Abstract
Here we demonstrate the rational design of allosterically controllable, metal-ion-triggered molecular switches. Specifically, we designed DNA sequences that adopt two low energy conformations, one of which does not bind to the target ion and the other of which contains mismatches sites serving as specific recognition sites for mercury(II) or silver(I) ions. Both switches contain multiple metal binding sites and thus exhibit homotropic allosteric (cooperative) responses. As heterotropic allosteric effectors we employ single-stranded DNA sequences that either stabilize or destabilize the non-binding state, enabling dynamic range tuning over several orders of magnitude. The ability to rationally introduce these effects into target-responsive switches could be of value in improving the functionality of DNA-based nanomachines.
Because of its easily predicted secondary structure, its low cost and its high stability, DNA has become the material of choice for the construction of complex nanometer-scale molecular structures1–3. Recently, the possibility of transforming these elegant nanostructures into active “addressable” nanomachines that respond to specific molecular inputs (analytes or even “fuels”) has been also demonstrated, opening up applications ranging from drug-release vehicles to autonomous molecular robots2b,4–5.
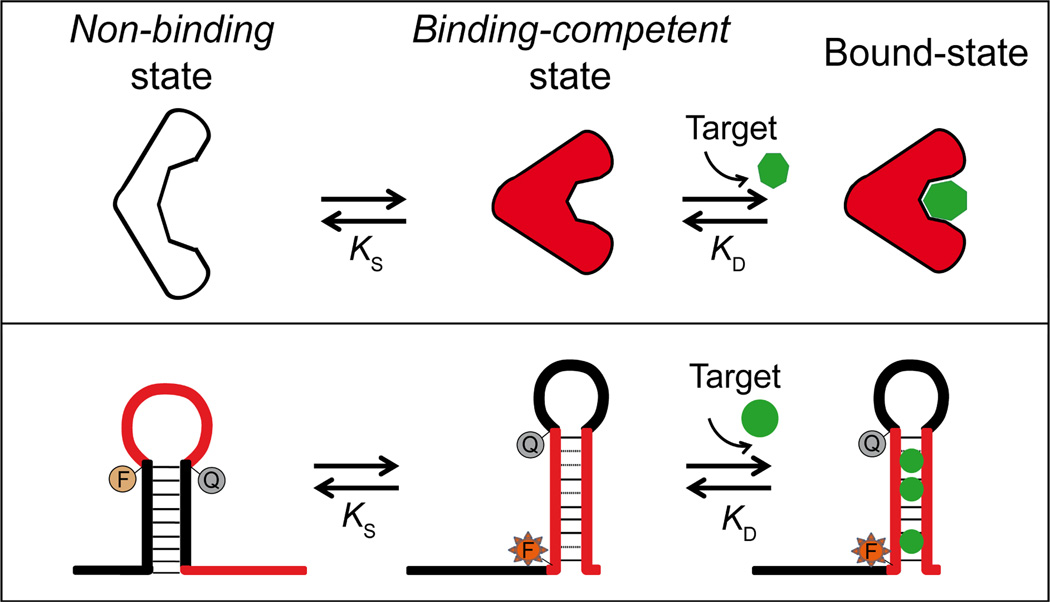
In order to couple input recognition to structural motion, which in turn can be coupled to a range of outputs (e.g. fluorescence, electrochemistry, drug release, catalysis), DNA switches are designed to flip from a non-binding conformation to a second, binding-competent conformation upon binding to a specific molecular input2a,2d,6 (Figure 1, top). An advantage of DNA-based switches is the wide range of effectors that can be used to trigger such switching, including complementary nucleic acid strands7 (binding through Watson-Crick base-pairing) as well as small molecule or protein targets (through the use of, for example, aptamer sequences or naturally occurring protein-binding sites8). A second advantage is the ease with which secondary effectors (ligands that bind distal sites on the switch) can be used to regulate their activity via an effect called “allostery”. This potentially valuable effect, however, has seen relatively less attention in the DNA-design literature7c,9. In response, we report here the rational design of allosterically tunable, conformation-linked DNA switches triggered by specific heavy metal ions.
Figure 1.
Top: Many naturally occurring chemo-receptors work via a population-shift mechanism, in which the receptor switches between a non-binding, non-signaling state and a binding-competent, signaling state. Target binding pushes the conformational equilibrium towards the latter state, leading to an increase in output signal6,10. Bottom: The predictability and modularity of DNA base pairing renders it easy to design switches that employ this same mechanism to couple target binding with a large-scale conformational change.
The affinity of binding-activated molecular switches (Figure 1) is generally well described by the population-shift model, in which both the intrinsic affinity, KD, of the binding-competent state and the switching equilibrium constant, KS, contribute to the overall, observed affinity11a, KD_obs:
| Eq. 1 |
Given this, we can tune the observed affinity, and thus the dynamic range over which the switch responds to changes in the concentration of its target, by tuning the switching equilibrium constant. This can be done during the design and fabrication of the switch via mutations that affect the switching equilibrium11–12. It can also be done on-the-fly via the addition of allosteric activators, which bind to and thus stabilize the binding-competent conformation, increasing KS, or allosteric inhibitors, which stabilize the non-binding state, reducing KS.
Recently, by tuning the useful dynamic range of molecular beacons13, a commonly employed bimolecular switch for the detection of specific nucleic acid sequences, and aptamers12, we have demonstrated that such allosteric control provides a rational, efficient, and reversible approach to modulate the affinity of a receptor. Here we employ this same mechanism to build tunable DNA-based switches triggered by specific heavy metal ions.
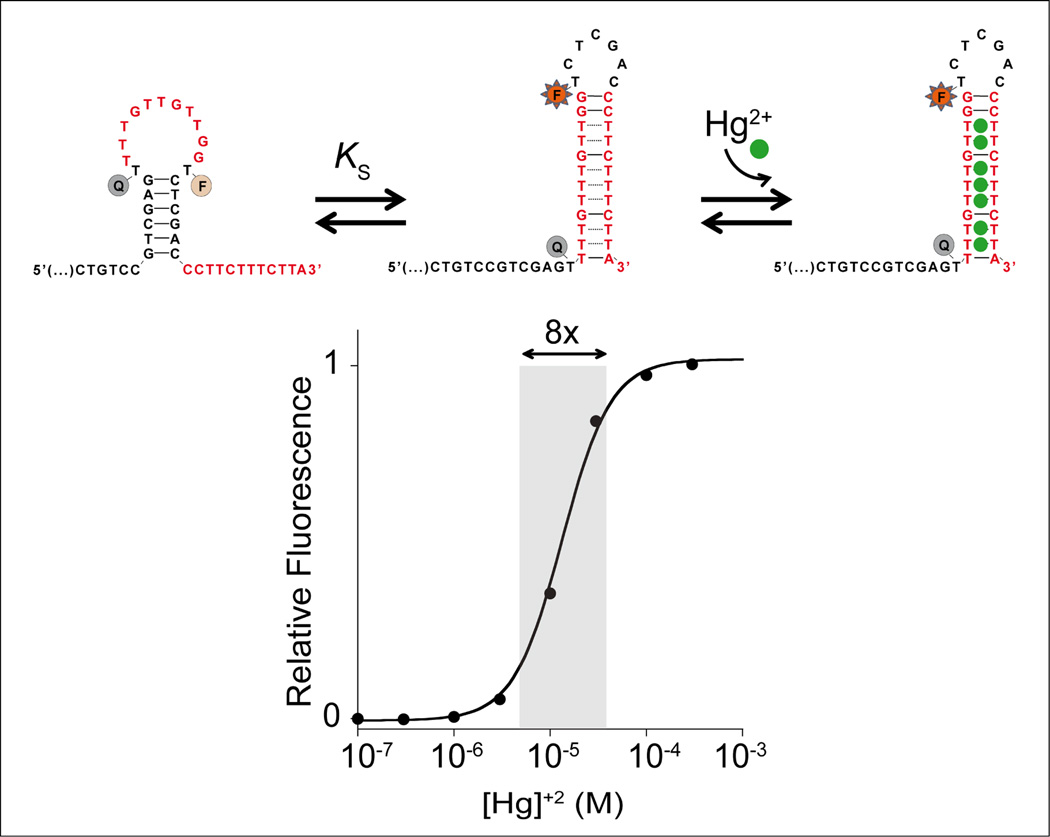
As the recognition elements in our switches we employ thymine-thymine (T-T) and cytosine-cytosine (C-C) mismatches, which specifically bind mercury(II)14 and silver(I)15 ions respectively. In our first example we introduced mercury(II) binding sites into a DNA sequence designed to adopt two low energy conformations, a non-binding conformation that lacks the mismatch pairs and a binding-competent conformation that contains multiple T-T mercury(II)-binding mismatches (Figure 2, top). The sequence is designed such that, in the absence of mercury(II) ions the non-binding state is more stable. In the presence of mercury(II) this equilibrium is then pushed towards the binding-competent conformation via a population-shift mechanism, coupling recognition with a large conformational change. Of note, however, the non-binding state should not be overstabilized because this would result in a lower affinity. More specifically, we previously demonstrated that optimal KS values are between 0.1 and 1 (e.g. 11a).
Figure 2.
Top: we have engineered DNA-based conformational switches triggered by specific heavy metal ions. In the example shown here we employed T-T mismatches to bind mercury(II) ions (Hg2+). To avoid over-stabilization of the non-binding state, which would harm affinity, the T-T mismatches are surrounded by Watson-Crick base pairs in the binding-competent state. Bottom: Because the binding of one target mercury(II) ion enhances the binding of subsequent ions, this switch exhibits positive homotropic allostery and the useful dynamic range is much narrower than for a usual single-site binding switch18. We confirmed the proposed switching mechanism by using a different “signal-off” dual labeled probe (with the fluorophore conjugated at the 3’ end) where, upon mercury(II) binding, the optical couple is brought closer and a signal suppression is observed (Figure S1). Such switching mechanism is also robust and performs well even in complex samples (Figure S2) unless of course there is mercury(II) complexant species that will shift the overall dynamic range of the switch (Figure S3).
In order to monitor binding-activated structure switching we have conjugated the sequence with a fluorophore/quencher pair (FAM/BHQ)16 such that, upon the conformational change, the two are segregated, resulting in increased fluorescence (Figure 2, top). Of note, this designed DNA-based switch differs from the numerous previous examples of Hg-triggered DNA probes17 in the fact that its affinity can be allosterically tuned with great control.
Our designed switches exhibit positive homotropic allostery, in which the binding of one copy of the target ligand facilitates the binding of subsequent copies of the target ligand. This mechanism, which is also known as positive cooperativity, narrows the useful dynamic range of the switch, leading to steeper, more responsive input-output behavior than those observed with single-site receptors18. This occurs because the switch is designed such that multiple heavy metal binding sites are present in the binding-competent state. As only the first binding event need to “pay the cost” associated with the unfavorable switching, subsequent binding events are made more favorable, leading to a steep, cooperative response. For example, whereas the dynamic range of a typical single-site receptor is 81-fold (this is the change in relative target concentration required to transit from 10% to 90% occupancy18–19), the dynamic range of our mercury(II) switch (which contains 8 target binding sites) is just 8-fold (Figure 2, bottom). Such behavior is of utility for applications, including logic gates or DNA computation where a more sensitive (larger change in response per unit change in target concentration) digital-like response curve is of value20.
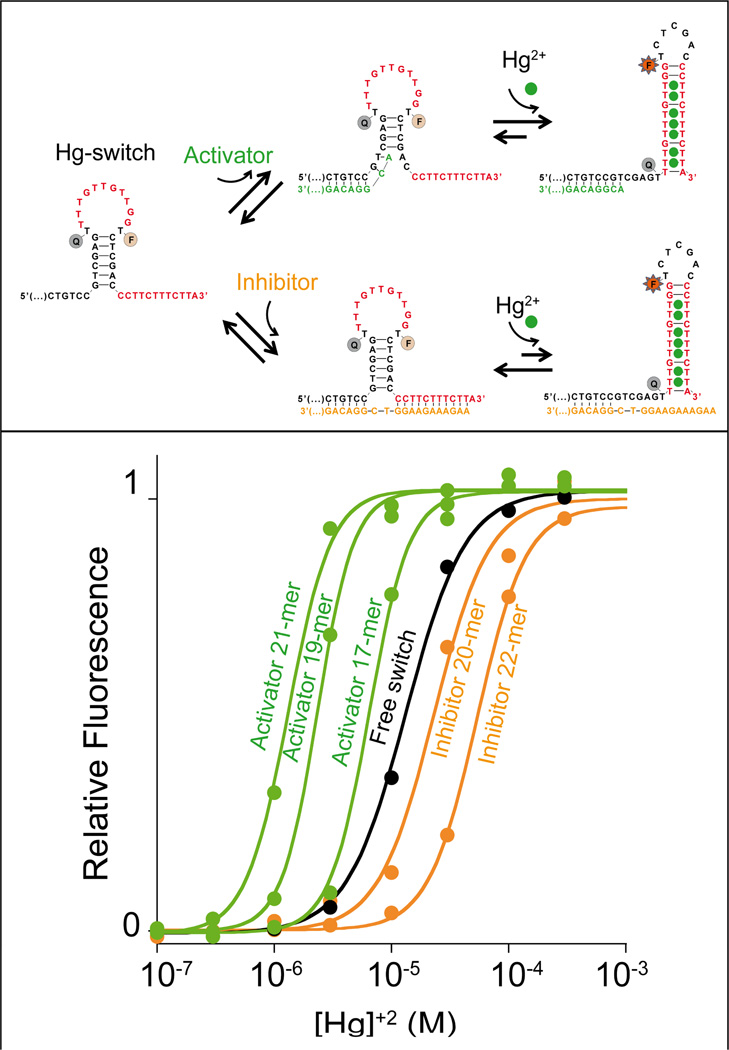
The useful dynamic range of the mercury(II)-binding switch can be easily controlled using heterotropic allostery, in which affinity is controlled by the binding of a non-target ligand (the “effector”). To create this situation, we introduced two heterotropic allosteric sites into the non-binding state (Figure 3). Once occupied, these change KS and thus change the sequence’s overall affinity for its target (Figure 3, top). As allosteric effectors we employ simple, single-stranded DNA sequences that, by binding to the allosteric sites, either destabilize (allosteric activator) or stabilize (allosteric inhibitor) the non-binding state. Specifically, activator binding destabilizes the non-binding state by partially disrupting the duplex stem that is broken upon the conformational switch (KS increases and affinity improves; Figure 3, top). Stabilization of the non-binding state, in contrast, is achieved with an inhibitor that increases the number of Watson-Crick base pairs that must be broken to perform the conformational switch (KS and affinity both decrease; Figure 3, top).
Figure 3.
We can tune the dynamic range of metal-activated DNA-switches using heterotropic allostery in which the addition of allosteric effectors (activator/inhibitor) pushes the useful dynamic range to higher or lower target concentrations. Top: As an allosteric activator we employ a single-stranded DNA that, when bound to a tail appended to the 5’ end of the switch, destabilizes the non-binding conformation and thus pushes the useful dynamic range of the switch to lower target concentrations (bottom, green curves). As an allosteric inhibitor, in contrast, we employed a sequence that binds the tails on both ends of the switch simultaneously, thus stabilizing the non-binding conformation and pushing the dynamic range of the switch to higher target concentrations (bottom, orange curves). Bottom: Increasing the length of the inhibitor from 20 to 22 bases increasingly stabilizes the non-binding state thus pushing the useful dynamic range to higher and higher target concentrations (orange curves). Increasing the length of the activator from 17 to 21 bases similarly increases the extent to which it destabilizes the non-binding state, pushing the useful range of the switch to lower target concentrations (green curves). The black curve (free switch) represents the binding of the switch in the absence of allosteric effectors. Binding curves are shown as normalized signals. We note, however, that, because activator binding destabilizes the non-binding conformation we observe an increase of the background signal. As a result, in the presence of activators, we obtain a lower signal gain in response to the target (Figure S4). We also note that no significant quenching of mercury(II) ions on the FAM signal was observed (Figure S5). Here the switch concentration was held at 10 nM and the activator or inhibitor concentration at 20 nM, a concentration that allows saturation of the switch (see figure S6 and SI for more details).
Using allosteric control we can tune the dynamic range of the mercury switch over ca. two orders of magnitude (Figure 3, bottom). For example, the KD_obs of the mercury(II) switch, 16 µM, can be pushed to 1.4 µM using the longest, most active activator that we have tested (21-base activator). Similarly, the most effective inhibitor that we have tested (22-base inhibitor) shifts the affinity to higher concentrations by a factor of six (KD_obs = 95.1 µM).
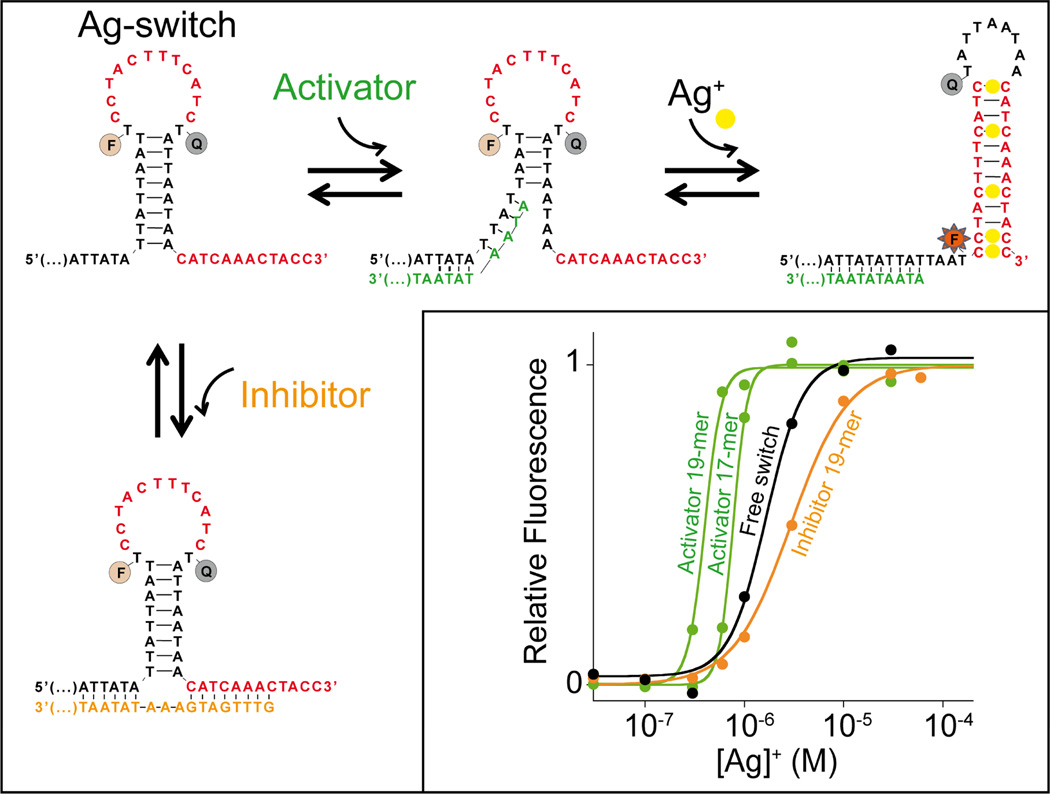
Using the same approach we have employed for our mercury(II) sensor, we have also designed a conformational switch that responds to the presence of silver(I) ions (Ag+). To do this we simply changed the portion of the switch sequence recognizing mercury(II) ions following the rules described above (avoid over-stabilization of non-binding state, alternation of mismatches with Watson-Crick base pairs in the binding-competent state) (Figure 4, top). Because it is also based on the population-shift mechanism, the silver(I) switch is likewise tunable through the use of allosteric activation and inhibition (Figure 4, bottom). Because structure-switching conformational change is specific and the two switches we have used are labelled with two distinct flurophores we can also measure both mercury(II) and silver(I) ions in the same solution containing a mixture of both their specific switches (Figure S7).
Figure 4.
To demonstrate the generality of this approach we have also designed a tunable, silver(I)-activated switch employing silver(I)-binding C-C mismatches as its recognition elements. Once again, single-stranded DNA sequences complementary to one or both of the allosteric tails on the non-binding state serve as allosteric effectors to activate (green curves) or inhibit (orange curve) the switch and tune its useful dynamic range. The black curve represents the binding of the switch in the absence of allosteric effectors. We note that with this switch the presence of the activators leads to a cooperative-like dose response curve. In the presence of the inhibitor this cooperative-like response is less pronounced21. Here the switch concentration was held at 10 nM and the effector concentrations at 20 nM (see SI for details).
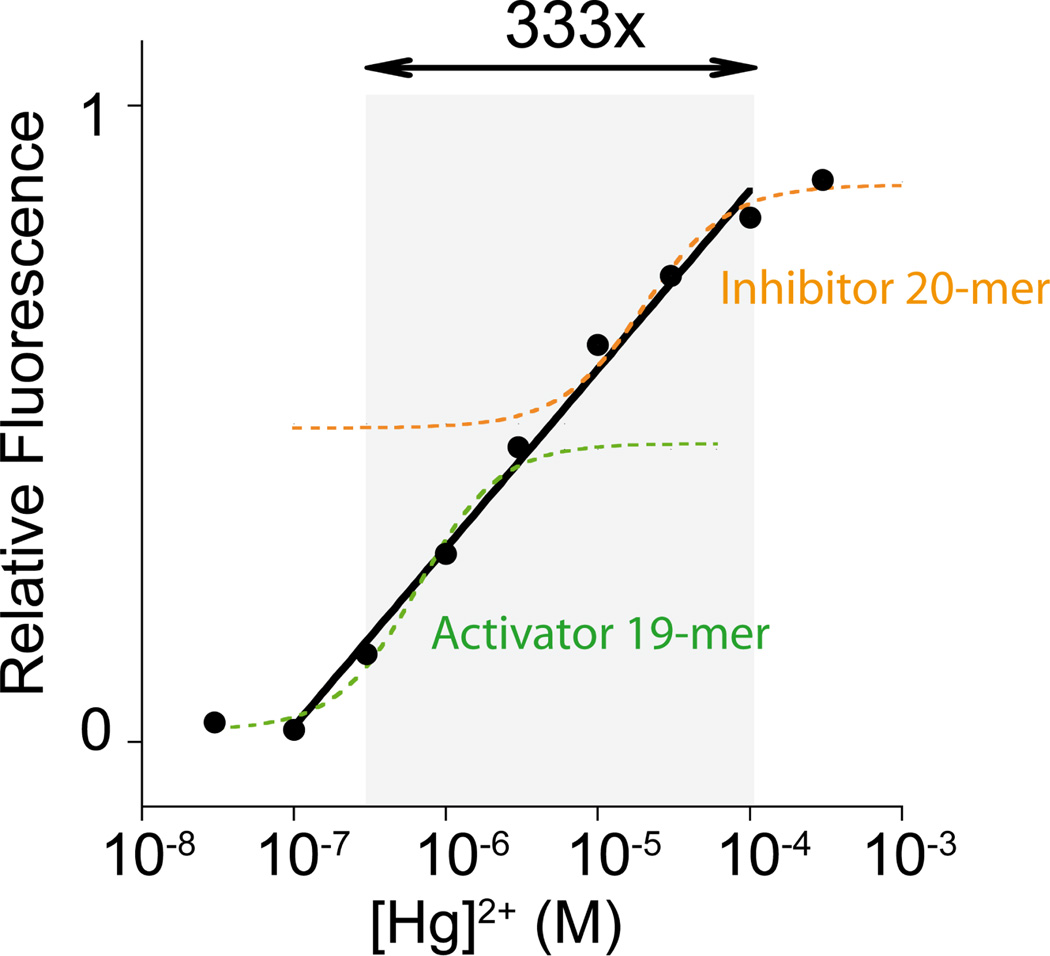
The allosteric control also provides a means of broadening the concentration range over which our sensors respond robustly to changes in the concentration of their target ligand. To generate such behavior we have used a mixture of two allosteric activators in the same tube. With each activator triggering its switch at a different ion concentration, this extends the switch’s dynamic range to ca. 2 orders of magnitude (Figure S8). Similarly, using an activator and an inhibitor in the same tube we have broadened the dynamic range to ca. 3 orders of magnitude (Figure 5).
Figure 5.
Extending the dynamic range of metal binding switches using a mixture of an allosteric activator and an allosteric inhibitor. While the switch in the presence of a single allosteric effector shows a limited dynamic range (only 8-fold in target concentration; see dashed curves), this same dynamic range is widened to almost 3 orders of magnitude in the presence of the activator/inhibitor mixture (solid line). Here the switch concentration was held at 10 nM and the activator (21-base) and inhibitor (20-base) concentrations at 7 and 3 nM respectively (see SI for details).
Here we have used the population-shift mechanism to develop metal-activated switches that couple target recognition with a large-scale conformational change. Compared to other previously reported DNA-based heavy metals triggered probes,14,15,17 the switches used here show a slightly poorer affinity for their targets, but their ability to support both homotropic and heterotropic allostery allow us to control both the width and placement of their useful dynamic ranges with unprecedented precision, a feature that can be of great utility in several fields. This ability to rationally introduce allosteric control into target-responsive switches may in fact play an important role in the construction of novel DNA-based nanomachines, improving their functionality in applications, such as targeted drug-release, DNA-based computation and theranostic approaches, in which tight control over input-output behavior would be of value2–5.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the MIUR (FIRB “Futuro in Ricerca”) (FR), by Bill & Melinda Gates Foundation through the Grand Challenges Explorations (OPP1061203) (FR), by the International Research Staff Exchange Scheme (IRSES) grant under the Marie Curie Actions program (FR), by the NIH through grant R01EB007689 (KWP) and by the Fonds de Recherche du Québec Nature et Technologies (FRQNT) (AVB). FR is supported by a Marie Curie Outgoing Fellowship (IOF) (Proposal No. 298491under FP7-PEOPLE-2011-IOF).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting methods, figures. This material is available free of charge at http://pubs.acs.org.
REFERENCES
- 1.(a) Seeman NC. Mol. Biotechnol. 2007;37:246. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chen J, Seeman NC. Nature. 1991;350:631. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]; (c) Shih WM, Quispe JD, Joyce GF. Nature. 2004;427:618. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]; (d) Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF. Science. 2005;310:1661. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]; (e) Rothemund PWK. Nature. 2006;440:297. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.(a) Simmel FC. Angew. Chem. Int. Ed. 2008;47:5884. doi: 10.1002/anie.200801982. [DOI] [PubMed] [Google Scholar]; (b) Ding B, Seeman NC. Science. 2006;314:1583. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. Science. 2011;332:342. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]; (d) Krishnan Y, Simmel FC. Angew. Chem. Int. Ed. 2011;50:3124. doi: 10.1002/anie.200907223. [DOI] [PubMed] [Google Scholar]
- 3.(a) Han D, Pal S, Liu Y, Yan H. Nat. Nanotechnol. 2010;5:712. doi: 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pinheiro AV, Han D, Shih WM, Yan H. Nat. Nanotechnol. 2011;6:763. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 4.(a) Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:7243. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]; (b) Goodman RP, Heilemann M, Doose S, Erben CM, Kapanidis AN, Turberfield AJ. Nat. Nanotechnol. 2008;3:93. doi: 10.1038/nnano.2008.3. [DOI] [PubMed] [Google Scholar]; (c) Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. Nature. 2000;406:605. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]; (d) Sherman WB, Seeman NC. Nano Lett. 2004;4:1203. [Google Scholar]
- 5.(a) Lund K, Manzo AJ, Dabby N, Michelotti N, Johnson-Buck A, Nangreave J, Taylor S, Pei R, Stojanovic M, Walter NG, Winfree E, Yan H. Nature. 2010;465:206. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Wickham SFJ, Endo M, Katsuda Y, Hidaka K, Bath J, Sugiyama H, Turberfield AJ. Nat. Nanotechnol. 2011;6:166. doi: 10.1038/nnano.2010.284. [DOI] [PubMed] [Google Scholar]
- 6.Vallée-Bélisle A, Plaxco KW. Curr. Opin. Struct. Biol. 2010;20:518. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Green SJ, Lubrich D, Turberfield AJ. Biophys. J. 2006;91:2966. doi: 10.1529/biophysj.106.084681. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Zhang DY, Winfree E. J. Am. Chem. Soc. 2008;130:13921. doi: 10.1021/ja803318t. [DOI] [PubMed] [Google Scholar]; (c) Zhang DY, Winfree E. J. Am. Chem. Soc. 2009;131:17303. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]; (d) Zhang Z, Zeng D, Ma H, Feng G, Hu J, He L, Fan C. Small. 2010;6:1854. doi: 10.1002/smll.201000908. [DOI] [PubMed] [Google Scholar]
- 8.(a) Nutiu R, Li Y. J. Am. Chem. Soc. 2003;125:4771. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]; (b) Wieland M, Benz A, Klauser B, Hartig JS. Angew. Chem. Int. Edit. 2009;48:2715. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]; (c) Vallée-Bélisle A, Bonham AJ, Reich NO, Ricci F, Plaxco KW. J. Am. Chem. Soc. 2011;133:13836. doi: 10.1021/ja204775k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Keefe AD, Pai S, Ellington A. Nat. Rev. Drug Discov. 2010;9:537. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Tang J, Breaker RR. Chem. Biol. 1997;4:453. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]; (b) Stojanovic MN, De Prada P, Landry DW. Chem. Biol. Chem. 2001;2(6):411. doi: 10.1002/1439-7633(20010601)2:6<411::AID-CBIC411>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; (c) Winkler WC, Breaker RR. Chem. Biol. Chem. 2003;4:1024. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]; (d) Famulok M, Hartig JS, Mayer G. Chem. Rev. 2007;107(9):3715. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]; (e) Teller C, Shimron S, Willner I. Anal. Chem. 2009;81:9114. doi: 10.1021/ac901773b. [DOI] [PubMed] [Google Scholar]; (f) Vinkenborg JL, Karnowski N, Famulok M. Nature Chem. Biol. 2011;7:519. doi: 10.1038/nchembio.609. [DOI] [PubMed] [Google Scholar]
- 10.(a) Ma B, Shatsky M, Wolfson HJ, Nussinov R. Protein Sci. 2002;11:184. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ma B, Kumar S, Tsai C, Nussinov R. Protein Eng. 1999;12:713. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]; (c) Tsai C, Kumar S, Ma B, Nussinov R. Protein Sci. 1999;8:1181. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kumar S, Ma B, Tsai C, Sinha N, Nussinov R. Protein Sci. 2000;9:10. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Vallée-Bélisle A, Ricci F, Plaxco KW. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13802. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vallée-Bélisle A, Ricci F, Plaxco KW. J. Am. Chem. Soc. 2012;134:2876. doi: 10.1021/ja209850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porchetta A, Vallée-Bélisle A, Plaxco KW, Ricci F. J. Am. Chem. Soc. 2012;134:20601. doi: 10.1021/ja310585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricci F, Vallée-Bélisle A, Porchetta A, Plaxco KW. J. Am. Chem. Soc. 2012;134:15177. doi: 10.1021/ja304672h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Ono A, Cao S, Togashi H, Tashiro M, Fujimoto T, Machinami T, Oda S. Chem. Comm. 2008;39:4825. doi: 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]; (b) Ono A, Togashi H. Angew. Chem. Int. Ed. 2004;43:4300. doi: 10.1002/anie.200454172. [DOI] [PubMed] [Google Scholar]; (c) Lee J, Han MS, Mirkin CA. Angew. Chem. Int. Ed. 2007;46:4093. doi: 10.1002/anie.200700269. (45) [DOI] [PubMed] [Google Scholar]; Li D, Wieckowska A, Willner I. Angew. Chem. Int. Ed. 2008;47:3927. doi: 10.1002/anie.200705991. [DOI] [PubMed] [Google Scholar]; (d) Liu S, Nie H, Jiang J, Shen G, Yu R. Anal. Chem. 2009;81:5724. doi: 10.1021/ac900527f. [DOI] [PubMed] [Google Scholar]; (e) Liu CW, Huang CC, Chang HT. Anal. Chem. 2009;81:2383. doi: 10.1021/ac8022185. [DOI] [PubMed] [Google Scholar]; (f) Liu J, Lu Y. Angew. Chem. Int. Ed. 2007;46:7587. doi: 10.1002/anie.200702006. [DOI] [PubMed] [Google Scholar]; (g) Shimron S, Elbaz J, Henning A, Willner I. Chem. Comm. 2010;46:3250. doi: 10.1039/b926003j. [DOI] [PubMed] [Google Scholar]
- 15.(a) Tanaka K, Yamada Y, Shionoya M. J. Am. Chem. Soc. 2002;124:8802. doi: 10.1021/ja020510o. [DOI] [PubMed] [Google Scholar]; (b) Clever GH, Kaul C, Carell T. Angew. Chem. Int. Ed. 2007;46:6226. doi: 10.1002/anie.200701185. [DOI] [PubMed] [Google Scholar]
- 16.(a) Marras SA. Methods Mol. Biol. 2006;335:3. doi: 10.1385/1-59745-069-3:3. [DOI] [PubMed] [Google Scholar]; (b) Marras SAE. Mol. Biotechnol. 2008;38(3):247. doi: 10.1007/s12033-007-9012-9. [DOI] [PubMed] [Google Scholar]
- 17.(a) Wang Z, Lee H, Lu Y. Chem. Comm. 2008;45:6005. doi: 10.1039/b812755g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu X, Sun C, Wu H, Zhang Y, Jiang J, Shen G, Yu R. Electroanal. 2010;22:2110. [Google Scholar]; (c) Li T, Dong S, Wang E. Anal. Chem. 2009;81:2144. doi: 10.1021/ac900188y. [DOI] [PubMed] [Google Scholar]; (d) Zhu Z, Su Y, Li J, Li D, Zhang J, Song S, Zhao Y, Li G, Fan C. Anal. Chem. 2009;81:7660. doi: 10.1021/ac9010809. [DOI] [PubMed] [Google Scholar]; (e) Li T, Shi L, Wang E, Dong S. Chem. Eur. J. 2009;15:3347. doi: 10.1002/chem.200900056. [DOI] [PubMed] [Google Scholar]
- 18.Koshland DE, Jr, Goldbeter A, Stock JB. Science. 1982;217:220. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]
- 19.(a) Ferrell JE., Jr Trends Biochem. Sci. 1996;21(12):460. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]; (b) Goldbeter A, Koshland DE., Jr Q. Rev. Biophys. 1982;15:555. doi: 10.1017/s0033583500003449. [DOI] [PubMed] [Google Scholar]
- 20.(a) Kim J, White KS, Winfree E. Mol. Syst. Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rafael PS, Vallée-Bélisle A, Fabregas E, Plaxco K, Palleschi G, Ricci F. Anal. Chem. 2012;84:1076. doi: 10.1021/ac202701c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ricci F, Vallée-Bélisle A, Plaxco KW. PLoS Comput. Biol. 2011;7:e1002171. doi: 10.1371/journal.pcbi.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Halamek J, Zavalov O, Halamkova L, Korkmaz S, Privman V, Katz E. J. Phys. Chem. B. 2012;116(15):4457. doi: 10.1021/jp300447w. [DOI] [PubMed] [Google Scholar]
- 21.In the presence of inhibitor the switch looses its cooperative-like dose-response curve probably because the five silver(I) binding sites of the switch are all forced to a lower affinity and cannot interact with each other. Thus, the binding of silver(I) ions to the first binding site does not increase the affinity of the other binding sites for the target.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.