Abstract
Antibody-overlay lectin microarray (ALM) has been used for targeted glycan profiling to identify disease-related protein glycoforms. In this context, high sensitivity is desired because it allows for the identification of disease-related glycoforms that are often present at low concentration. We describe a new Tyramide Signal Amplification (TSA) for Antibody-overlay Lectin Microarray procedure for sensitive profiling of glycosylation patterns. We demonstrated that TSA increased the sensitivity of the microarray over 100 times for glycan profiling using the model protein Prostate Specific Antigen (PSA). The glycan profile of PSA enriched from LNCAP cells, obtained at a sub-nanogram level with the aid of TSA, was consistent with the previous reports. We also established the glycan profile of Prostate Specific Membrane Antigen (PSMA) using the TSA and ALM. Thus, the Tyramide Signal Amplification for Antibody-overlay Lectin Microarray is a sensitive, rapid, comprehensive, and high-throughput method for targeted glycan profiling and can potentially be used for the identification of disease-related protein glycoforms.
Introduction
Changes in glycan structures are hallmarks of cancer. This reflects cancer-specific changes in glycan biosynthesis pathways which includes expressions of glycosyltransferases and glycosidases1–5. The increased activity of sialyltransferases leading to over-expression of sialylated glycans (e.g., sialyl Lewis x, sialyl Tn, Globo H, Lewis Y, and polysialic acid) have been demonstrated in malignant tissues throughout the body, including brain, breast, colon, and prostate6–12. Increased β1-6 branching of N-glycans resulting from the enhanced expression of UDP-GlcNAc:N-glycan GlcNAc transferase V (GlcNAcT-V) have also been strongly correlated with metastatic potential of cancer cells13. These observations suggest that glycoproteins accompanying substantial tumor-specific structural changes in glycan moieties may be used as cancer biomarkers to improve specificity. In this context, antibody-overlay lectin microarray has been used for the analysis of a target glycoprotein to identify disease-related protein glycoforms14. A target glycoprotein, often enriched from crude samples by immunoprecipitation, is incubated with lectins on the array. A glycan profile of this protein is acquired with the aid of a specific antibody against the protein and the quantitative detection of the antibody.
Antibody-overlay lectin microarray allows for rapid, comprehensive, and high-throughput profiling of complex glycans of a target glycoprotein. However, sensitivity may be an issue when only a small amount of the target glycoprotein (less than 20 ng) is available. Low sensitivity is largely due to the weak lectin-glycan interaction (dissociation constant, Kd > 10−6 M)15. This could lead to the missing identification of some disease-related protein glycoforms. To increase the sensitivity, Kuno et al. developed an evanescent-field fluorescence-assisted lectin microarray for in situ detection of lectin-glycan interactions under equilibrium conditions14–15. Although this procedure has an advantage of real-time detection of weak lectin-glycan interactions, it requires a specialized evanescent-field fluorescence scanner that may not be readily available. Here, we describe an alternative Tyramide Signal Amplification (TSA) for Antibody-overlay Lectin Microarray (TSA-ALM) to increase the sensitivity of glycan profiling. TSA is a horseradish peroxidase (HRP)-mediated signal amplification method often used in immunohistochemistry and in situ hybridization protocols, but has not been applied for lectin microarray. TSA does not require specialized instruments and can be easily integrated into the workflow of antibody-overlay lectin microarray.
Methods
Reagents and Cell Culture
Lectins (supplementary Table 1) were purchased from EY Laboratories (San Mateo, CA) and Vector Labs (Burlingame, CA). Stock solutions of these lectins were prepared in PBS buffer at a concentration of 1, 2, or 5 mg/mL. Aliquots of 240 µL of these lectins were placed into a dry ice bath (snap frozen), and then immediately stored at −80° C up to a year without loss of activity. Human seminal fluid PSA (100% free PSA) was from Lee Biosolutions, Inc. (St. Louis, MO). Recombinant Human PSMA (rhPSMA) produced by a Chinese Hamster Ovary (CHO) cell line was purchased from R&D Systems (Minneapolis, MN, Catalog#4234-ZN). Sulfo-NHA-LC biotin, Dylight amine-reactive fluor 594, and Dylight 549 conjugated streptavidin were from Thermo Scientific (Rockford, IL). Tyramide Signal Amplification (TSA) Biotin kit was from PerkinElmer (Shelton, CT). Mouse anti-PSA monoclonal antibody (Clone BP001) was from Scripps Laboratory (San Diego, CA). Mouse anti-PSMA monoclonal antibody (J-591) was kindly provided by Dr. Neil H Bander from Cornell University. Bovine serum albumin (BSA) was purchased from Roche Diagnostics (Indianapolis, IN). PSA and PSMA antibodies and BSA were biotinylated using the Sulfo-NHA-LC biotin following the manufacturer’s instructions. Mouse polyclonal IgG and ethanolamine were from Sigma (St. Louis, MO). LNCAP cell line (Clone FGC) was purchased from ATCC (Manassas, VA) and cultured according the manufacturer’s instructions. α1-2 fucosidase was purchased from the New England BioLabs (Ipswich, MA).
Lectin Microarray Fabrication and Printing Quality Check
We fabricated 38-lectin microarrays with varying density following the procedures by Hus et al. 16 with modifications. The printing buffer contained PBS with 0.01% Tween 20. Each lectin was prepared in three concentrations (1, 0.5, and 0.25 mg/mL) using the printing buffer. We also included BSA, Cy5-labeled BSA, and 5 concentrations of biotinylated BSA (0.4, 1.2, 3.6, 10.8, 32.4 µg/mL) as controls. One hundred and fourteen lectin solutions (n=38×3) and controls were loaded into 384-well plates in a desired order, and spotted on Nexterion H slides (Schott, Jena, Germany) using a BioRobotics MicroGrid II 600 arrayer (Digilab, Holliston, MA) in triplicates. Twelve identical 19×20 arrays were printed on each slide. After printing, the slides were left in the printing chamber overnight at 50% relative humidity to ensure maximum coupling efficiency before being stored at −20° C for future use.
To monitor the quality of spotting, the first and last slides of each printing cycle were selected for protein staining by incubating them at room temperature (RT) for 1 hour with Dylight amine-reactive fluor 594 diluted 100 times in PBS. Before the staining, 50mM ethanolamine in 40mM sodium borate buffer (pH 8.5) was used to block the slides at RT for 1 hour. Then the slides were washed three times using PBST 0.1 (PBS + 0.1% Tween 20) and one time using water, and dried by centrifugation at 200 rpm for 5 mins. After the staining, the slides were also washed and dried as previously described and scanned with a GenePix 4100B scanner (Sunnyvale, CA) at 10µm resolution, 350 mV laser power and 100% PMT value in the Cy3 channel, and 450 mV laser power and 100%PMT value in the Cy5 channel. These same scanning conditions were used for all other slides in this study.
Immunoprecipitation of LNCAP PSA
We incubated 100 µL of paramagnetic beads coated with mouse monoclonal anti-PSA antibody (Beckman Coulter, Brea, CA) with concentrated LNCAP cell culture supernatant at RT for 4 hours. Then the beads were washed 3 times with 500 µL PBST 0.1, and the bound PSA was eluted using 100 µL of 100mM Glycine, pH 2.7, overnight at 4°C. The eluate was then neutralized using 38 µL of 1M Tris-HCl, pH 8.0, before measuring the PSA concentration using the Beckman Hybritech PSA assay. PSA concentrations of all the eluate from the tissue specimens were normalized to 100 ng/mL using PBST0.1 buffer before analyses by the lectin microarray.
Tyramide Signal Amplification for Antibody-overlay Lectin Microarray
The 12 microarrays on the slide were divided by placing a Whatman® FAST slide with 16 wells on the top of the slide and assembled into a FAST Frame. Sample (e.g., seminal PSA, enriched PSA from LNCAP cells, rbPSMA) of 100 µL in volume was applied to each lectin microarray. After overnight incubation at RT, 20 µg of mouse polyclonal IgG was added to each array followed by 30-mins incubation. This step was to mask the residual lectin sites to eliminate binding to the biotinylated PSA antibody. Then the incubation solution was discarded, and the slide was washed three times with PBST0.1. A 100 µL 4 µg/mL of biotinylated antibody (e.g., PSA or PSMA) in PBST0.1 was applied to the array and incubated at RT for 1h. The array was washed 3 times with PBST0.1, and 100 µL of streptavidin conjugated Dylight 549 (diluted 500 times in PBST0.1) was added to the array and incubated at RT for 30 mins before the array was washed, dried, and scanned.
With TSA, however, additional steps were performed according to the manufacturer’s instruction with modification, after antibody incubation. First, the array was washed three times with TNT wash buffer (100 mM Tris-HCl, 150 mM NaCl, and 0.05% Tween 20, pH 7.4). Second, 100 µL of TNB blocking buffer (100 mM Tris-HCl, 150 mM NaCl, and 0.5% Blocking Reagent supplied in the TSA kit) was incubated with the array for 30 mins. Third, 100 µL of 11 µg/mL unlabeled horseradish peroxidase (HRP) in the TNB buffer was incubated with the array for 30 mins and then washed 2 times using the TNT buffer. Fourth, 100 µL of 3% H2O2 in the TNB buffer was incubated with the array for 10 minutes and washed 2 times using the TNT buffer. Then, 100 µL of streptavidin-conjugated HRP (in the TSA kit), diluted 400 times in the TNB buffer, were incubated with the array for 30 mins, followed by 3 washes using the TNT buffer and incubation of 50 µL of biotin-tyramide (diluted in the amplification buffer with the TSA kit) for 5 mins. Last, the array was washed 3 times before incubation of streptavidin-conjugated Dylight 549, as described without TSA, before scanning.
Data Analysis
Lectins on the arrays were stained using the amine-reactive Dylight 594 to check printing quality. Total Coefficient of Variation (CV) of labeled fluorescent intensities across all 24 arrays on the first and last slides of each batch (n=72) was calculated for each lectin at the highest density (1 mg/mL). All the lectins, except PWM, bound to the Nexterion H slides under the printing condition as described. Reproducibility of these arrays in glycosylation profiling of PSA was evaluated using 100 ng/mL of immunoprecipitated LNCAP PSA by total CV of the fluorescent intensities of lectins at the highest density across 12 arrays within one slide. Fluorescent intensities from biotinylated BSA were compared among arrays to normalize the variation of protein printing.
Results and Discussion
TSA increases sensitivity of ALM for glycan profiling
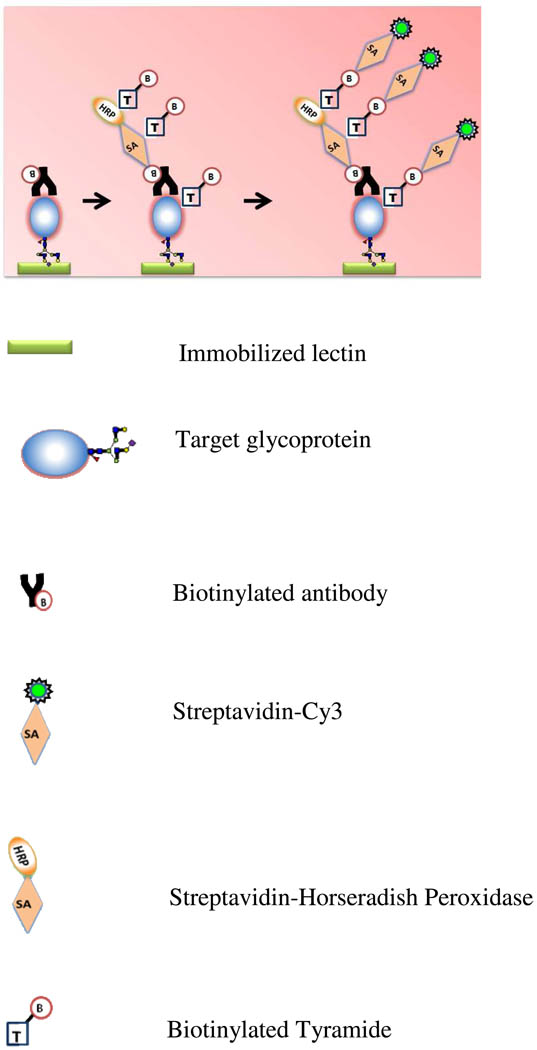
We developed a new TSA-ALM procedure to increase the sensitivity of targeted glycan profiling. Figure 1 shows the principle of detection of the target glycoprotein on lectin microarray using TSA. After incubation of the biotinylated detection antibody, the array is incubated with streptavidin-labeled HRP (SA-HRP). The enzyme portion of the SA-HRP catalyzes localized deposition of biotin tyramide, resulting in higher levels of biotin which will be detected by streptavidin-labeled Cy3.
Figure 1.
The principle of detection of a target glycoprotein on antibody-overlay lectin microarray using TSA. After incubation of the biotinylated detection antibody, the array is incubated with streptavidin-labeled HRP (SA-HRP). The enzyme portion of the SA-HRP catalyzes localized deposition of biotin tyramide, resulting in higher levels of biotin signals that will be detected by streptavidin-labeled Cy3 (SA-Cy3).
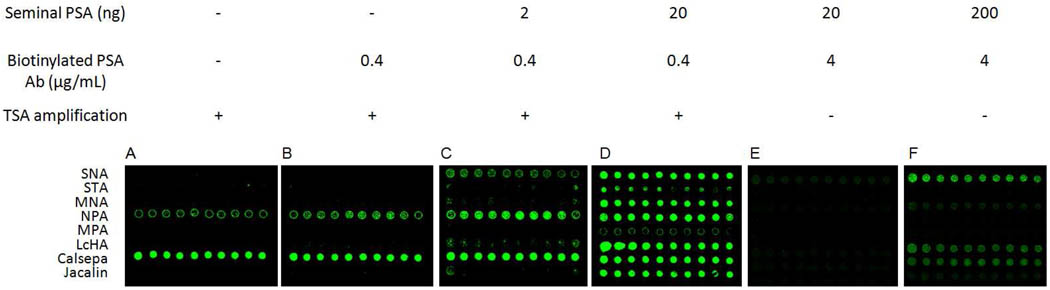
We compared the sensitivity of lectin microarray for PSA purified from human seminal fluid (sfPSA) with TSA to that without TSA. As a model, we made an 8-lectin microarray with 10 spots of the same concentration (1 mg/mL), including SNA that has well-defined specificity to sfPSA. By measuring the spotting reproducibility in a separate experiment, we determined that the CV was in the range of 14–28%. We compared the dose-dependent fluorescence signals of the ALM for sfPSA (2–200 ng/100µL) with and without TSA (Figure 2). No apparent signals were observed for 20 ng of sfPSA (Figure 2E), and only weak signals for 200 ng of sfPSA without TSA (Figure 2F). However, with TSA, strong signals were observed for 20 ng of sfPSA (Figure 2D). Specific dose-dependent fluorescent signals were obtained for SNA, MNA, LcHA, and Jacalin (data not shown). By comparing the sensitivity of these lectins to sfPSA, calculated as the slopes of their dose-response curves, we demonstrated that overall TSA increased the sensitivity of these lectins to sfPSA over 100 times (Table 1). MNA and Jacalin have weaker interactions with sfPSA than SNA and LcHA, as evidenced by their lower sensitivity to sfPSA. Nevertheless, their increases in sensitivity were more significant (513 and 530 times) than SNA and LcHA (140 and 282 times). Therefore, TSA allowed for the detection of weak interactions of sfPSA to MNA and Jacalin that were not detectable without TSA (Figure 2E). Using the model lectin microarray, we demonstrated that the TSA increased the sensitivity of glycosylation profiling using ALM over 100 times and allowed the detection of weak lectin-glycan interactions.
Figure 2.
TSA increases the sensitivity for lectin microarray for glycan profiling of seminal PSA. A, background fluorescent signals of TSA on NPA and Calsepa; B–D, dose-dependent fluorescent signals of the seminal PSA on the array with TSA; E–F, dose-dependent fluorescent signals of the seminal PSA on the array without TSA.
Table 1.
Comparison of the sensitivity of SNA, MNA, LcHA, and Jacalin to sfPSA with and without TSA.
| TSA Sensitivitya |
No TSA Sensitivitya |
# times increase in sensitivity |
|
|---|---|---|---|
| SNA | 244 | 1.6 | 140 |
| MNA | 154 | 0.3 | 513 |
| LcHA | 254 | 0.9 | 282 |
| Jacalin | 106 | 0.2 | 530 |
Sensitivity, defined as the degree of fluorescence response to a change in ng of sfPSA, was calculated as the slope of the dose-response curve.
Optimization of the TSA-ALM
We observed high background signals with the TSA on lectins NPA and Calsepa (Figure 2A). This was likely due to these two lectins interacting with glycans on SA-HRP, especially on HRP, because SA lacks carbohydrate. Since HRP has also been shown to bind strongly to other lectins such as AAL, GNA, HHL, and UDA15, this background problem would potentially affect more lectins than just NPA and Calsepa and it may prevent the use of TSA for glycan profiling of more complex and heterogeneous features of glycoproteins. However, this problem was similar to that of using antibody for glycoprotein detection on lectin microarray, which could also interact with lectins. Kuno et al. overcame the antibody problem by masking the residual binding sites on lectins using non-labeled polyclonal IgG that have substantially the same glycans as those of the detection antibody14. We may be able to overcome the TSA background problem using a similar approach. To this end, we fabricated a new lectin microarray containing 38 lectins. Every lectin was spotted three times at three concentrations (1, 0.5, and 0.25 mg/mL). Specificity for most of the lectins had been determined in detail by frontal affinity chromatography (FAC)17. We also spotted biotinylated bovine serum albumin (BSA) three times at five different concentrations (0.4, 1.2, 3.6, 10.8, and 32.4 µg/mL) as controls for the TSA and SA-Cy3 fluorescence detection. This new design of array would help (1) investigate the severity of the background problem and (2) determine the feasibility of the TSA-ALM in studying more complex and heterogeneous features of glycoproteins.
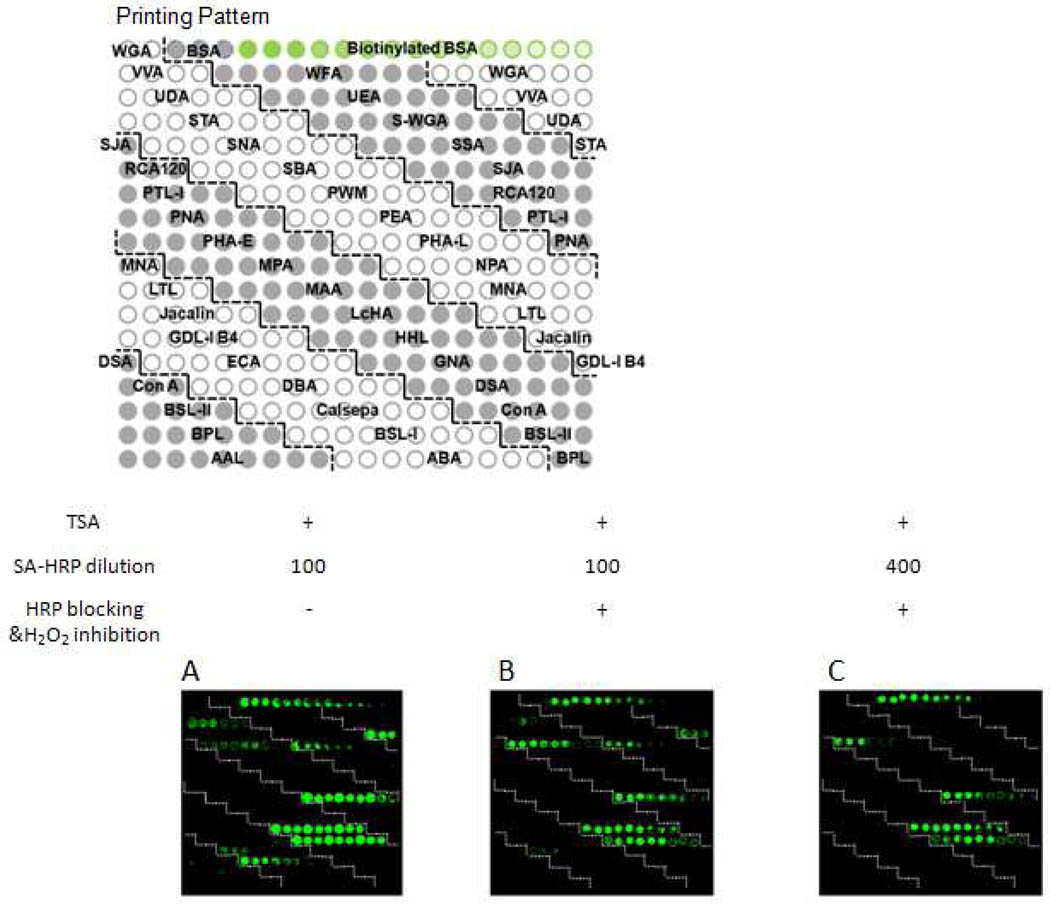
Using the new array, we found additional lectins UDA, SSA, HHL, GNA, and DBA with high background signals (Figure 3A). To suppress the undesirable background and eliminate the false positives, we blocked the binding sites using 11 µg/mL excessive, non-labeled HRP (in a 30-min incubation) that essentially has the same glycoforms as those of SA-HRP but lacks the biotin epitope. Then we permanently inhibited the enzymatic activity of HRP by treating the non-labeled HRP-bound microarray with 10% H2O2 (in a-10-min incubation) prior to the TSA. We further optimized the concentration of SA-HRP to minimize the exchange between non-labeled HRP and SA-HRP during the 30-min SA-HRP incubation. We determined that 400 times dilution of the SA-HRP stock solution from the TSA kit gave the fewest false positives without sacrificing the amplification power of the TSA, as indicated by the fluorescence intensities of the biotin controls spotted on the array. As the result of the optimization of experimental conditions, the false positives on UDA, DBA, SSA, and Calsepa were significantly diminished (Figure 3). For SNA, NPA, HHL, and GNA in which the false positive signals were still present after the optimization, we excluded them from glycan profiling using the TSA-ALM.
Figure 3.
Optimization of the experimental conditions for the TSA-ALM. Scan images of background of the TSA lectin microarray when SA-HRP is diluted 100 times without HRP blocking (A), 100 times with HRP blocking (B), and 400 times with HRP blocking (C).
The TSA-ALM of LNCAP PSA
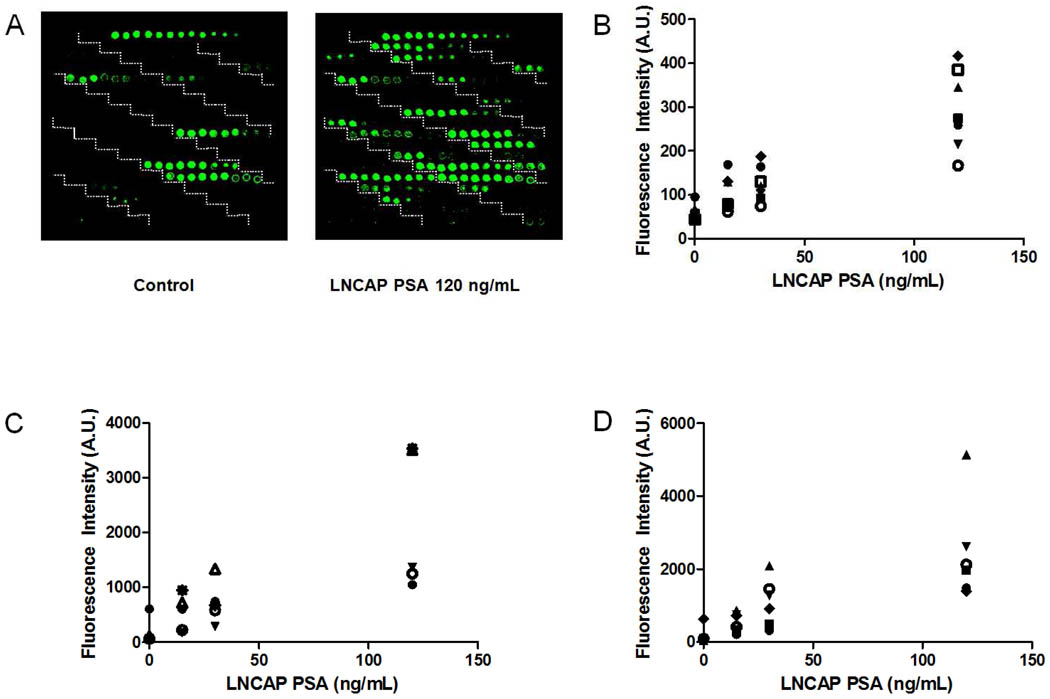
To validate the TSA-ALM for comprehensive profiling of complex glycan structures, we chose PSA enriched from the LNCAP cell culture supernatant as a model. The glycan profile of the LNCAP PSA was well-established using the evanescent-field fluorescent-assisted ALM14. Various concentrations of LNCAP PSA (1.5–12 ng/100µL) was applied to the lectin microarray followed by incubation of 4 µg/mL of biotinylated mouse anti-PSA monoclonal antibody, HRP blocking, H2O2 inhibition, SA-HRP, biotin tyramide, and detection using SA-Cy3. Eighteen lectins demonstrated specific signals in a dose-dependent manner for LNCAP PSA (Figure 4). They can be categorized into 2 groups based on their interactions with LNCAP PSA: (1) weak interactions with less than 1000 A.U. fluorescence intensities, which include BSL-II, LTL, MAA, PHA-L, SBA, SJA, and VVA (Figure 4B); (2) strong interactions with the highest fluorescence intensities more than 1000 A.U, which include DBA, LcHA, MPA, PEA, WFA, Jacalin, ECA, MNA, PHA-E, UDA, and UEA (Figure 4C and 4D). Disappearance of the signals on UEA after treating LNCAP PSA with α1-2 fucosidase, a highly specific exoglycosidase that cut the terminal α1-2 fucose off glycans, indicated that the signals on UEA was specific for terminal α1-2 fucose (supplementary Figure 1).
Figure 4.
TSA-ALM glycan profiling of the LNCAP PSA. A, scan images of background of the TSA lectin microarray (control) and the TSA lectin microarray for LNCAP PSA (right); B, dose-dependent fluorescent signals of the LNCAP PSA for BSL II (●), LTL (■), MAA (▲), PHA-L (▼), SBA (◆), SJA ( ), and VVA (
), and VVA ( ); C, dose-dependent fluorescent signals of the LNCAP PSA for DBA(●), LcHA(▼), MPA (
); C, dose-dependent fluorescent signals of the LNCAP PSA for DBA(●), LcHA(▼), MPA ( ), PEA (△), and WFA (
), PEA (△), and WFA ( ); D, dose-dependent fluorescent signals of the LNCAP PSA for Jacalin (●), ECA (■), MNA (▲), PHA-E (▼), UDA (◆), and UEA (
); D, dose-dependent fluorescent signals of the LNCAP PSA for Jacalin (●), ECA (■), MNA (▲), PHA-E (▼), UDA (◆), and UEA ( ).
).
The glycosylation profile of LNCAP PSA comprised of these 18 lectins provides the following information: (1) modification of biantennary N-glycan with Core α1-6 fucosylation as evidenced by strong signals on PEA and LcHA; (2) galactosylated tri or tetra-antenna glycan that associated with DSA, PHA-L, ECA, and BSL-II; (3) LacdiNAc structures associated with signals on WFA, SBA, and VVA; (4) terminal α1-2 fucosylation associated with signals on LTL and UEA; (5) bisecting GlcNAc associated with PHA-E; (6) α2-3 sialylation associated with MAA. These structural features correlate well with results in previous reports12, 18–21. Thus, we validated that the TSA-ALM provided sensitive glycan profiling of complex and heterogeneous features of a target protein even with low quantity (less than 20 ng).
TSA-ALM of rbPSMA
Prostate-specific membrane antigen (PSMA) is a 750-amino acid, type II transmembrane zinc metallopeptidase that is most highly expressed in the nervous system, prostate, kidney, and small intestine22–23. It is a glycoprotein with 10 potential N-glycosylation sites24–26. A recent report suggests that PSMA is also O-glycosylated27. Increased expression of PSMA has been shown to be related to prostate cancer, particularly in hormone-refractory metastatic disease22. Given its property as a prostate-cancer-related cell membrane antigen, PSMA has been exploited as a target for antibody-based therapeutic strategies28–31. In this context, it is important to analyze glycan structures of PSMA since enzymatic activity of PSMA, folding, antibody recognition may be influenced by differential glycosylation in tumor and normal cells25, 27.
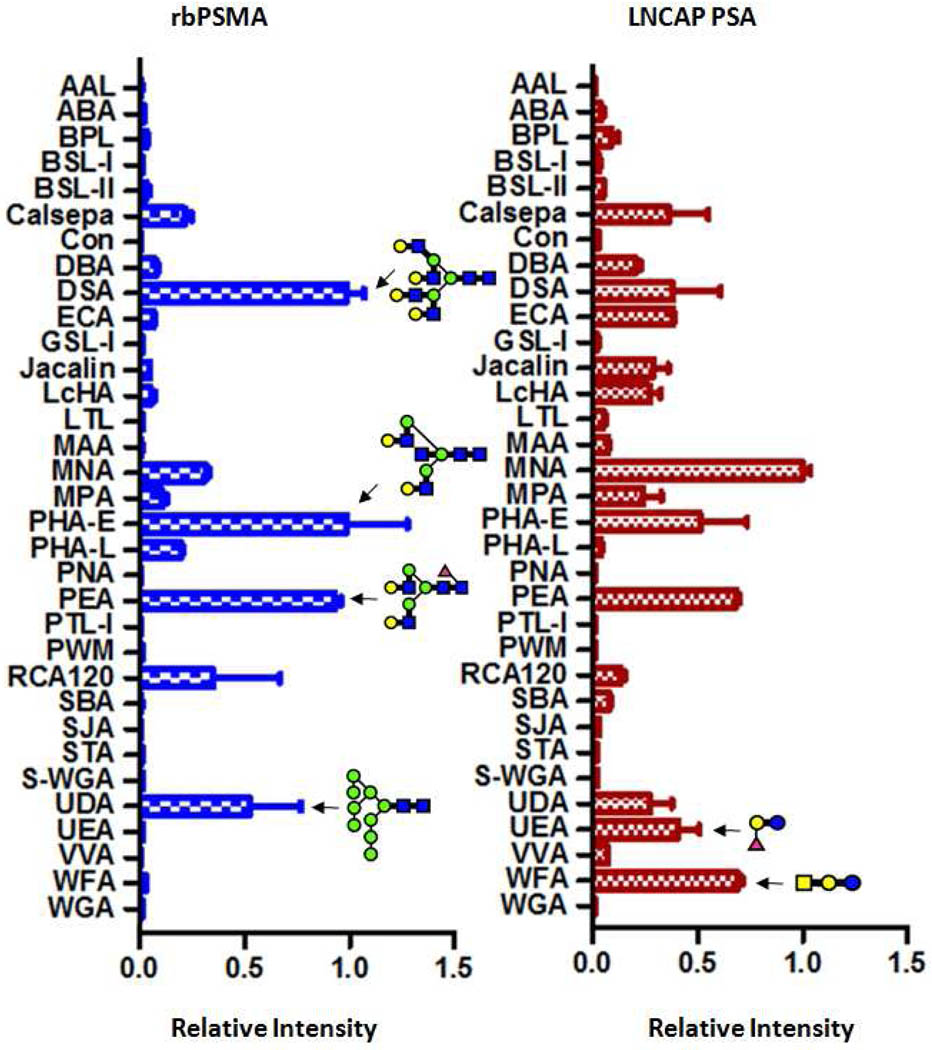
Previous reports on glycan structures of PSMA were carried out through a variety of exo- and endoglycosidase treatments followed by analysis of PSMA by Western blots or analysis of glycans by mass spectrometry24–26, 32. We were the first to report the glycosylation profile of PSMA using a lectin microarray. We used recombinant human PSMA (rhPSMA) produced by a Chinese Hamster Ovary (CHO) cell line (R&D Systems, Catalog#4234-ZN) as the model. This rbPSMA has amino acids from Lys44 to Ala750 and an N-terminal 6-His tag. Twelve lectins demonstrated specific signals in a dose-dependent manner (Figure 5), which provides the following information on the glycan structures of the rhPSMA: (1) bisecting GlcNAc associated with PHA-E; (2) modification of biantennary N-glycan with Core α1-6 fucosylation as evidenced by signals on PEA; (3) galactosylated tri or tetra-antenna glycans associated with DSA, PHA-L and RCA120; (4) high-mannose type glycans that associated with UDA. Previous reports on glycan analysis of the rhPSMA derived from the CHO cells suggested that the rhPSMA was composed of core-fucosylated hybrid mannose type of sugars25. While in agreement with the previous reports, our results indicated that the glycan structures of rhPSMA may be more heterogeneous. We also observed distinct glycosylation patterns between LNCAP PSA and CHO rbPSMA (Figure 6). LNCAP PSA gave much stronger signals on WFA and UEA, indicating a higher amount of LacdiNAc structures and terminal α1-2 fucosylation. In contrast, CHO rbPSMA gave much stronger signals on DSA, UDA, and PHA-E, indicating a higher amount of high-mannose type glycans, tri- or tetra-antenna glycans, and bisecting GlcNAc, respectively. We demonstrated that the TSA antibody-overlay lectin microarray is a useful tool to study the glycosylation profile of PSMA, and that this approach may be used for the identification of altered PSMA glycoforms.
Figure 5.
TSA-ALM glycan profiling of the rbPSMA. A, scan images of background of the TSA lectin microarray (control) and the TSA lectin microarray for the rhPSMA (right); B, dose-dependent fluorescent signals of the PSMA for 12 lectins: BSL II ( ), Jacalin (▼), LcHA (◆), MPA (▽), ECA (▲), PHA-L (★), MNA (
), Jacalin (▼), LcHA (◆), MPA (▽), ECA (▲), PHA-L (★), MNA ( ), RCA120 (●), UDA (
), RCA120 (●), UDA ( ), PEA (◇), DSA (■), and PHA-E (△).
), PEA (◇), DSA (■), and PHA-E (△).
Figure 6.
Glycan profiles of the LNCAP PSA and the rbPSMA. Relative fluorescent intensities of 33 lectins determined from the ratio to the maximal fluorescent intensity on the array are indicated with bars. Typical glycan epitopes (structural units) relevant to the present analysis are indicated by the Consortium for Functional Glycomics format.
Conclusions
We developed a TSA-ALM approach for sensitive, rapid, comprehensive, and high-throughput profiling of complex and heterogeneous glycan structure of glycoproteins. This approach does not require a specialized fluorescence scanner and can be easily incorporated into the workflow of the ALM for increased sensitivity. We validated this approach in targeted glycan profiling using PSA enriched from LNCAP cells. We also applied it to study the glycan profile of the rhPSMA. This is the first report on PSMA glycosylation using a lectin microarray. We believe that the TSA-ALM is a sensitive, rapid, comprehensive, versatile, and high-throughput technique for targeted glycan profiling and can potentially be used to identify disease-related protein glycoforms.
Supplementary Material
Acknowledgment
We thank Dr. Neil H. Bander from Cornell University for providing J-591 PSMA antibody, NIH/NCI/EDRN and Mr. David Koch for supporting the project.
Footnotes
Supporting Information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1473(1):21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- 2.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat Rev Cancer. 2005;5(7):526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 3.Dube DH, Bertozzi CR. Glycans in cancer and inflammation--potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 4.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin Chem. 2010;56(2):223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda M. Possible roles of tumor-associated carbohydrate antigens. Cancer Res. 1996;56(10):2237–2244. [PubMed] [Google Scholar]
- 6.Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20(2):362–371. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Schulz G, Cheresh DA, Varki NM, Yu A, Staffileno LK, Reisfeld RA. Detection of ganglioside GD2 in tumor tissues and sera of neuroblastoma patients. Cancer Res. 1984;44(12 Pt 1):5914–5920. [PubMed] [Google Scholar]
- 8.Chiricolo M, Malagolini N, Bonfiglioli S, Dall'Olio F. Phenotypic changes induced by expression of beta-galactoside alpha2,6 sialyltransferase I in the human colon cancer cell line SW948. Glycobiology. 2006;16(2):146–154. doi: 10.1093/glycob/cwj045. [DOI] [PubMed] [Google Scholar]
- 9.Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H, Miles D, Taylor-Papadimitriou J. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9(12):1307–1311. doi: 10.1093/glycob/9.12.1307. [DOI] [PubMed] [Google Scholar]
- 10.Leivonen M, Nordling S, Lundin J, von Boguslawski K, Haglund C. STn and prognosis in breast cancer. Oncology. 2001;61(4):299–305. doi: 10.1159/000055337. [DOI] [PubMed] [Google Scholar]
- 11.Sewell R, Backstrom M, Dalziel M, Gschmeissner S, Karlsson H, Noll T, Gatgens J, Clausen H, Hansson GC, Burchell J, Taylor-Papadimitriou J. The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. J Biol Chem. 2006;281(6):3586–3594. doi: 10.1074/jbc.M511826200. [DOI] [PubMed] [Google Scholar]
- 12.Fukushima K, Satoh T, Baba S, Yamashita K. alpha1,2-Fucosylated and beta-N-acetylgalactosaminylated prostate-specific antigen as an efficient marker of prostatic cancer. Glycobiology. 2010;20(4):452–460. doi: 10.1093/glycob/cwp197. [DOI] [PubMed] [Google Scholar]
- 13.Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6(3):306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 14.Kuno A, Kato Y, Matsuda A, Kaneko MK, Ito H, Amano K, Chiba Y, Narimatsu H, Hirabayashi J. Focused Differential Glycan Analysis with the Platform Antibody-assisted Lectin Profiling for Glycan-related Biomarker Verification. Mol Cell Proteomics. 2009;8(1):99–108. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: a new strategy for glycan profiling. Nat Methods. 2005;2(11):851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 16.Hsu KL, Mahal LK. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protoc. 2006;1(2):543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 17.Tateno H, Nakamura-Tsuruta S, Hirabayashi J. Frontal affinity chromatography: sugar-protein interactions. Nat Protoc. 2007;2(10):2529–2537. doi: 10.1038/nprot.2007.357. [DOI] [PubMed] [Google Scholar]
- 18.Peracaula R, Tabares G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13(6):457–470. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 19.Ohyama C, Hosono M, Nitta K, Oh-eda M, Yoshikawa K, Habuchi T, Arai Y, Fukuda M. Carbohydrate structure and differential binding of prostate specific antigen to Maackia amurensis lectin between prostate cancer and benign prostate hypertrophy. Glycobiology. 2004;14(8):671–679. doi: 10.1093/glycob/cwh071. [DOI] [PubMed] [Google Scholar]
- 20.Tabares G, Radcliffe CM, Barrabes S, Ramirez M, Aleixandre RN, Hoesel W, Dwek RA, Rudd PM, Peracaula R, de Llorens R. Different glycan structures in prostate-specific antigen from prostate cancer sera in relation to seminal plasma PSA. Glycobiology. 2006;16(2):132–145. doi: 10.1093/glycob/cwj042. [DOI] [PubMed] [Google Scholar]
- 21.Tajiri M, Ohyama C, Wada Y. Oligosaccharide Profiles of the Prostate Specific Antigen in Free and Complexed Forms from the Prostate Cancer Patient Serum and in Seminal Plasma: a Glycopeptide Approach. Glycobiology. 2008;18(1):2–8. doi: 10.1093/glycob/cwm117. [DOI] [PubMed] [Google Scholar]
- 22.Chang SS, Heston WD. The clinical role of prostate-specific membrane antigen (PSMA) Urol Oncol. 2002;7(1):7–12. doi: 10.1016/s1078-1439(01)00124-7. [DOI] [PubMed] [Google Scholar]
- 23.Murphy GP, Elgamal AA, Su SL, Bostwick DG, Holmes EH. Current evaluation of the tissue localization and diagnostic utility of prostate specific membrane antigen. Cancer. 1998;83(11):2259–2269. [PubMed] [Google Scholar]
- 24.Barinka C, Sacha P, Sklenar J, Man P, Bezouska K, Slusher BS, Konvalinka J. Identification of the N-glycosylation sites on glutamate carboxypeptidase II necessary for proteolytic activity. Protein Sci. 2004;13(6):1627–1635. doi: 10.1110/ps.04622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh A, Heston WD. Effect of carbohydrate moieties on the folate hydrolysis activity of the prostate specific membrane antigen. Prostate. 2003;57(2):140–151. doi: 10.1002/pros.10289. [DOI] [PubMed] [Google Scholar]
- 26.Holmes EH, Greene TG, Tino WT, Boynton AL, Aldape HC, Misrock SL, Murphy GP. Analysis of glycosylation of prostate-specific membrane antigen derived from LNCaP cells, prostatic carcinoma tumors, and serum from prostate cancer patients. Prostate Suppl. 1996;7:25–29. [PubMed] [Google Scholar]
- 27.Castelletti D, Fracasso G, Alfalah M, Cingarlini S, Colombatti M, Naim HY. Apical transport and folding of prostate-specific membrane antigen occurs independent of glycan processing. J Biol Chem. 2006;281(6):3505–3512. doi: 10.1074/jbc.M509460200. [DOI] [PubMed] [Google Scholar]
- 28.Murphy GP, Tjoa BA, Simmons SJ, Jarisch J, Bowes VA, Ragde H, Rogers M, Elgamal A, Kenny GM, Cobb OE, Ireton RC, Troychak MJ, Salgaller ML, Boynton AL. Infusion of dendritic cells pulsed with HLA-A2-specific prostate-specific membrane antigen peptides: a phase II prostate cancer vaccine trial involving patients with hormone-refractory metastatic disease. Prostate. 1999;38(1):73–78. doi: 10.1002/(sici)1097-0045(19990101)38:1<73::aid-pros9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 29.Feneley MR, Jan H, Granowska M, Mather SJ, Ellison D, Glass J, Coptcoat M, Kirby RS, Ogden C, Oliver RT, Badenoch DF, Chinegwundoh FI, Nargund VH, Paris AM, Britton KE. Imaging with prostate-specific membrane antigen (PSMA) in prostate cancer. Prostate Cancer Prostatic Dis. 2000;3(1):47–52. doi: 10.1038/sj.pcan.4500390. [DOI] [PubMed] [Google Scholar]
- 30.Bander NH. Technology insight: monoclonal antibody imaging of prostate cancer. Nat Clin Pract Urol. 2006;3(4):216–225. doi: 10.1038/ncpuro0452. [DOI] [PubMed] [Google Scholar]
- 31.Fishman M. A changing world for DCvax: a PSMA loaded autologous dendritic cell vaccine for prostate cancer. Expert Opin Biol Ther. 2009;9(12):1565–1575. doi: 10.1517/14712590903446921. [DOI] [PubMed] [Google Scholar]
- 32.Christiansen JJ, Rajasekaran SA, Inge L, Cheng L, Anilkumar G, Bander NH, Rajasekaran AK. N-glycosylation and microtubule integrity are involved in apical targeting of prostate-specific membrane antigen: implications for immunotherapy. Mol Cancer Ther. 2005;4(5):704–714. doi: 10.1158/1535-7163.MCT-04-0171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.