes over the same circular fragment, generate a relatively low number of high quality reads, allowing the RS to be used as a cheaper variant validation tool over Sanger sequencing. For true single molecule sequencing, no amplification should occur on the sample to avoid amplification bias. Native DNA contains modified bases like 5-methyl cytosine that can be measured directly based on signature pausing signals with PacBio sequencing. For now, the lack of amplification may be the biggest drawback of single molecule sequencing, because some samples are just too low in material. This highlights the importance of creating new tools that can manipulate smaller reactions and use less input material.
Each sequencing instrument is limited as to the number of rounds it can record as well as the accuracy of the recording. In addition, there is a limit on the number of template molecules that can be read on each sequencing cell. Table 1 summarizes some of the main sequence platforms on the market today in terms of their relative costs, sequencing yields, quality scores, and sequencing times.
Table 1. Comparison of the current next generation sequencing platforms.
A cross comparison of Illumnia (Hiseq2500, Miseq), Ion Torrent (PGM 318, Proton I), PacBio (RS), and Roche 454 (FS FLX+, GS Junior) next generation platforms is presented. Each instrument was compared to show the specifications provided by the vendors including the cost, speed, accuracy, primary error type, and size of the data set that can be expected from each instrument. From those specifications, a cost per MB index was calculated. As each instrument needs to be maintained over time, the instrument maintenance cost was also provided by the vendors.
| Platform | Cost/run | Unique library molecules sequenced |
Benchmark Accuracy |
Read Length at Benchmark Accuracy |
Total Output at Specified Accuracy (GB) |
Cost ($/Benchmark MB) |
Sequencing time (not including library prep) |
Single Molecule (Yes/No) |
Primary Error Type |
Annual cost of maintenance contract |
|---|---|---|---|---|---|---|---|---|---|---|
| Hiseq2500 2×100bp Rapid Mode (2 flow cells) | $5,830 | 1.2 billion | 99.90% | 80% @ 100bp PE, 80×80 (160+bp total) | 100 GB | $0.06 | 27 hours | No | Substitution | $56,925 |
| Hiseq2500 2×100bp High Output Run Mode (2 flow cells) | $5,830 | 1.5 billion | 99.90% | 80% @ 100bp PE, 80×80 (160+bp total) | 540 GB | $0.01 | 11 days | No | Substitution | $56,925 |
| Miseq 2×250bp | $995 | 15–17 million | 99.90% | 70% @ 250bp PE 200×150 (350+bp total) | 5.6GB | $0.18 | 39 hours | No | Substitution | $11,250 |
| Ion Torrent PGM 318 chip | $749 Including 318 chip OneTouch 2 template kit Sequencing Kit | 3–6 million | 99% raw | Up to 400bp | 2Gb | $0.38 | 3.5 hours | No | Insertion Deletion | $8000/PGM System (includes torrent server and onetouch) |
| Ion Torrent Proton I | $1,000 including Ion PI Chip PI template kit for OneTouch 2 | 60–80 million | 99% raw | Up to 200bp | 10Gb | $0.10 | 2–4 hours | No | Insertion Deletion | $24,400 for Ion Proton System (includes OneTouch and TorrentServer) |
| PacBio RS 1, 120min video XL Binding kit and C2 chemistry | $110 | 23 thousand | 87% raw | Up to 21,000 bp (4,500 average) | 100MB | $1.10 | 12 hours | Yes | Insertion | $85,000 |
| PacBio RS 2, 55min videos, XL Binding kit and C2 chemistry | $110 | 62 thousand | 87% raw | Up to 13,000 (4,200 average) | 260MB | $0.42 | 12 hours | Yes | Insertion | $85,000 |
| 454 GS FLX + | $2,500 | Over 1 million | 99.90% | Up to 1000 (700 average) | 700 MB | $3.57 | 6 hours | No | Insertion Deletion | $50,000 |
| 454 GS Junior | $800 | 70–100 thousand | 99.90% | Up to 600 (400 average) | 40 MB | $20.00 | 3.5 hours | No | Insertion Deletion | $12,600 |
Common Applications in Next Generation Sequencing
DNA Sequencing
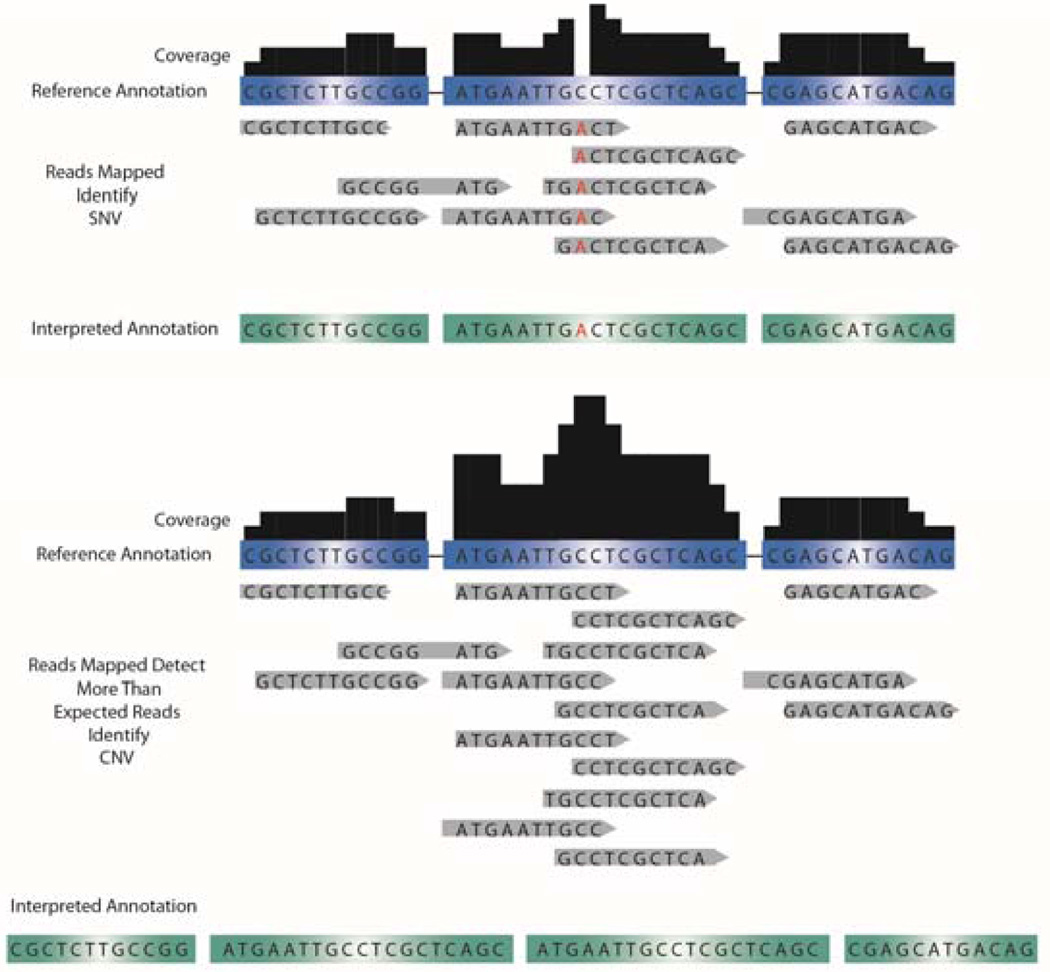
The order of DNA sequence and its variation dictates human developmental processes, uniquely identify each person, and encodes our susceptibility to diseases 23–25. By using high throughput DNA sequencing (DNA-seq) technologies, it is possible to identify genetic variants that play a role in human health. In whole genome sequencing, sequencing information of both the exons and introns is obtained 15, 26, 27, which may provide critical information on enhancer regions, promoters, and cis/trans regulatory elements that reside in the intronic regions along with structural variants such as copy number variants (CNV), inversions, and translocations affecting the exonic regions. To give a confidence level of how accurate the sequencing information is, the term “depth” is used to define the average number of times each nucleotide in the genome is observed 28. For example, if each nucleotide of the genome is reported an average of 10 times during sequencing, a depth level of 10× is obtained. Read depth is important to interpret structural variations, because for a given interval, an increase in the amount of reads at a given read depth may indicate an increase in copy number, whereas a decrease in the amount of reads at a given read depth may indicate deletions 29 (Figure 3). In general, as the read depth level increases, the sequencing information becomes more confident. It is recommended that an average read depth of 30× produces an adequate coverage level (the amount of times a nucleotide is reported within an assembled sequence) for whole-genome analysis and at a 50× read depth, approximately 94.9% of single nucleotide variants can be observed 30–32. Read depth is important in considering the experimental goals. To observe small changes (i.e., point mutations) associated with complex diseases, a high read depth and sequencing multiple individuals are required. However, observing large structural changes in comparison to a reference genome could be achieved with a very low read depth. In most cases, increasing read depth is more costly and alternative, targeted, and more cost-effective methods may be preferred over HTS (i.e., customized hybridization chips). In addition, given that in each sequencer run only a limited number of sequence fragments can be read, it may be more cost effective to only analyze the sequence of the genetic material that is transcribed into mRNA (exons), depending on the experimental needs. In this regard, whole exome sequencing uses sequence capture methods to enrich a subset of genomic DNA 33–35 using commercially available capture arrays (Roche NimbleGen, Agilent SureSelect, and Illumina 62MB). These arrays work with a set of binding oligonucleotides complementary to the human exome bound to magnetic beads. Further magnetic bead isolation enriches exome DNA, but may also introduce sequencing bias since some capture methods may not uniformly capture target DNA. However, isolating the exome provides relatively inexpensive 100× coverage of the genome coding regions and may be particularly useful for identifying rare genomic variants from a population of cells.
Figure 3. Detection of SNVs and CNVs.
Mapping HTS reads to control annotations (unaffected family member, reference annotation) is used to identify single nucleotide changes up to large structural DNA changes. Discrepancies between the reference annotation and the mapped sequence can be called to annotate SNV. More reads mapped (i.e., higher read depth) results in a higher confidence level to the called SNV in question. If a disproportionate amount of reads are mapped to a gene for a given read depth, this region of DNA may be interpreted as a CNV.
Bioinformatics analysis of DNA sequencing has come a long way since the original chromosome walking technique employed at the beginning of the Human Genome Project. Shotgun sequencing was developed to utilize high throughput short read technologies to assemble large genomes de novo. Large contiguous regions (contigs) are assembled from shorter ones using overlapping regions to link contigs, or reads, and the overhangs are used to extend the contig. Once a reference genome has been assembled for a species (or sometimes an individual), alignment within the reference is possible. For example, one could align reads to the reference genome (using programs such as BWA 36, SOAP3 37, and BFAST 38) or call single nucleotide variants (using GATK 39, MAQ 40 or SAMtools 41) and compare whole genomes through consensus with some flexibility allowed for variants/differences from a reference. Difficulties in DNA sequencing analysis lie in obtaining coverage in regions of extreme GC/AT content, discerning sequencing and amplification errors from actual variants (especially in the case of heterozygous variants, and rare variants from a population of cells) and being able to utilize short reads to assemble large repeats and/or large structural variants such as inversions. In addition, errors in mapping short reads can also occur given the ambiguous highly repetitive genomic regions and highly homologous gene families. To overcome these difficulties, newer software algorithms, longer read lengths, and/or higher coverage will need to be developed.
HTS has the potential to play an important role in cardiovascular research because many diseases have an unknown underlying genetic component. For example, arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C) is caused by mutations associated with desmosomes. However, non-desmosomal mutations in TGFβ3 RYR2 and TMEM43 have also been implicated in the disease phenotype of ARVD 42. While mutations in multiple genes have been identified to cause ARVD/C, only in 50–60% of ARVD/C patients could an underlying genetic mutation be found (reviewed in 43). In addition, some clinical presentations of ARVD/C are very similar to Brugada syndrome (predominantly exhibited by males, associated with familial inheritance, and exhibits idiopathic ventricular fibrillation) 44. Histopathological or advanced imaging modalities are required to distinguish between these two diseases 43. Whole genome and exome sequencing will lead to the discovery of previously unknown mutations that cause cardiovascular diseases as well as aid in the distinction between diseases that share very similar clinical presentations.
High throughput DNA sequencing will be instrumental in the screening and diagnostics of heart diseases related to larger structural genomic changes such as Down syndrome 45, DiGeorge syndrome 46, 4q- Syndrome 47, and 8p- Syndrome 48, as well as complex diseases related to copy number variants 49 and single nucleotide changes (single-nucleotide polymorphisms (SNPs), single-nucleotide variants (SNVs), and mutations). SNVs are variable regions of the DNA in which single nucleotide differences have been identified in the genetic code, whereas a SNP is a variant that appears with a >1% minor allele frequency in the population 15, 50. These observed polymorphisms may help predict the susceptibility of a patient cohort to develop heart disease. This is exemplified in the study by Matkovich et al.,51 where pooled sequencing data from four cardiac signaling genes identified a greater representation of specific SNPs within the cardiovascular heat shock protein gene HSPB7 from patients with heart failure. In addition, while one SNP was found to be within an intron of HSPB7, no differences were observed in the splicing or mRNA levels of this gene. Sequencing the adjacent renal chloride channel CLCNKA gene, however, identified a SNP in an exon of this gene, which demonstrated linkage disequilibrium with the intronic SNP in HSPB7. Further functional characterization of the renal chloride demonstrated an approximate 50% loss-of-function of the variant channel 52. In summary, HTS performed in these studies led to the identification of a common genetic risk factor for heart failure.
Given that SNV analysis can determine which genetic regions may influence a patient’s susceptibility to develop heart disease, SNV analysis could also be used to determine which drug therapy will be best suited for a particular patient. For instance, warfarin is an anticoagulant drug often prescribed to prevent thrombosis, and patients with common SNPs in the CYP2C9 and VKORC1 genes have been suggested to successfully predict a patient’s response to the anticoagulant effects of warfarin 53, 54. Further clinical studies will be required to warrant the use of SNP data to predict warfarin treatment. In addition, SNP analysis is being used to identify which SNPs can be either cardioprotective versus cardiotoxic to the effects of doxorubicin 55–57. Future SNP analysis studies will be important for optimizing patient specific treatment to existing cardiovascular drugs and for determining the effectiveness and safety of drugs under development 58.
Chromatin Immunoprecipitation Sequencing (ChIP-seq)
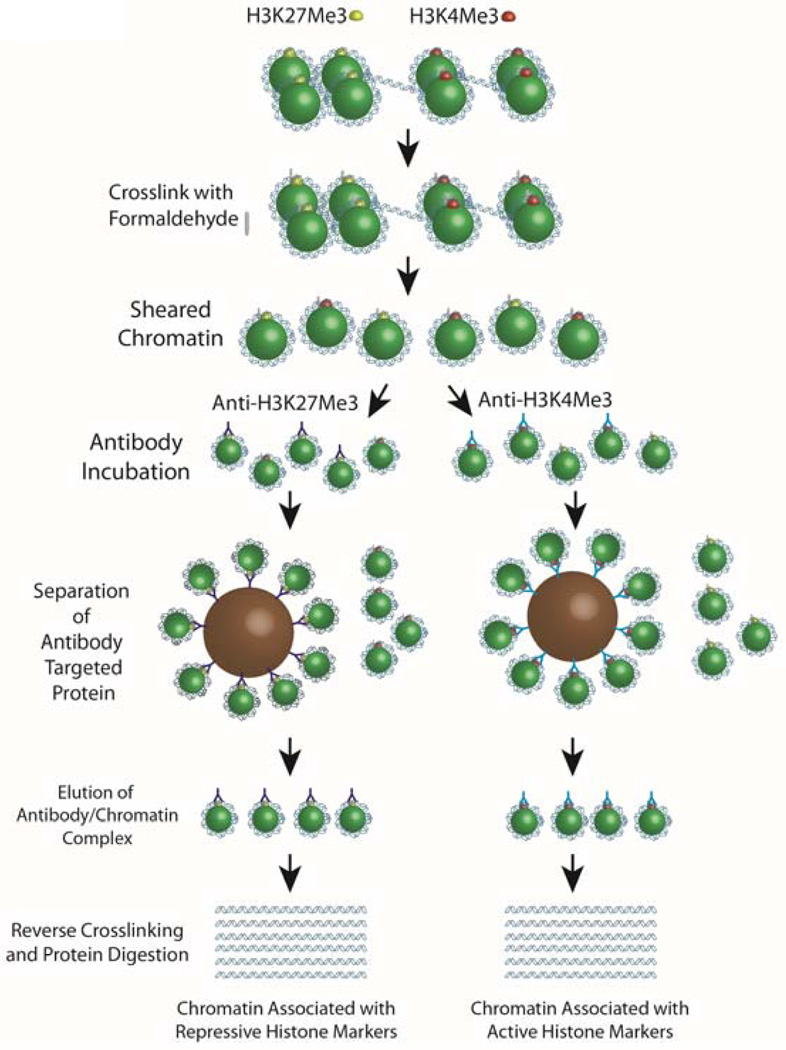
Gene expression can be influenced by epigenetic modifications which can be assessed by ChIP-seq. DNA in the nucleus is divided into actively transcribed regions called euchromatin, or transcriptionally silenced regions called heterochromatin 59. These regions represent loosely or tightly compact DNA regions and these different states are influenced by histone protein modifications 60, 61. Histone acetylation and methlyation are two modifications for histones, and depending on the histone modification, genes may be actively transcribed or repressed during these processes. For example, H3K27Me3 modification represses gene expression 61 whereas H3K4Me3 modification enhances gene activity 62. By performing chromatin immunoprecipitations with antibodies towards various histone modification states and sequencing the resulting immunopreciptated DNA, it is possible to assess different regions of DNA that may be actively transcribed or are transcriptionally silent. In ChIP-seq, formaldehyde is first used to covalently bond DNA to proteins with which they are interacting (Figure 4). The DNA-protein complex is fragmented and immunoglobulins specific for the protein of interest are used to pull down the fragment of DNA to which they are attached 63. From here the target DNA is isolated and a sequencing library is made using a standard library preparation method. Sequencing of a ChIP-seq library generates reads that align near the genomic regions associated with the target protein. Controls include a negative control antibody library and an input DNA library. Given that some antibodies are better at pulling down the target protein-DNA interactions, the use of a ChIP certified antibody greatly improves the signal-to-noise ratio in downstream analysis. In addition, under- or over-paraformaldehyde crosslinking as well as under/over DNA sheering can affect downstream ChIP-seq analysis. Bioinformatic analysis of ChIP-seq data involves mapping reads to a reference genome and using peak detection software (currently over 31 open source programs)11 to identify regions that have enriched mapping frequency. Therefore, the peak detection of immunoprecipitation from H3K27Me3 and H3K4Me3 histone modification states can provide information on which regions of the DNA are in an active or open chromosome state versus transcriptionally silent.
Figure 4. Chromatin immunoprecipitation for ChIP-seq.
To determine sequences of DNA that may be “open” for transcription, ChIP-seq uses the immunoprecipitation of chromatin bound to different histone modification states [transcriptionally silent (H3K27me3) and transcriptionally active (H3K4Me3) markers]. Crosslinking chromatin to the histones is first performed using formaldehyde, and chromatin is sheared into smaller fragments. Immunoglobulins specific for each histone modification state are incubated in separate reaction tubes, and magnetic or Sepherose beads known to bind to immunoglobulins are used to isolate bound immunoglobulin/chromatin complexes. Chromatin is separated from the histones by reverse crosslinking (high salt conditions) and protein digestion (proteinase K). DNA libraries are then made from the isolated DNA and sequencing of these DNA fragments is processed using one of the next generation sequencing platforms.
An additional important application of ChIP-seq is to determine which genes are bound by transcription factors and to determine which enhancer regions are active in the heart. In a study by Blow et al.,64 ChIP-seq of the transcriptional co-activator protein p300 on embryonic day 11.5 heart tissue was used to elucidate which enhancer regions are active in the developing mouse heart. ChIP-seq can also be used to determine how aberrations in transcription factor binding can disrupt gene expression and lead to cardiovascular diseases. Given that NKX2-5 mutations have been shown to cause hypoplastic left heart syndrome, atrial septal defects, and patent foramen ovale 65, immunoprecipitation of control and mutant NKX2-5 could be used to identify how NKX2-5 mutations affect transcription.
Identifying gene and protein interactions can also be determined using chromosome confirmation capture (3C) sequencing. 3C sequencing is important to identify functional associations among distal chromosomal regions (such as enhancers) 66, 67. These DNA interactions can be determined by first crosslinking DNA/protein complexes using formaldehyde and then using restriction enzymes to digest DNA into smaller fragments, leaving the crosslinked DNA fragments connected. DNA is then intramolecularly ligated and reverse crosslinked with heat. From here adaptors can be ligated on to generate a sequencing library. In a study by Korostowski et al.,68 3C sequencing was used to demonstrate how changes in chromosomal interactions occur with the promoter of the potassium channel Kcnq1 and interactions with the Kcnq1 promoter was demonstrated to influence the transition of a monoallelic to a biallelic expression of this gene during development of the heart. Future studies using 3C sequencing will not only be important to determine which distal chromosomal regions interact but also to study how these interactions occur temporally or in a tissue specific manner. By delineating which regions of DNA interact spatially, a deeper and more complete understanding into the mechanisms causing cardiovascular disease can be achieved.
Methylation Sequencing (Methyl-seq)
Methylation of DNA is another epigenetic modification that can influence gene expression and the methylation status of DNA can be determined by Methyl-seq. 5-methyl cytosine is the most common modified base in humans and generally methylation of cytosine occurs when cytosine neighbors a guanine nucleotide called cytosine guanine dinucleotides (CpGs) 24. Areas of the genome high in CpG concentration have increased methyl transferase activity and may be referred to as CpG islands. Methylation at CpG islands decreases the activity of promoters and generally decreases gene expression. DNA methylation in CpG islands has been shown to strongly suppress promoter activity and seems to occur as a function of age, causing loss-of-function phenotypes, and may be a target in many disorders including heart disease 69. One method to sequence methylated regions of DNA (Methyl-seq) involves first isolating DNA, fragmenting the DNA, and then separating this sample into two reactions. One reaction is treated with bisulfite and the other portion is left untreated. Bisulfite treatment changes cytosine nucleotides to uracil nucleotides, leaving methylated-cytosine nucleotides unchanged because they are resistant to bisulfite treatment 70.
Methyl-seq data analysis involves sequencing bisulfite-treated DNA and comparing this with the fraction that did not undergo bisulfite treatment, producing differences that can identify the regions that did not convert to uracil; an uracil is “read” as a thymidine during sequencing and is reported as a methylation site. Incomplete bisulfate conversion can be problematic because these regions will be detected as a methylated site. One method to determine if bisulfate conversion went to completion is the addition of spike in control DNA in which the methylation status is known. Methylation rich regions can also be immunoprecipitated using an anti-methylated cytosine antibody. In a study by Movassagh et al.,71 immunoprecipitation of methylation regions led to genome-wide DNA methylation patterns that were quite similar between control and end-stage cardiomyopathy hearts. Moreover, differences in methylation could be observed when analyzing the methylation pattern at the single gene level. Identification of the gene promoters that are hypermethylated versus hypomethylated may therefore be useful in predicting which gene will become active during various stages of heart disease.
Transcriptome Sequencing (RNA-seq)
RNA-seq is particularly useful in assessing the current state of a cell or tissue as well as the possible effects of disease states or treatment conditions on the transcriptome. RNA-seq also provides information on the differences between the transcriptome and the exome that result from RNA editing. While microarrays have revolutionized the study of transcriptomics and proved useful in determining gene expression profiles, RNA-seq by comparison is more sensitive, provides absolute quantity levels, is not affected by on-chip sequence biases, and gives additional information in gene expression levels and splice junction variants 72, 73.
In RNA-seq, RNA is commonly first converted to a more stable complementary DNA (cDNA) through a combination of reverse transcription and the selection process to isolate the RNA from the abundant rRNA. The input RNA quality is very important in RNA-seq preparation because RNAse enzymes are ubiquitous and extremely stable and fragmentation can also occur simply when a divalent cation is present. Library preparation and sequencing of cDNA follows the same sequencing procedure as DNA-seq. However, numerous variations of RNA-seq library preparations have been developed, each with its benefits and limitations in terms of relative costs and input requirements. The main differences in these various library preparations are the methods of purifying and isolating RNA of interest (mRNA, uRNA, full length transcripts etc.). RNA-seq libraries can be made using polyadenylated tail selection, not-so-random primers (for reverse transcription), and ribosomal depletion 74–76. Isolating polyadenylated mRNA and then reverse transcribing is the conventional method of preparing an RNA-seq sample, but it favors the 3' end of transcripts and does not work well with low-quality or degraded samples or provide any information about non-coding RNA. A commercially available kit (Clontech SMARTer) is available to generate full-length cDNA from high-quality, low-input RNA samples by using the 3’ poly-A tail as the priming site for first strand cDNA synthesis and by enzymatically adding on a specific primer hybridization site on to the 5’ end after first strand synthesis for the second strand. Two other general methods have been commercially developed to selectively remove rRNA. For example, a method for selectively amplifying non-ribosomal RNA offered in a sequencing preparation kit (NuGEN Ovation) involves the use of a designed set of reverse transcription primers that contain all variants of random oligonucleotides (random primers), excluding the ones that would amplify ribosomal RNA (non-random primers). Ribosomal depletion immobilizes ribosomal RNA to remove it before reverse transcription. The use of one of these methods can recover additional RNA signals that would not be otherwise obtained via a poly-A selection due to the degraded and non-coding RNA. To annotate de novo discovery, strand specific RNA sequencing is used to determine which strand of RNA was the original template in reverse transcription. By pre-processing RNA to select for polyadenylated mRNA, or by selectively removing ribosomal RNA, a greater sequencing depth can be achieved. Depending on the experimental design, a greater sequencing depth may be required when complex genomes are being studied or if information on low abundant transcripts or splice variants is required.
In general, bioinformatic analysis consists of aligning the sequence reads to a reference genome, assembling the reads into transcripts, and detecting differences in transcript expression between or among groups. The Tuxedo Suite consisting of Bowtie 77, Tophat 78, and Cufflinks 79 can be used as open-source software packages to perform these operations and multiple updates to these software packages have increased the speed and accuracy in RNA-seq analysis. Additional splice variant detection and alternative exon usage can be identified using software packages such as MISO 80 and DEXseq 81, which can quantify reads to individual exons. While many of these software packages give a probabilistic framework to identify changes in transcript splicing patterns, false discovery cutoffs are necessary to identity true splicing events.
Transcriptome changes assessed by RNA-seq play a valuable role in cardiovascular medicine, because transcriptome changes can identify how cardiovascular diseases change with time. Lee et al. 82 used RNA-seq to study how murine hearts change during heart failure, whereas Song et al. 83 employed RNA-seq to decipher the transcriptome differences between physiological hypertrophy and pathological hypertrophy. In addition, Hu et al. 84 studied the mRNA and microRNA transcriptome changes that occur during pressure overloading hypertrophy in mice hearts by HTS. By identifying which mRNAs and microRNAs changed during hypertrophy, and which microRNA–mRNA interaction occurred with immunoprecipations of argonaute 2 RNA-Induced Silencing Complexes (termed RISC-seq), they demonstrated that small changes in microRNA expression could lead to global mRNA changes during heart stress. Other results show that the use of RNA-seq with ChIP-seq information has provided significant advances in the study of how transcription factor binding can influence changes in gene expression. To demonstrate the potential of this approach, RNA-seq was performed on the hearts from Tbx20 knockout mice that had rapidly developed heart failure. By combining the transcriptome changes that occur from the loss of Tbx20 with the putative transcription factor binding sites of Tbx20 previously identified with ChIP-seq 85, a comprehensive analysis of how the loss of Tbx20 leads to heart failure was achieved 86. In addition, combining ChIP-seq along with RNA-seq was also used to successfully identify genes and chromatin marks involved in the progression of cardiomyocyte differentiation from human induced pluripotent stem cells (iPSCs) 87.
Ribosomal Sequencing (Ribo-seq)
The RNA content within the cell does not automatically lead to the production of functional proteins. While the analysis of the total RNA can give an overview of the current RNA fragments present in the cell, selecting the RNA that is bound to ribosomes can offer a better indication of which RNA fragments are in the state of translation. Studying the sequence of RNA bound to ribosomes is called ribosomal footprinting and sequencing these short RNA fragments is called “Ribo-seq” 88, 89. Ribo-seq is particularly useful when studying transient transcriptional events that are tightly controlled, including mitosis 90. To perform Ribo-seq, cycloheximide treatment is first used to block the elongation phase of eukaryotic translation 91. Cells are lysed and fragmentation of RNA is performed (RNase I treatment). Ribosomes containing short fragments of RNA are then separated by ultracentrifugation and the short RNA fragments are separated from the ribosomes using proteases (proteinase K). These isolated short RNA fragments are sequenced to indicate which RNA fragments were being actively translated. While this technique can provide valuable information on how the cell’s translational machinery operates, it is also more technically challenging, and consequently only a few studies have used Ribo-seq to study the processes of cardiovascular diseases thus far.
The Future of Sequencing Technology
Some observers have compared the pace of advancements in DNA sequencing to that seen in the computer industry, which has been able to reduce costs and processing time exponentially since its inception. However, genetic sequencing is less mature as a field and faces many more technical challenges ahead. For instance, accuracy is paramount for sequencing to become more widely applicable in the clinic, a goal that may be amenable with better algorithms that can correct for reading errors and advanced molecular biology techniques and applications. One application that may prove useful in cardiovascular medicine is cell free DNA and RNA sequencing. Given that one of the components of circulating whole blood is cell free nucleic acids, sequencing these cell free nucleic acids may indicate the current state of the cardiovascular system. RNA is short lived in the presence of RNAses and this makes RNA present in the blood a good temporal measure of what is occurring in the body at the time of extraction. Therefore, DNA and RNA sequencing from cell free nucleic acids could prove to be a relatively noninvasive measure of cardiovascular health.
Recent advances have led to more precise control of picoliter scale volumes and chemical reactions. The next milestone may be to sequence unamplified, unmodified native nucleic acids. There are single molecule technologies such as the Pacific Bioscience RS, but they do require adaptor ligation to modify the sample before sequencing can take place. The future direction of sequencing technologies will likely involve methods of directly sequencing single molecules of DNA or RNA in native form from low input starting material (e.g., a few cells containing approximately 50 pg of DNA/cell), without sacrificing accuracy or cost 92. Automated microfluidic sample prep methods are being developed that can isolate a single cell’s genetic material and process it into a sequencing library all in one closed system.
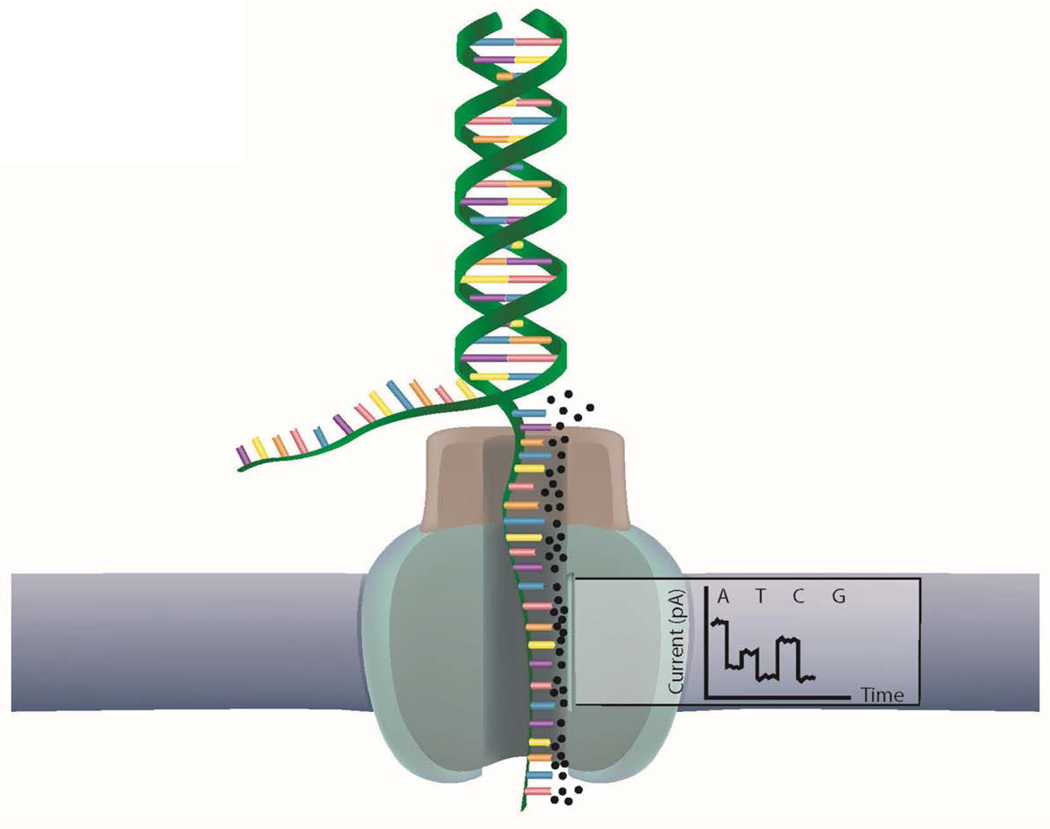
Nanopore Sequencing Technologies
Oxford Nanopore (http://www.nanoporetech.com/) is developing a nanopore technology that someday may be capable of sequencing unmodified miRNA and mRNA molecules. Nanopore sequencing technologies comprise a relatively newer set of techniques being developed by companies such as Oxford Nanopore and NABsys (http://www.nabsys.com/), which are working on massively parallel sequencers not based on sequencing by synthesis. The concept is to electrophorese molecules through a pore of a membrane and then measure the electrical current through the pore as molecules pass through 93 (Figure 5). By characterizing the current of a pore over time, nanopore technology may be able to determine exactly what has traversed the pore and in which order. These companies hope to develop products that can determine the entire genome, sense and antisense, from small amounts of unmodified input in a short amount of time 94. This technology will also be used to measure RNA and proteins directly someday, and a major challenge will be to control the flow of DNA through the pore and to decipher the resulting message 95, 96. To tackle this problem, Oxford Nanopore is using an approach in which an exonuclease releases one nucleotide at a time, whereas NABsys has adopted an approach using a series of oligonucleotide probes hybridized to the denatured sample that subsequently can be "seen" crossing the pore and positioning along the template molecule.
Figure 5. Nanopore technology.
Third generation sequencing is expected to measure the change in ion flow (current) within a membrane as small molecules are passed through a small pore inside the membrane. Different current profiles will therefore indicate which nucleotide passed through and in which order.
Conclusion
In summary, advances in HTS technologies are enabling a more accurate and comprehensive representation of cardiac development and disease processes. While more researchers are using HTS to study cardiovascular medicine, the full potential of current HTS sequencing platforms in cardiovascular medicine has yet to be realized. HTS will be essential in identifying biomarkers of disease, staging disease progression, and linking genotypic to phenotypic outcomes. Given the rapid pace of development in sequencing technology during the last decade, future sequencing technologies promise to further help us understand the roles that the genome, transcriptome, and proteome play in the cell by identifying cellular mechanisms. This may also lead to deeper and more comprehensive insights into disease mechanisms at a subcellular level, possibly connecting causation effects to gene expression levels. As different platforms improve depth by increasing in read length and decreasing in sequencing time, and decrease in cost, improvements in HTS applications will provide a more complete molecular picture into the functionality of biological processes. Ultimately, improved understanding of these biological processes may lead to dramatically safer and more effective therapies for cardiovascular diseases.
Acknowledgments
Sources of funding
We gratefully acknowledge funding support from Burroughs Wellcome Foundation, NIH New Innovator Award DP2OD004437, R01 HL113006, U01 HL099776 (JCW), and P01 GM099130 (MPS).
Nonstandard Abbreviations and Acronyms
- ARVD/C
arrhythmogenic right ventricular dysplasia/cardiomyopathy
- CCS
circular consensus sequencing
- cDNA
complementary DNA
- ChIP-seq
chromatin immunoprecipitation sequencing
- CLCNKA
chloride channel, voltage-sensitive Ka
- CNV
copy number variants
- Contig
large contiguous regions
- CpGs
cytosine guanine dinucleotides
- CYP2C9
cytochrome P450, family 2, subfamily C, polypeptide 9
- ddNTPs
2′,3′-dideoxynucleotides
- dNTPs
2′-deoxynucleotides
- ENCODE
ENCyclopedia Of DNA Elements
- HSPB7
heat shock protein family, member 7
- HTS
high throughput sequencing
- iPSCs
induced pluripotent stem cells
- Kcnq1
potassium voltage-gated channel, KQT-like subfamily, member 1
- MB
megabase
- Methyl-seq
methylation sequencing
- NKX2-5
NK2 homeobox 5
- Ribo-seq
ribosome sequencing
- RISC-seq
RNA-Induced Silencing Complex sequencing
- RNA-seq
RNA sequencing
- RYR2
Ryanodine receptor 2
- SNP
single-nucleotide polymorphisms
- SNV
single-nucleotide variants
- SMRT
single-molecule real-time
- TGFβ3
Transforming growth factor beta 3
- TMEM43
Transmembrane protein 43
- KORC1
vitamin K epoxide reductase complex, subunit 1
- ZMW
Zero Mode Waveguides
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 2.Temin HM. Reverse transcription in the eukaryotic genome: Retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol Biol Evol. 1985;2:455–468. doi: 10.1093/oxfordjournals.molbev.a040365. [DOI] [PubMed] [Google Scholar]
- 3.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, Khatun J, Lajoie BR, Landt SG, Lee BK, Pauli F, Rosenbloom KR, Sabo P, Safi A, Sanyal A, Shoresh N, Simon JM, Song L, Trinklein ND, Altshuler RC, Birney E, Brown JB, Cheng C, Djebali S, Dong X, Ernst J, Furey TS, Gerstein M, Giardine B, Greven M, Hardison RC, Harris RS, Herrero J, Hoffman MM, Iyer S, Kelllis M, Kheradpour P, Lassman T, Li Q, Lin X, Marinov GK, Merkel A, Mortazavi A, Parker SC, Reddy TE, Rozowsky J, Schlesinger F, Thurman RE, Wang J, Ward LD, Whitfield TW, Wilder SP, Wu W, Xi HS, Yip KY, Zhuang J, Bernstein BE, Green ED, Gunter C, Snyder M, Pazin MJ, Lowdon RF, Dillon LA, Adams LB, Kelly CJ, Zhang J, Wexler JR, Good PJ, Feingold EA, Crawford GE, Dekker J, Elinitski L, Farnham PJ, Giddings MC, Gingeras TR, Guigo R, Hubbard TJ, Kellis M, Kent WJ, Lieb JD, Margulies EH, Myers RM, Starnatoyannopoulos JA, Tennebaum SA, Weng Z, White KP, Wold B, Yu Y, Wrobel J, Risk BA, Gunawardena HP, Kuiper HC, Maier CW, Xie L, Chen X, Mikkelsen TS, Gillespie S, Goren A, Ram O, Zhang X, Wang L, Issner R, Coyne MJ, Durham T, Ku M, Truong T, Eaton ML, Dobin A, Lassmann T, Tanzer A, Lagarde J, Lin W, Xue C, Williams BA, Zaleski C, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Batut P, Bell I, Bell K, Chakrabortty S, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Li G, Luo OJ, Park E, Preall JB, Presaud K, Ribeca P, Robyr D, Ruan X, Sammeth M, Sandu KS, Schaeffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Hayashizaki Y, Reymond A, Antonarakis SE, Hannon GJ, Ruan Y, Carninci P, Sloan CA, Learned K, Malladi VS, Wong MC, Barber GP, Cline MS, Dreszer TR, Heitner SG, Karolchik D, Kirkup VM, Meyer LR, Long JC, Maddren M, Raney BJ, Grasfeder LL, Giresi PG, Battenhouse A, Sheffield NC, Showers KA, London D, Bhinge AA, Shestak C, Schaner MR, Kim SK, Zhang ZZ, Mieczkowski PA, Mieczkowska JO, Liu Z, McDaniell RM, Ni Y, Rashid NU, Kim MJ, Adar S, Zhang Z, Wang T, Winter D, Keefe D, Iyer VR, Sandhu KS, Zheng M, Wang P, Gertz J, Vielmetter J, Partridge EC, Varley KE, Gasper C, Bansal A, Pepke S, Jain P, Amrhein H, Bowling KM, Anaya M, Cross MK, Muratet MA, Newberry KM, McCue K, Nesmith AS, Fisher-Aylor KI, Pusey B, DeSalvo G, Parker SL, Balasubramanian S, Davis NS, Meadows SK, Eggleston T, Newberry JS, Levy SE, Absher DM, Wong WH, Blow MJ, Visel A, Pennachio LA, Elnitski L, Petrykowska HM, Abyzov A, Aken B, Barrell D, Barson G, Berry A, Bignell A, Boychenko V, Bussotti G, Davidson C, Despacio-Reyes G, Diekhans M, Ezkurdia I, Frankish A, Gilbert J, Gonzalez JM, Griffiths E, Harte R, Hendrix DA, Hunt T, Jungreis I, Kay M, Khurana E, Leng J, Lin MF, Loveland J, Lu Z, Manthravadi D, Mariotti M, Mudge J, Mukherjee G, Notredame C, Pei B, Rodriguez JM, Saunders G, Sboner A, Searle S, Sisu C, Snow C, Steward C, Tapanan E, Tress ML, van Baren MJ, Washieti S, Wilming L, Zadissa A, Zhengdong Z, Brent M, Haussler D, Valencia A, Raymond A, Addleman N, Alexander RP, Auerbach RK, Bettinger K, Bhardwaj N, Boyle AP, Cao AR, Cayting P, Charos A, Cheng Y, Eastman C, Euskirchen G, Fleming JD, Grubert F, Habegger L, Hariharan M, Harmanci A, Iyenger S, Jin VX, Karczewski KJ, Kasowski M, Lacroute P, Lam H, Larnarre-Vincent N, Lian J, Lindahl-Allen M, Min R, Miotto B, Monahan H, Moqtaderi Z, Mu XJ, O'Geen H, Ouyang Z, Patacsil D, Raha D, Ramirez L, Reed B, Shi M, Slifer T, Witt H, Wu L, Xu X, Yan KK, Yang X, Struhl K, Weissman SM, Tenebaum SA, Penalva LO, Karmakar S, Bhanvadia RR, Choudhury A, Domanus M, Ma L, Moran J, Victorsen A, Auer T, Centarin L, Eichenlaub M, Gruhl F, Heerman S, Hoeckendorf B, Inoue D, Kellner T, Kirchmaier S, Mueller C, Reinhardt R, Schertel L, Schneider S, Sinn R, Wittbrodt B, Wittbrodt J, Jain G, Balasundaram G, Bates DL, Byron R, Canfield TK, Diegel MJ, Dunn D, Ebersol AK, Frum T, Garg K, Gist E, Hansen RS, Boatman L, Haugen E, Humbert R, Johnson AK, Johnson EM, Kutyavin TM, Lee K, Lotakis D, Maurano MT, Neph SJ, Neri FV, Nguyen ED, Qu H, Reynolds AP, Roach V, Rynes E, Sanchez ME, Sandstrom RS, Shafer AO, Stergachis AB, Thomas S, Vernot B, Vierstra J, Vong S, Weaver MA, Yan Y, Zhang M, Akey JA, Bender M, Dorschner MO, Groudine M, MacCoss MJ, Navas P, Stamatoyannopoulos G, Stamatoyannopoulos JA, Beal K, Brazma A, Flicek P, Johnson N, Lukk M, Luscombe NM, Sobral D, Vaquerizas JM, Batzoglou S, Sidow A, Hussami N, Kyriazopoulou-Panagiotopoulou S, Libbrecht MW, Schaub MA, Miller W, Bickel PJ, Banfai B, Boley NP, Huang H, Li JJ, Noble WS, Bilmes JA, Buske OJ, Sahu AO, Kharchenko PV, Park PJ, Baker D, Taylor J, Lochovsky L. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 8.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolata GB. The 1980 nobel prize in chemistry. Science. 1980;210:887–889. doi: 10.1126/science.7001629. [DOI] [PubMed] [Google Scholar]
- 10.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 11.Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in chip-seq peak detection. PLoS One. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karger BL, Guttman A. DNA sequencing by ce. Electrophoresis. 2009;30(Suppl 1):S196–S202. doi: 10.1002/elps.200900218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kircher M, Kelso J. High-throughput DNA sequencing--concepts and limitations. Bioessays. 2010;32:524–536. doi: 10.1002/bies.200900181. [DOI] [PubMed] [Google Scholar]
- 14.Stein LD. Human genome: End of the beginning. Nature. 2004;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 16.Consortium IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 17.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 18.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 19.Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: Comparison of ion torrent, pacific biosciences and illumina miseq sequencers. BMC Genomics. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minoche AE, Dohm JC, Himmelbauer H. Evaluation of genomic high-throughput sequencing data generated on illumina hiseq and genome analyzer systems. Genome Biol. 2011;12:R112. doi: 10.1186/gb-2011-12-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 22.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, Dewinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 23.Ziegler A, Koch A, Krockenberger K, Grosshennig A. Personalized medicine using DNA biomarkers: A review. Hum Genet. 2012;131:1627–1638. doi: 10.1007/s00439-012-1188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabory A, Attig L, Junien C. Developmental programming and epigenetics. Am J Clin Nutr. 2011;94:1943S–1952S. doi: 10.3945/ajcn.110.000927. [DOI] [PubMed] [Google Scholar]
- 26.Chou J, Ohsumi TK, Geha RS. Use of whole exome and genome sequencing in the identification of genetic causes of primary immunodeficiencies. Curr Opin Allergy Clin Immunol. 2012;12:623–628. doi: 10.1097/ACI.0b013e3283588ca6. [DOI] [PubMed] [Google Scholar]
- 27.Snyder M, Du J, Gerstein M. Personal genome sequencing: Current approaches and challenges. Genes Dev. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alex Buerkle C, Gompert Z. Population genomics based on low coverage sequencing: How low should we go? Mol Ecol. 2012 doi: 10.1111/mec.12105. [DOI] [PubMed] [Google Scholar]
- 29.Mir KU. Sequencing genomes: From individuals to populations. Brief Funct Genomic Proteomic. 2009;8:367–378. doi: 10.1093/bfgp/elp040. [DOI] [PubMed] [Google Scholar]
- 30.Koboldt DC, Ding L, Mardis ER, Wilson RK. Challenges of sequencing human genomes. Brief Bioinform. 2010;11:484–498. doi: 10.1093/bib/bbq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ajay SS, Parker SC, Abaan HO, Fajardo KV, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21:1498–1505. doi: 10.1101/gr.123638.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam HY, Clark MJ, Chen R, Natsoulis G, O'Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ, Ji HP, Snyder M. Performance comparison of whole-genome sequencing platforms. Nat Biotechnol. 2012;30:78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, O'Connor BD, Merriman B, Funari VA, Homer N, Chen Z, Cohn DH, Nelson SF. Improving the efficiency of genomic loci capture using oligonucleotide arrays for high throughput resequencing. BMC Genomics. 2009;10:646. doi: 10.1186/1471-2164-10-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci U S A. 2008;105:9296–9301. doi: 10.1073/pnas.0803240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu CM, Wong T, Wu E, Luo R, Yiu SM, Li Y, Wang B, Yu C, Chu X, Zhao K, Li R, Lam TW. Soap3: Ultra-fast gpu-based parallel alignment tool for short reads. Bioinformatics. 2012;28:878–879. doi: 10.1093/bioinformatics/bts061. [DOI] [PubMed] [Google Scholar]
- 38.Homer N, Merriman B, Nelson SF. Bfast: An alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and samtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lombardi R, Marian AJ. Molecular genetics and pathogenesis of arrhythmogenic right ventricular cardiomyopathy: A disease of cardiac stem cells. Pediatr Cardiol. 2011;32:360–365. doi: 10.1007/s00246-011-9890-2. [DOI] [PubMed] [Google Scholar]
- 43.Murray B. Arrhythmogenic right ventricular dysplasia/cardiomyopathy (arvd/c): A review of molecular and clinical literature. J Genet Couns. 2012;21:494–504. doi: 10.1007/s10897-012-9497-7. [DOI] [PubMed] [Google Scholar]
- 44.Ohe T. Idiopathic ventricular fibrillation of the brugada type: An atypical form of arrhythmogenic right ventricular cardiomyopathy? Intern Med. 1996;35:595. doi: 10.2169/internalmedicine.35.595. [DOI] [PubMed] [Google Scholar]
- 45.Al-Aama JY, Bondagji NS, El-Harouni AA. Congenital heart defects in down syndrome patients from western saudi arabia. Saudi Med J. 2012;33:1211–1215. [PubMed] [Google Scholar]
- 46.Kobayashi D, Sallaam S, Humes RA. Tetralogy of fallot with complete digeorge syndrome: Report of a case and a review of the literature. Congenit Heart Dis. 2012 doi: 10.1111/j.1747-0803.2012.00694.x. [DOI] [PubMed] [Google Scholar]
- 47.Strehle EM, Gruszfeld D, Schenk D, Mehta SG, Simonic I, Huang T. The spectrum of 4q- syndrome illustrated by a case series. Gene. 2012;506:387–391. doi: 10.1016/j.gene.2012.06.087. [DOI] [PubMed] [Google Scholar]
- 48.Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA, Kang SH. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2009;149A:1661–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, Gonzalez JR, Gratacos M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 51.Matkovich SJ, Van Booven DJ, Hindes A, Kang MY, Druley TE, Vallania FL, Mitra RD, Reilly MP, Cappola TP, Dorn GW., 2nd Cardiac signaling genes exhibit unexpected sequence diversity in sporadic cardiomyopathy, revealing hspb7 polymorphisms associated with disease. J Clin Invest. 2010;120:280–289. doi: 10.1172/JCI39085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cappola TP, Matkovich SJ, Wang W, van Booven D, Li M, Wang X, Qu L, Sweitzer NK, Fang JC, Reilly MP, Hakonarson H, Nerbonne JM, Dorn GW., 2nd Loss-of-function DNA sequence variant in the clcnka chloride channel implicates the cardio-renal axis in interindividual heart failure risk variation. Proc Natl Acad Sci U S A. 2011;108:2456–2461. doi: 10.1073/pnas.1017494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poe BL, Haverstick DM, Landers JP. Warfarin genotyping in a single pcr reaction for microchip electrophoresis. Clin Chem. 2012;58:725–731. doi: 10.1373/clinchem.2011.180356. [DOI] [PubMed] [Google Scholar]
- 55.Jamieson D, Cresti N, Bray J, Sludden J, Griffin MJ, Hawsawi NM, Famie E, Mould EV, Verrill MW, May FE, Boddy AV. Two minor nqo1 and nqo2 alleles predict poor response of breast cancer patients to adjuvant doxorubicin and cyclophosphamide therapy. Pharmacogenet Genomics. 2011;21:808–819. doi: 10.1097/FPC.0b013e32834b6918. [DOI] [PubMed] [Google Scholar]
- 56.Bains OS, Karkling MJ, Grigliatti TA, Reid RE, Riggs KW. Two nonsynonymous single nucleotide polymorphisms of human carbonyl reductase 1 demonstrate reduced in vitro metabolism of daunorubicin and doxorubicin. Drug Metab Dispos. 2009;37:1107–1114. doi: 10.1124/dmd.108.024711. [DOI] [PubMed] [Google Scholar]
- 57.Jamieson D, Boddy AV. Pharmacogenetics of genes across the doxorubicin pathway. Expert Opin Drug Metab Toxicol. 2011;7:1201–1210. doi: 10.1517/17425255.2011.610180. [DOI] [PubMed] [Google Scholar]
- 58.Wells QS, Delaney JT, Roden DM. Genetic determinants of response to cardiovascular drugs. Curr Opin Cardiol. 2012;27:253–261. doi: 10.1097/HCO.0b013e32835220e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 60.Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA. Identification of higher-order functional domains in the human encode regions. Genome Res. 2007;17:917–927. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3k27me3 forms blocs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattout A, Biran A, Meshorer E. Global epigenetic changes during somatic cell reprogramming to ips cells. J Mol Cell Biol. 2011;3:341–350. doi: 10.1093/jmcb/mjr028. [DOI] [PubMed] [Google Scholar]
- 63.Furey TS. Chip-seq and beyond: New and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. Chip-seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, Feneley M, Harvey RP. Cardiac homeobox gene nkx2-5 mutations and congenital heart disease: Associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41:2072–2076. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- 66.Tanizawa H, Noma K. Unravelling global genome organization by 3c–seq. Semin Cell Dev Biol. 2012;23:213–221. doi: 10.1016/j.semcdb.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 68.Korostowski L, Raval A, Breuer G, Engel N. Enhancer-driven chromatin interactions during development promote escape from silencing by a long non-coding rna. Epigenetics Chromatin. 2011;4:21. doi: 10.1186/1756-8935-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Friso S, Lotto V, Choi SW, Girelli D, Pinotti M, Guarini P, Udali S, Pattini P, Pizzolo F, Martinelli N, Corrocher R, Bernardi F, Olivieri O. Promoter methylation in coagulation f7 gene influences plasma fvii concentrations and relates to coronary artery disease. J Med Genet. 2012;49:192–199. doi: 10.1136/jmedgenet-2011-100195. [DOI] [PubMed] [Google Scholar]
- 70.Krueger F, Kreck B, Franke A, Andrews SR. DNA methylome analysis using short bisulfite sequencing data. Nat Methods. 2012;9:145–151. doi: 10.1038/nmeth.1828. [DOI] [PubMed] [Google Scholar]
- 71.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, Siggens L, Vujic A, Simeoni I, Penkett C, Goddard M, Lio P, Bennett MR, Foo RS. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Labaj PP, Leparc GG, Linggi BE, Markillie LM, Wiley HS, Kreil DP. Characterization and improvement of rna-seq precision in quantitative transcript expression profiling. Bioinformatics. 2011;27:i383–i391. doi: 10.1093/bioinformatics/btr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matkovich SJ, Zhang Y, Van Booven DJ, Dorn GW., 2nd Deep mrna sequencing for in vivo functional analysis of cardiac transcriptional regulators: Application to galphaq. Circ Res. 2010;106:1459–1467. doi: 10.1161/CIRCRESAHA.110.217513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoeijmakers WA, Bartfai R, Stunnenberg HG. Transcriptome analysis using rna-seq. Methods Mol Biol. 2013;923:221–239. doi: 10.1007/978-1-62703-026-7_15. [DOI] [PubMed] [Google Scholar]
- 75.Sanchez-Pla A, Reverter F, Ruiz de Villa MC, Comabella M. Transcriptomics: Mrna and alternative splicing. J Neuroimmunol. 2012;248:23–31. doi: 10.1016/j.jneuroim.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 76.Raz T, Causey M, Jones DR, Kieu A, Letovsky S, Lipson D, Thayer E, Thompson JF, Milos PM. Rna sequencing and quantitation using the helicos genetic analysis system. Methods Mol Biol. 2011;733:37–49. doi: 10.1007/978-1-61779-089-8_3. [DOI] [PubMed] [Google Scholar]
- 77.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trapnell C, Pachter L, Salzberg SL. Tophat: Discovering splice junctions with rna-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of rna sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anders S, Reyes A, Huber W. Detecting differential usage of exons from rna-seq data. Genome Res. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee JH, Gao C, Peng G, Greer C, Ren S, Wang Y, Xiao X. Analysis of transcriptome complexity through rna sequencing in normal and failing murine hearts. Circ Res. 2011;109:1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song HK, Hong SE, Kim T, Kim do H. Deep rna sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS One. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu Y, Matkovich SJ, Hecker PA, Zhang Y, Edwards JR, Dorn GW., 2nd Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac micrornas. Proc Natl Acad Sci U S A. 2012;109:19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, Boogerd CJ, Cai CL, Peterson K, Chen J, Nobrega MA, Evans SM. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakabe NJ, Aneas I, Shen T, Shokri L, Park SY, Bulyk ML, Evans SM, Nobrega MA. Dual transcriptional activator and repressor roles of tbx20 regulate adult cardiac structure and function. Hum Mol Genet. 2012;21:2194–2204. doi: 10.1093/hmg/dds034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- 89.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian micrornas predominantly act to decrease target mrna levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kalisky T, Blainey P, Quake SR. Genomic analysis at the single-cell level. Annu Rev Genet. 2011;45:431–445. doi: 10.1146/annurev-genet-102209-163607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thompson JF, Oliver JS. Mapping and sequencing DNA using nanopores and nanodetectors. Electrophoresis. 2012;33:3429–3436. doi: 10.1002/elps.201200136. [DOI] [PubMed] [Google Scholar]
- 94.Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boersma AJ, Brain KL, Bayley H. Real-time stochastic detection of multiple neurotransmitters with a protein nanopore. ACS Nano. 2012;6:5304–5308. doi: 10.1021/nn301125y. [DOI] [PubMed] [Google Scholar]
- 96.Ayub M, Bayley H. Individual rna base recognition in immobilized oligonucleotides using a protein nanopore. Nano Lett. 2012;12:5637–5643. doi: 10.1021/nl3027873. [DOI] [PMC free article] [PubMed] [Google Scholar]