Abstract
Background
Associations of polymorphisms from dopaminergic neurotransmitter pathway genes have been reported in Caucasian ancestry schizophrenia (SZ) samples. As studies investigating single SNPs with SZ have been inconsistent, more detailed analyses utilizing multiple SNPs with the diagnostic phenotype as well as cognitive function may be more informative. The analyses were conducted in a north Indian sample.
Methods
Indian SZ case-parent trios (n = 601 families); unscreened controls (n= 468) and an independent set of 118 trio families were analyzed. Representative SNPs in the Dopamine D3 receptor (DRD3), dopamine transporter (SLC6A3), vesicular monoamine transporter 2 (SLC18A2), catechol-o-methyltransferase (COMT) and dopamine beta hydroxylase (DBH) were genotyped using SNaPshot/SNPlex assays (n=59 SNPs). The Trail Making Test (TMT) was administered to a subset of the sample (n=260 cases and_n=302 parents).
Results
Eight SNPs were nominally associated with SZ in either case-control or family based analyses (p<0.05, rs7631540 and rs2046496 in DRD3; rs363399 and rs10082463 in SLC18A2; rs4680, rs4646315 and rs9332377 in COMT). rs6271 at DBH was associated in both analyses. Haplotypes of DRD3 SNPs incorporating rs7631540-rs2134655-rs3773678-rs324030-rs6280-rs905568 showed suggestive associations in both case-parent and trio samples. At SLC18A2, rs10082463 was nominally associated with psychomotor performance and rs363285 with executive functions using the TMT but did not withstand multiple corrections.
Conclusions
Though suggestive associations with dopaminergic genes were detected in this study, but convincing links between dopaminergic polymorphisms and SZ or cognitive function were not observed.
Keywords: Schizophrenia, Dopamine genes, SNPs, association, Haplotypes, cognition
INTRODUCTION
Schizophrenia (SZ) is a common, severe disorder with a lifetime prevalence of approximately 1% worldwide (Gottesman, 1982; Saha, et al., 2005). Its prevalence was estimated at 4/1000 in India and likely represents an underestimate (Ganguli, 2000). Interactions between genetic and environmental etiological factors provide the most plausible explanations for the relatively high heritability estimates of 70-80% (Owen, 2002; Sullivan, et al., 2003;Sullivan, 2005; Lichtenstein et al., 2009). Prior gene mapping studies have identified multiple putative susceptibility loci with genes underlying neurotransmitter pathways being implicated frequently (Allan, et al., 2008; Ng et al., 2009; Seeman & Kapur, 2000; Seeman, 2002; Staddon et al., 2005; Dominguez et al., 2007; Talkowski et al., 2006; Talkowski et al., 2008; Srivastava et al., 2010). Further, neuroimaging studies in SZ patients reveal neuronal disorganization in cortical and limbic regions of the brain and increased dopamine D2 receptor binding (Keshavan etal., 2008). We have previously reported consistent associations with the gene encoding the dopamine D3 receptor (DRD3) and related haplotypes in Indian and US Caucasian samples (Talkowski et al., 2006). Indeed, associations at DRD3 have been reported recently in two other independent Caucasian samples (Staddon et al., 2005; Dominguez et al., 2007), but were not replicated in a Japanese cohort (Nunokawa et al., 2010).
The dopamine gene associations do not feature prominently in recent genome wide association studies (GWAS)(Lencz et al., 2007; Pearson et al., 2007;Wellcome Trust Case Control Consortium, 2007; Sklar et al., 2008; Sullivan et al., 2008; Shi et al., 2009; Stefansson et al., 2008; Stefansson et al., 2009; The International Schizophrenia Consortium 2009). On the other hand, the available genome-wide significant associations, such as human zinc finger protein 804A (ZNF804A), neurogranin (NRGN) and transcription factor 4 (TCF4), suggest common alleles of small effect, rare alleles of large /small effect, and complementary analysis of association signals from various genes grouped according to their interactions and pathways may contribute to some risk towards disease etiology (Jia et al., 2010). Thus pathway based candidate gene approach still holds true for genetic studies of SZ. Recently, systematic analyses of eighteen DA genes in US Caucasian samples revealed significant associations with SNPs at DRD3, dopamine transporter DAT (alias, SLC6A3), catechol-O-methyltransferase (COMT), dopamine beta-hydroxylase (DBH) and vesicular monoamine transporter 2 (SLC18A2). Some epistatic interactions between pairs of SNPs across these genes were also significant; these associations were replicated in a Bulgarian family based sample. Simulation studies suggested that the replicable associations were unlikely to be due to chance (Talkowski et al., 2008).
Cross population studies could help identify genuine genetic associations and enable fine mapping of disease associated loci. Such studies are infrequent in SZ research (O’Donovan et al., 2009). On the other hand, associations may not be detected consistently across ethnic groups for a number of reasons. Population structure may impact success in gene mapping studies (Novembre et al., 2008), and divergent linkage disequilibrium (LD) patterns may explain “flip-flop” associations, i.e., associations with different alleles of SNPs (Lin et al., 2007). On the other hand, the choice of markers investigated could also contribute to the inability to replicate some associations. Single marker association analyses are inefficient as they may reflect only the localized effect of an individual SNP or the polymorphism being analyzed may not be the risk variant and/or in LD with the functional variant (Johnson et al., 2001; Gabriel et al., 2002), or it may have low polymorphism information content. In such situations, approaches considering LD based/multi-marker haplotype analysis could be more informative (Akey et al., 2001; Kamatani et al., 2004; Lin et al., 2004; de Bakker et al., 2005; Dominguez et al., 2007). The multi-marker analyses may also help overcome some differences in LD structure across populations.
Against this background, we tested several SNPs from pharmacologically relevant DA pathway genes mentioned above as well as additional selected SNPs in a large Indian sample. We adopted a multi-marker analysis approach because such analyses may indicate associations even if the causative SNP is not genotyped. Though the LD structure in outbred north Indian populations is reportedly similar to Caucasian populations (Pemberton et al., 2008), the precise LD structure at the genes of interest has not been analyzed extensively in Indian samples. To maximize the chance of capturing the susceptibility locus identified in Caucasians, we employed variable-sized sliding window (VSW) analysis encompassing 2-6 SNPs. A case-control as well as family based study design was employed with the cases being common for both strategies. The dual design was utilized because each type of control has distinct advantages and shortcomings (Bacanu, et al., 2000).
Dopamine is integral to cognition, learning and memory, and dysfunctions of the frontal cortical dopamine system have been implicated in both transcript and protein levels during postnatal development (Rothmond et al., 2012). Variations in the DRD2 gene have been associated with working memory performance (Kellendonk et al., 2006; Bertolino et al., 2010). Selective blockade of dopamine D3 receptors reverses the visual recognition memory deficit and hyperactivity produced by isolation rearing support the potential use of dopamine D3 receptor antagonists to treat schizophrenia (Watson et al., 2011). A role for COMT in regulating executive functions and selective attention (Barnett et al., 2007; Solis-Ortiz et al., 2010; Watson et al., 2011) has been reported. Hence we evaluate the DA gene polymorphisms in relation to cognitive functions. The A and B tasks of TMT evaluates the psychomotor and executive functions respectively. The cognitive functions measured by TMT test are highly heritable (Quinones et al., 2009) and relate mainly to executive and psychomotor functions (Bhatia et al., 2007; Bhatia et al., 2009; Quinones et al., 2009).
METHODS
Samples
The study was approved by the Institutional Ethics Committees at Dr Ram Manohar Lohia Hospital, New Delhi, the Lok Nayak Hospital, New Delhi, Delhi University, South campus, New Delhi and the University of Pittsburgh, Pittsburgh IRB. Written informed consent was obtained from all participants (maternal consent for neonatal samples). The Hindi version of the Diagnostic Interview for Genetic Studies (DIGS), a structured, validated diagnostic interview was administered to each patient as described (Deshpande et al., 1998; Chowdari et al., 2002; Bhatia et al., 2009). Based on this information, consensus diagnoses were established by certified psychiatrists and psychologists using DSM IV criteria. Inter-rater and inter-site diagnostic reliability was checked throughout the study and Kappa values of 0.8 or greater were aimed for (Bhatia et al., 2006).
Participants recruited in the study were confirmed to be of north Indian origin based on language/mother tongue and the geographical location for three generations and were largely drawn from Delhi and neighboring states of Uttar Pradesh, Punjab, Bihar, Haryana, Himachal Pradesh, Uttaranchal, Rajasthan, etc. They included cases, their parents and unrelated community based controls. The latter were composed of neonatal cord blood samples from live births at Lok Nayak Hospital, New Delhi. Mental Illness was evaluated among parents of schizophrenia patients using the Family Interview for Genetic studies (FIGS; (Maxwell, 1992).
Sample characteristics
A total of 601 case-parent trios (n = 1800 participants) and 468 controls were analyzed. The sample included patients diagnosed with schizophrenia using DSM IV criteria. Some of the participants (n=123 families) in this study overlapped with those used in our previous genetic association studies (Talkowski et al., 2006). Another 208 samples were shared with another study (Srivastava et al., 2010). An additional 119 north Indian trio sample set was used as an independent sample for analysis of the DRD3 associations only.
The cognitive functions of the probands, and parents were measured by administering TMT in a family to a subset of cases (n=260) and their healthy parents (n=302).
Genetic Analysis
Genomic DNA was isolated from venous blood samples using the phenol chloroform extraction method and was quantified using the picogreen method. A total of 59 SNPs were analyzed. They were distributed across five DA genes, namely DRD3 (3q13.3), SLC6A3 (alias DAT, 5p15.3), SLC18A2 (alias VMAT2, 10q25), COMT (22q11.21) and DBH (9q34). A list of all the SNPs is provided in Supplementary Table I. Of these, 46 SNPs were obtained from a earlier report on dopamine genes in schizophrenia (Talkowski et al., 2008). These included the four commonly investigated exonic SNPs (rs6280 from DRD3, rs4680 from COMT, and rs1108580 and rs6271 from DBH). We also included 13 additional SNPs from the HapMap phase II release in this study to achieve more comprehensive coverage. SNPs were assayed using either SnaPshot (Mansour et al., 2005) or SNPlex assays, ABI Biosystems (Tobler et al., 2005) with CEPH samples as positive controls.
Statistical Analysis
The power to detect associations in the study cohort was evaluated using Quanto software (Gauderman, 2006; http://hydra.usc.edu/GxE), assuming an additive model and a disease frequency of 1%. Hardy Weinberg equilibrium (HWE) was examined for each SNP using PLINK (Purcell et al., 2007; http://pngu.mgh.harvard.edu/purcell/plink/). Only the SNPs conforming to HWE (p > 0.01) were included in the association analyses. LD values (r2>0.8) were estimated for the genotyped data using the Tagger algorithm in Haploview version 4.1 (Barrett et al., 2005; http://www.broad.mit.edu/mpg/haploview/). Transmission distortion in families was assessed using FBAT software (Laird et al., 2000). The Armitage Trends test (SAS software) was used for case-control analysis. Sliding window haplotypes for 2-6 SNPs were generated for both cases-control and family based data using UNPHASED 3.1.5 (Dudbridge, 2008). A global or omnibus test of haplotype-based association was performed in UNPHASED. This tests log-likelihood ratios under a log-linear model for global p values. Since both common and rare variants and their haplotypes have been reported for association with schizophrenia (Li et al., 2005; Agim et al., 2013) and also in age related macular degeneration (AMD) (Raychaudhuari et al. 2011) the haplotypes frequency cutoff was set at 1% for haplotypes analysis. In the first step, all the markers were analyzed using the default Davidon-Fletcher-Powell (DFP) method in UNPHASED. All the p-values till this step were subjected to multiple corrections and haplotypes that retained significance were reassessed using Nelder & Mead’s (NM) method in PLINK (Minor Haplotype ≥1%) for likelihood maximization, since DFP can sometimes converge to error-prone solutions (www.rfcgr.mrc.ac.uk/~fdudbrid/software/unphased/). There were 505 tests in all, so an alpha value of 0.0001 (0.05/505) or lower was considered to be significant. PLINK was used to estimate the actual haplotypes and frequencies, since TDT-like counts of transmitted and untransmitted alleles/haplotypes and resulting TDT statistic are not available in UNPHASED. The p-values for individual marker associations and sliding window haplotypes were graphed separately for case-control and family based data for each gene using Graphical Assessment of Sliding P-values (GrASP v.0.82 beta) to present and assess p-values from multiple tests (Mathias et al., 2006; http://research.nhgri.nih.gov/GrASP/).
Linear regression analyses were conducted for both A and B task separately using the Statistical Package for Social Sciences (SPSS Version 16) to test associations with cognitive variables. Since the parent group was older in comparison with the patients, the cognitive scores were adjusted for age. The dependent variables, namely Task A and Task B scores evaluated separately as the dependent variables, with genotypes for individual SNPs as the predictor variables, with gender and diagnosis as covariates.
RESULTS
Quality Control
Over all, more than, 90% genotype data was available for each of the markers in the entire sample set (601 case-parent trios and 468 controls). All markers were in Hardy-Weinberg equilibrium (p>0.01). Four SNPs (rs2134655, rs324030, rs6280 and rs905568 in DRD3 ) were genotyped twice (with different methods) in 123 families reported in the present and a previously published study (Talkowski et al., 2006) and two other SNPs (rs1108580 in DBH and rs4680 in COMT) were common for 208 samples analyzed in another study (Srivastava et al., 2010). The overall genotyping call discordance rate for these SNPs was 3.3%. For the discrepant genotypes, the genotypes generated by SNPlex methodology were checked using Sanger sequencing and were found to be correct.
Linkage disequilibrium (LD)
Using a cutoff value of r2 > 0.8, eight out of eleven tag SNPs for COMT, 15 out of 18 tag SNPs for DBH, six out of seven tag SNPs for DRD3, nine out of ten tag SNPs for SLC18A2 and 13 tag SNPs for SLC6A3 were determined to be non-redundant. These SNPs (n=51, see below) were used for multi-marker/ haplotype analysis tests. Pattern of LD for DRD3 for the north Indian controls used in this study, as well as all Phase II hapmap populations (http://www.hapmap.org/) is provided in Supplementary Figure I. The patterns of LD for all the five genes were generally similar in the Indian and Caucasian datasets (Basu et al., 2003).
Single SNP associations
Allelic associations were observed with four SNPs namely rs363399 C/T (p = 0.05) in SLC18A2, rs6271C/T (p = 0.004) in DBH and rs4680 A/G (p = 0.05) [OR(95%CI) 1.19[1.0 −1.42]; rs9332377 T/C (p = 0.02) [OR (95%CI) 1.34[1.06-1.71], both in COMT, in case-control analysis. Following family based analysis, allelic associations were observed with five SNPs (rs7631540 and rs2046496 in DRD3; rs10082463 in SLC18A2; rs4646315 in COMT and rs6271 in DBH). Thus, for rs6271 in DBH, nominal associations were noted with both types of analyses. Trends for association were observed at six other SNPs. SNPs with nominally significant associations (uncorrected p<0.05) or trends of association (p<0.1) are presented in Table I, but none remained associated after corrections for multiple comparisons. Allele frequencies for all the analyzed SNPs are presented in Supplementary Table I.
Table I. SNPs in dopamine genes with suggestive associations in family based and case-control analyses.
| Chr | GENE | SNP | position | N | F_ca | F_co | Z1 | P1 | Z2 | P2 |
|---|---|---|---|---|---|---|---|---|---|---|
| 3 | DRD3 | rs7631540 | 115313209 | C | 0.555 | 0.554 | 0.052 | 0.959 | 2.216 | 0.027 |
| 3 | DRD3 | rs2046496 | 115317621 | C | 0.557 | 0.553 | 0.165 | 0.868 | 2.765 | 0.006 |
| 3 | DRD3 | rs2134655 | 115340891 | G | 0.760 | 0.758 | 0.097 | 0.923 | 1.812 | 0.070 |
| 3 | DRD3 | rs324030 | 115364131 | C | 0.616 | 0.653 | −1.722 | 0.084 | −1.134 | 0.257 |
| 5 | SLC6A3 | rs403636 | 1491354 | G | 0.876 | 0.848 | 1.872 | 0.058 | −0.187 | 0.852 |
| 9 | DBH | rs6271 | 135512095 | C | 0.998 | 0.988 | 2.686 | 0.004 | 2.309 | 0.021 |
| 10 | SLC18A2 | rs363399 | 118998861 | T | 0.677 | 0.633 | 2.036 | 0.047 | 1.837 | 0.066 |
| 10 | SLC18A2 | rs363338 | 118999379 | T | 0.599 | 0.560 | 1.798 | 0.075 | 1.249 | 0.212 |
| 10 | SLC18A2 | rs10082463 | 119011397 | A | 0.810 | 0.841 | −1.864 | 0.067 | −2.085 | 0.037 |
| 10 | SLC18A2 | rs363285 | 119029149 | A | 0.844 | 0.817 | 1.627 | 0.093 | −0.260 | 0.795 |
| 22 | COMT | rs4680 | 18331271 | G | 0.543 | 0.586 | −1.966 | 0.049 | 0.785 | 0.433 |
| 22 | COMT | rs4646315 | 18331897 | G | 0.766 | 0.753 | 0.685 | 0.483 | 2.756 | 0.006 |
| 22 | COMT | rs9332377 | 18335692 | C | 0.824 | 0.863 | −2.447 | 0.016 | −1.025 | 0.305 |
N: major allele in north Indian cohort
F_ca, F_co: frequency of cases and controls respectively
Z1, Z2: z scores for case-control comparisons and family based associations, respectively
P1, P2: p values for case-control comparisons and family based associations, respectively
Position: Nucleotide location based on NCBI build 36.3
Multi-marker associations
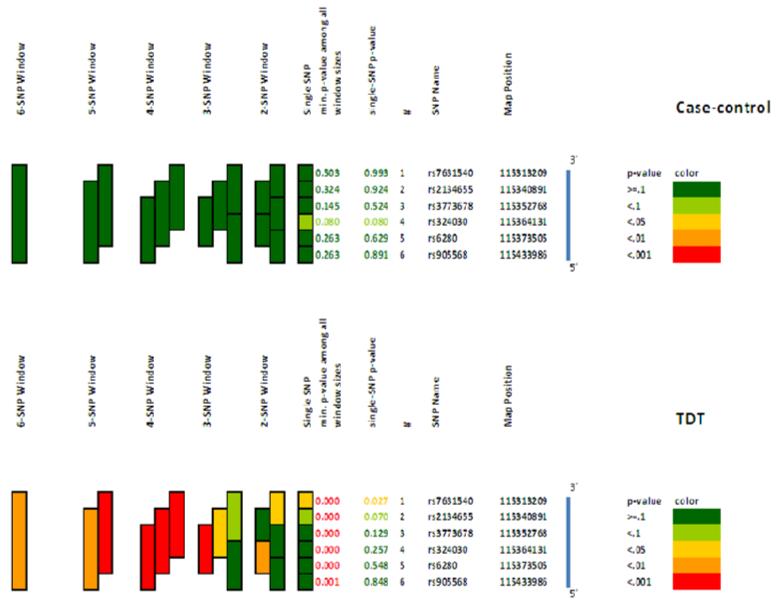
Following sliding window analysis, several trends were observed for global p-values using UNPHASED (Table II, Supplementary Table II). However, only family based haplotype associations at DRD3 remained significant following corrections for multiple comparisons (Table II). They included 2, 3, 4, 5 and 6 SNP haplotypes beginning with rs7631540, a SNP that was also associated with SZ when analyzed individually (Tables II). For each of the associated windows, estimated haplotype frequencies, the number of transmissions and TDT statistics generated using PLINK and significant (p<0.05) transmissions for each of the windows are presented in Supplementary Table III. To evaluate consistency in nomenclature of SNPs, in this disease associated gene, selected control individuals (n=50) were sequenced across the following SNPs: rs7631540, rs2134655, rs3773678, rs324030, rs6280 and rs905568. Rs2046496 was not sequenced as it is tagged to rs7631540 (r2= 0.99) which is further in the regulatory 3′ region.
TABLE II. Sliding window based haplotypes of DRD3 markers showing significant associations in Case-control and TDT analyses.
| C-C(601/468) | Trio (n=601 ) | Addl. Trio (n=118) | Pooled Trio (n=719) | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # | Map Information | SNP | p,df=1* | 2 – mhap , df=3 | 3 – mhap, p(df) | 4 – mhap , p(df) | 5 – mhap , p(df) | 6 - mhap, p(df) | ,df=1* | 2 – mhap (df) | 3 – mhap , p(df) | 4 – mhap , p(df) | 5 – mhap , p(df) | 6 – mhap , p(df) | ,df=1* | 2 – mhap df=3 | 3 - mhap, p(df) | 4 - mhap, p(df) | 5 - mhap, p(df) | 6 - mhap, p(df) | ,df=1* | 2 – mhap df=3 | 3 - mhap, p(df) | 4 - mhap, p(df) | 5 - mhap, p(df) | 6 - mhap, p(df) |
| DRD3 | ||||||||||||||||||||||||||
| 1 M | 115313209 | rs7631540 | 0.99 | 0.93(2) | 0.554) | 0.44(7) | 0.60(9) | 0.47(13) | 0.03 | 0.01(2) | 0.06(4) | 6×10−4(8) | 5×10− 7(13)** |
0.008(17) | 0.93 | 4×10−3(3) | 0.03(5) | 0.21(9) | 0.20(10) | 0.37(18) | 0.04 | 1×10− 4(3)** |
0.004(5) | 0.23(8) | 2×10− 4(17) |
0.01(21) |
| 2 M | 115340891 | rs2134655 | 0.92 | 0.83(2) | 0.26(4) | 0.31(5) | 0.40(11) | 0.07 | 0.11(2) | 0.03(5) | 1×10−4(9)** | 0.007(13) | 0.62 | 0.20(3) | 0.50(6) | 0.41(8) | 0.36(15) | 0.07 | 0.12(2) | 0.46(5) | 0.13(11) | 0.14(18) | ||||
| 3 M | 115352768 | rs3773678 | 0.52 | 0.14(3) | 0.16(4) | 0.40(9) | 0.13 | 0.28(3) | 1×10− 5(6)** |
7×10− 4(12)** |
0.83 | 0.89(3) | 0.07(7) | 0.25(11) | 0.15 | 0.38(3) | 0.14(7) | 0.09(13) | ||||||||
| 4 M | 115364131 | rs324030 | 0.08 | 0.29(3) | 0.31(6) | 0.26 | 4×10− 3(3) |
0.13(6) | 0.77 | 0.006(3) | 0.03(7) | 0.25 | 0.17(3) | 0.30(7) | ||||||||||||
| 5 M | 115373505 | rs6280 | 0.63 | 0.85(3) | 0.55 | 0.82(3) | 0.57 | 0.15(3) | 0.75 | 0.56(3) | ||||||||||||||||
| 6 M | 115433986 | rs905568 | 0.89 | 0.85 | 0.18 | 0.67 | ||||||||||||||||||||
C-C: case-control
Trio: family consisting of one proband and two parents
Addl. Trio: a replicate set of trio families
Pooled Trio: total of trios and addition trios
2 – mhap: two marker haplotypes generated using UNPHASED. P-values given; 3-mhap: three marker haplotypes generated using UNPHASED. P-values (degree of freedom) given. Similarly, mhap-3, -4, -5 and -6 denote haplotypes incorporating the respective number of SNPs. Haplotypes with a frequency lower than 1% were not included in the analysis.
p values based on analysis of individual SNPs (see Table 1).
p values significant after Bonferroni correction of global p value for the respective haplotype (alpha value= 0.0001 for 505 tests)
Analysis of additional independent trio sample (n=119) for DRD3
In view of the highly significant haplotypic associations of DRD3 markers in family analysis, the results were further tested in a smaller family sample set available to us from the same population. Though no significant associations with individual SNPs were observed, associations in the 2, 3 SNP windows comprising the 3′ region SNPs was observed, (Table II).
Comparison with published analyses of Caucasian samples
Comparison of results from this study with that reported for two US Caucasian samples in a prior study (Talkowski et al., 2008) showed 22 SNPs which were associated in either of the populations but none of them showed consistent association in the Caucasian and Indian populations (Supplementary Table IV). However, only with four SNPs namely rs2134655 and rs324030 in DRD3; rs403636 in SLC6A3 and rs363338 in SLC18A2, associated in Caucasian samples, a trend towards association in the Indian samples was observed (Supplementary Table IV). Notably except for rs363338, the other three markers showed association with the opposite allele (Supplementary Table IV).
Power of the study
For the trio, as well as the case-control sample, the power exceeds 80% for an odds ratio of 1.4 if the SNP frequency is between 20-50%.
Cognitive analyses
Taking all the associated SNPs from table I as covariates and Task A and B as outcome variables separately, rs363285 (p=0.025) from SLC18A2 was associated with Task B of TMT. We also analyzed SNPs having a p<0.05 in only family data for cognitive analysis. These included rs2046496 (in LD to rs7631540) from DRD3; rs6271 from DBH; rs10082463 from SLC18A2 and rs4646315 from COMT. Only rs10082463 (p=0.027) from SLC18A2 showed significant association with Task A (Supplementary Table V), but was not significant following multiple corrections.
DISCUSSION
We conducted case-control and family based association analyses of key dopamine genes in a north Indian SZ sample set. Family based analyses overcome artifacts related to population sub-structure as they have an inherent correction for the confounding effects of population stratification since the cases are compared to family based controls (Seltman et al., 2001; Lange et al., 2008). On the other hand, case-control analyses can have greater power in certain settings (Bacanu et al., 2000). Therefore, associations detected with each method can be credible, provided replications are available and plausible function can be attributed. Following individual analysis of each of 59 SNPs, significant associations were noted in both case-control and family based analyses at rs6271 (+1603C→T), a relatively infrequent non-synonymous SNP located in exon 11 of DBH which encodes a non-conservative amino acid change (arg535cys). In view of the low frequency of the minor allele at this SNP, there are relatively few informative transmissions from heterozygous parents, thus diminishing power for the TDT analysis (supplementary Table I). Hence the results should be interpreted with caution. In the same vein, a marginal association with rs4680, an exonic SNP at COMT was noted in our case-control sample and has also been reported by several other investigators (Handoko et al., 2005;Nunokawa et al., 2010;Liao et al., 2009; Okochi et al., 2009; Bhakta et al., 2012; Singh et al., 2012). Apart from these exonic variants, nominally significant SNP associations were also noted at all the other genes tested, except at SLC6A3 (Table I) in either case-control or family based analysis. A larger Indian sample may be required for confirmation.
Occasionally, distinct genetic associations of single SNPs may not be observed but subtle contributions by a combination or a set of SNPs may be found using haplotype based analysis (Haig, 2011). Though a potential disadvantage of this strategy is increased number of multiple tests resulting in type II error (Hunter & Kraft, 2007), haplotype based analyses in north Indian samples are important since in this study, a “best guess selection” was made for tag SNPs based on available data from Hapmap CEU populations (Pemberton et al., 2008; Indian Genome Variation Consortium, 2009; Reich et al., 2009). Using the multi-SNP sliding window approach, notable haplotype based association was observed at DRD3 (Figure 1). Since rare alleles and haplotypes have shown association with Schizophrenia (Li et al., 2005; Agim et al., 2013) the rare haplotypes frequency was set at 1% for the global haplotypes. Haplotype windows beginning rs3773678, rs2134655 and rs7631540 were significantly associated, suggesting the presence of risk factors in the 3′ region of this gene. Four of the five associated haplotype windows comprised rs6280, an exonic SNP previously nominated as a SZ risk factor (Jonsson et al., 2003; Ma et al., 2008; Hwang et al., 2010; Utsunomiya et al., 2012) was observed for DRD3. However, none of the associated haplotypes commenced with rs6280 (Table II). Since we used HapMap Caucasian data to select tag SNPs, it is possible that there may be some key SNP present in the Indian sample, which were excluded due to population differences. Nevertheless, our results are credible because we used a family based approach that is not prone to artifacts following sub-structure. We also applied conservative corrections for multiple comparisons, and similar results were obtained by an independent NM method of analysis. Furthermore, similar trends of haplotype based associations were observed in another smaller, independent sample ascertained identically (n=119 trio families, Table II). The pooled (n=719, 601+119) family data for DRD3 showed similar associations in all sliding window sizes (1-6), comprising the 3′ SNPs. The 2 and 3 SNP windows starting with rs7631540 showed stronger association (P= 0.0001; p= 0.004 respectively), but the association is weaker in the 4 SNP window after pooling the families for DRD3 (Table II).
Figure I.
Haplotype based associations at DRD3 illustrated using Graphical Assessment of sliding p-values (GrASP)
Analogous haplotype based associations have been reported at DRD3 in a Spanish Galician sample (Dominguez et al., 2007). This group reported a 3-SNP haplotype comprising rs7631540 as being the most significantly associated after corrections in case-control analysis. Notably, nominally significant associations with haplotypes comprising rs2134655 & rs7631540 were also noted in our family sample set. Based on case-control analyses, Costas and colleagues recently proposed an operative positive selection for a common DRD3 haplotype in three independent schizophrenia samples of European origin (Costas et al., 2009). They also concluded that this particular “common but protective haplotype in Europeans” is at intermediate frequencies in other populations, being at the lowest in Sub-Saharan African populations. Significant association was not observed in an updated meta analysis (Nunokawa et al., 2010) for the haplotype T–T–T–G for the SNPs rs7631540–rs1486012–rs2134655–rs963468 reported by Costas and colleagues but found it less frequently in patients than in control subjects (26.5% vs. 28.6%). Differences between our sample and the European samples may reflect population differences or sample selection.
Of the 59 SNPs studied here, comparable association data were available for 46 SNPs, from our prior analyses for US and Bulgarian (Caucasian) samples (Talkowski et al., 2008). Single SNP associations reported in US sample were not detected in the Indian samples, apart from suggestive trends for rs2134655 and rs324030 at DRD3; rs403636 at SLC6A3 and rs363338 at SLC18A2. Indeed, different alleles were over-represented among cases in the Indian and the Caucasian data sets for the first three SNPs (Supplementary Table IV). These differences were confirmed by resequencing rs7631540, rs2134655, rs3773678, rs324030, rs6280 and rs905568 at DRD3 in 100 chromosomes in our Indian control samples. Such differences and the lack of consistent cross-national replication may indicate a lack of significant SZ risk variants at these loci. Factors such as differences in power and specific differences in LD structure may also contribute to the inconsistency. Other variables such as age dependent associations or subtle differences in allele frequencies could also impact on replicability (Lasky-Su et al., 2008; Liu et al., 2008; Greene et al., 2009). It is also possible that as yet undetected common or rare alleles confer risk for SZ, but were not genotyped in any of the populations. This possibility is supported by our haplotype based analyses at DRD3 and locus overlap observed with reports from other populations (Staddon et al., 2005; Dominguez et al., 2007; Costas et al., 2009).
While the influence of dopamine on cognition is clearly complex (Takahashi et al., 2008) and may not provide a complete account for the full range of cognitive deficits in schizophrenia, the importance of dopamine to cognition has been demonstrated by studies in which dopamine levels and activity were modulated using different types of pharmacological agents (Condray & Yao, 2011). Administration of amphetamine, improves cognitive performance working memory, attention, and language production processes (Barch & Carter, 2005) and was associated with the significant changes in regional cerebral blood flow (rCBF) among schizophrenia patients receiving antipsychotic medication (Daniel et al., 1991). Administration of the dopamine precursor L-3,4-dihydroxyphenylalanine (L-DOPA) to healthy volunteers produced more focused activation of semantic memory, reflected in reduced indirect priming (Kischka et al., 1996), and increased the rate of learning and long-term retention of an artificial vocabulary (Knecht et al., 2004).
Previously COMT Val-Met polymorphism has most widely been studied in relation to cognitive functions (Alfimova et al., 2007; Rosa et al., 2010; Solis-Ortiz et al., 2010) with mixed results. On the other hand, an exonic SNP at DRD3 (rs6280) has been reported to be associated with executive function, but only in individuals with psychoses (Bombin et al., 2008). This association was not observed in our sample. Only two SNPs, namely rs363285 and rs10082463 of SLC18A2 showed significant association in our samples with task B and A respectively. This may be partly due to the relatively small sample. Another SNP rs363227 from SLC18A2 which was not associated in our sample set has been shown to be associated for Psychotic disorder in a Dutch population and their unaffected siblings (p=0.04). SNP rs10082463 is in LD to rs363227 (D’0.87; r2=0.61).
In conclusion, we report on a systematic analysis of SNPS at five DA genes with SZ. An exonic SNP at DBH (rs6271) was nominally in both the case-control and family based analyses in our population and several others were nominally significant. We also report on suggestive haplotype based associations at DRD3 in our ethnically distinct population, which is in consonance with other independent studies. Two SNPs (rs10082463 & rs363285) of SLC18A2 were nominally associated with cognition. None of the associations remained significant following corrections for multiple comparisons except a DRD3 2-Marker haplotypes (rs324030-rs6280; p=0.00004). Analysis of larger samples is warranted.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr Michael Talkowski for helpful advice in designing the study and Dr. Pushplata Prasad with analysis of the additional 100 trio families. We are grateful for the sample collection and DNA isolation by trained and dedicated staff at RML hospital and at University of Delhi, South Campus respectively. Genotyping and sequencing facilities at Central Instrumentation Facility, University of Delhi, South Campus is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akey J, Jin L, Xiong M. Haplotypes vs single marker linkage disequilibrium tests: what do we gain? European Journal of Human Genetics. 2001;9:291–300. doi: 10.1038/sj.ejhg.5200619. [DOI] [PubMed] [Google Scholar]
- Alfimova MV, Golimbet VE, Gritsenko IK, Lezheiko TV, Abramova LI, Strel’tsova MA, Khlopina IV, Ebstein R. Interaction of dopamine system genes and cognitive functions in patients with schizophrenia and their relatives and in healthy subjects from the general population. Neuroscience and Behavioral Physiology. 2007;37:643–650. doi: 10.1007/s11055-007-0064-x. [DOI] [PubMed] [Google Scholar]
- Allan CL, Cardno AG, McGuffin P. Schizophrenia: from genes to phenes to disease. Current Psychiatry Reports. 2008;10:339–343. doi: 10.1007/s11920-008-0054-x. [DOI] [PubMed] [Google Scholar]
- Bacanu SA, Devlin B, Roeder K. The power of genomic control. The American Journal of Human Genetics. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barch DM, Carter CS. Amphetamine improves cognitive function in medicated individuals with schizophrenia and in healthy volunteers. Schizophrenia Research. 2005;77:43–58. doi: 10.1016/j.schres.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Barnett JH, Jones PB, Robbins TW, Müller U. Effects of the catechol-O-methyl transferase Val158Met polymorphism on executive function: a meta-analysis of the Wisconsin Card Sort Test in schizophrenia and healthy controls. Mol Psychiatry. 2007;12:502–509. doi: 10.1038/sj.mp.4001973. [DOI] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S, Majumder PP. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Research. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino A, Taurisano P, Pisciotta NM, Blasi G, Fazio L, Romano R, Gelao B, Lo Bianco L, Lozupone M, Di Giorgio A, Caforio G, Sambataro F, Niccoli-Asabella A, Papp A, Ursini G, Sinibaldi L, Popolizio T, Sadee W, Rubini G. Genetically determined measures of striatal D2 signaling predict prefrontal activity during working memory performance. PLoS One. 2010;5:e9348. doi: 10.1371/journal.pone.0009348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakta SG, Zhang JP, Malhotra AK. The COMT Met158 allele and violence in schizophrenia: a meta-analysis. Schizophr Res. 2012;140:192–197. doi: 10.1016/j.schres.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia T, Garg K, Pogue-Geile M, Nimgaonkar VL, Deshpande SN. Executive functions and cognitive deficits in schizophrenia: comparisons between probands, parents and controls in India. Journal of Postgraduate Medicine. 2009;55:3–7. doi: 10.4103/0022-3859.43546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia T, Shriharsh V, Adlakha S, Bisht V, Garg K, Deshpande SN. The trail making test in India. Indian Journal of Psychiatry. 2007;49:113–116. doi: 10.4103/0019-5545.33258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia T, Thomas P, Semwal P, Thelma BK, Nimgaonkar VL, Deshpande SN. Differing correlates for suicide attempts among patients with schizophrenia or schizoaffective disorder in India and USA. Schizophrenia Research. 2006;86:208–214. doi: 10.1016/j.schres.2006.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombin I, Arango C, Mayoral M, Castro-Fornieles J, Gonzalez-Pinto A, Gonzalez-Gomez C, Moreno D, Parellada M, Baeza I, Graell M, Otero S, Saiz PA, Patino-Garcia A. DRD3, but not COMT or DRD2, genotype affects executive functions in healthy and first-episode psychosis adolescents. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B:873–879. doi: 10.1002/ajmg.b.30710. [DOI] [PubMed] [Google Scholar]
- Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, B KT, Ferrell RE, Middleton FA, Devlin B, Levitt P, Lewis DA, Nimgaonkar VL. Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Human Molecular Genetics. 2002;11:1373–1380. doi: 10.1093/hmg/11.12.1373. [DOI] [PubMed] [Google Scholar]
- Condray R, Yao JK. Cognition, dopamine and bioactive lipids in schizophrenia. Frontiers in bioscience (Scholar edition) 2011;3:298–330. doi: 10.2741/s153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas J, Carrera N, Dominguez E, Vilella E, Martorell L, Valero J, Gutierrez-Zotes A, Labad A, Carracedo A. A common haplotype of DRD3 affected by recent positive selection is associated with protection from schizophrenia. Human Genetics. 2009;124:607–613. doi: 10.1007/s00439-008-0584-7. [DOI] [PubMed] [Google Scholar]
- Daniel DG, Weinberger DR, Jones DW, Zigun JR, Coppola R, Handel S, Bigelow LB, Goldberg TE, Berman KF, Kleinman JE. The effect of amphetamine on regional cerebral blood flow during cognitive activation in schizophrenia. Journal of Neuroscience. 1991;11:1907–1917. doi: 10.1523/JNEUROSCI.11-07-01907.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genetics. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Deshpande SN, Mathur MN, Das SK, Bhatia T, Sharma S, Nimgaonkar VL. A Hindi version of the Diagnostic Interview for Genetic Studies. Schizophrenia Bulletin. 1998;24:489–493. doi: 10.1093/oxfordjournals.schbul.a033343. [DOI] [PubMed] [Google Scholar]
- Dettling M, Cascorbi I, Opgen-Rhein C, Schaub R. Clozapine-induced agranulocytosis in schizophrenic Caucasians: confirming clues for associations with human leukocyte class I and II antigens. Pharmacogenomics J. 2007 Oct;7(5):325–32. doi: 10.1038/sj.tpj.6500423. [DOI] [PubMed] [Google Scholar]
- Dominguez E, Loza MI, Padin F, Gesteira A, Paz E, Paramo M, Brenlla J, Pumar E, Iglesias F, Cibeira A, Castro M, Caruncho H, Carracedo A, Costas J. Extensive linkage disequilibrium mapping at HTR2A and DRD3 for schizophrenia susceptibility genes in the Galician population. Schizophrenia Research. 2007;90:123–129. doi: 10.1016/j.schres.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Human Heredity. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Ganguli HC. Epidemiological findings on prevalence of mental disorders in India. Indian Journal of Psychiatry. 2000;42:14–20. [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Morrison JM. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydra.usc.edu/gxe.
- Gottesman II, Shields J. Schizophrenia: The Epigenetic Puzzle. Cambridge University Press; Cambridge, UK.: 1982. [Google Scholar]
- Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4:e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Does heritability hide in epistasis between linked SNPs? European Journal of Human Genetics. 2011;19:123. doi: 10.1038/ejhg.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko HY, Nyholt DR, Hayward NK, Nertney DA, Hannah DE, Windus LC, McCormack CM, Smith HJ, Filippich C, James MR, Mowry BJ. Separate and interacting effects within the catechol-O-methyltransferase (COMT) are associated with schizophrenia. Molecular Psychiatry. 2005;10:589–597. doi: 10.1038/sj.mp.4001606. [DOI] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P. Drinking from the fire hose--statistical issues in genomewide association studies. New England Journal of Medicine. 2007;357:436–439. doi: 10.1056/NEJMp078120. [DOI] [PubMed] [Google Scholar]
- Hwang R, Zai C, Tiwari A, Müller DJ, Arranz MJ, Morris AG, McKenna PJ, Munro J, Potkin SG, Lieberman JA, Meltzer HY, Kennedy JL. Effect of dopamine D3 receptor gene polymorphisms and clozapine treatment response: exploratory analysis of nine polymorphisms and meta-analysis of the Ser9Gly variant. Pharmacogenomics J. 2010;10:200–218. doi: 10.1038/tpj.2009.65. [DOI] [PubMed] [Google Scholar]
- Indian Genome Variation Consortium Genetic landscape of the people of India: a canvas for disease gene exploration. Journal of Genetics. 2008;87:3–20. doi: 10.1007/s12041-008-0002-x. [DOI] [PubMed] [Google Scholar]
- Jia P, Wang L, Meltzer HY, Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophrenia Research. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA. Haplotype tagging for the identification of common disease genes. Nature Genetics. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Flyckt L, Burgert E, Crocq MA, Forslund K, Mattila-Evenden M, Rylander G, Asberg M, Nimgaonkar VL, Edman G, Bjerkenstedt L, Wiesel FA, Sedvall GC. Dopamine D3 receptor gene Ser9Gly variant and schizophrenia: association study and meta-analysis. Psychiatric Genetics. 2003;13:1–12. doi: 10.1097/00041444-200303000-00001. [DOI] [PubMed] [Google Scholar]
- Kamatani N, Sekine A, Kitamoto T, Iida A, Saito S, Kogame A, Inoue E, Kawamoto M, Harigai M, Nakamura Y. Large-scale single-nucleotide polymorphism (SNP) and haplotype analyses, using dense SNP Maps, of 199 drug-related genes in 752 subjects: the analysis of the association between uncommon SNPs within haplotype blocks and the haplotypes constructed with haplotype-tagging SNPs. The American Journal of Human Genetics. 2004;75:190–203. doi: 10.1086/422853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellendonk C, Simpson EH, Polan HJ, Malleret G, Vronskaya S, Winiger V, Moore H, Kandel ER. Transient and selective overexpression of dopamine D2 receptors in the striatum causes persistent abnormalities in prefrontal cortex functioning. Neuron. 2006;49:603–615. doi: 10.1016/j.neuron.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Keshavan MS, Tandon R, Boutros NN, Nasrallah HA. Schizophrenia, “just the facts”: what we know in 2008 Part 3: neurobiology. Schizophrenia Research. 2008;106:89–107. doi: 10.1016/j.schres.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Kischka U, Kammer T, Maier S, Weisbrod M, Thimm M, Spitzer M. Dopaminergic modulation of semantic network activation. Neuropsychologia. 1996;34:1107–1113. doi: 10.1016/0028-3932(96)00024-3. [DOI] [PubMed] [Google Scholar]
- Knecht S, Breitenstein C, Bushuven S, Wailke S, Kamping S, Floel A, Zwitserlood P, Ringelstein EB. Levodopa: faster and better word learning in normal humans. Annals Of Neurology. 2004;56:20–26. doi: 10.1002/ana.20125. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genetic Epidemiology. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lange EM, Sun J, Lange LA, Zheng SL, Duggan D, Carpten JD, Gronberg H, Isaacs WB, Xu J, Chang BL. Family-based samples can play an important role in genetic association studies. Cancer Epidemiology, Biomarkers & Prevention. 2008;17:2208–2214. doi: 10.1158/1055-9965.EPI-08-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky-Su J, Anney RJ, Neale BM, Franke B, Zhou K, Maller JB, Vasquez AA, Chen W, Asherson P, Buitelaar J, Banaschewski T, Ebstein R, Gill M, Miranda A, Mulas F, Oades RD, Roeyers H, Rothenberger A, Sergeant J, Sonuga-Barke E, Steinhausen HC, Taylor E, Daly M, Laird N, Lange C, Faraone SV. Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B:1355–1358. doi: 10.1002/ajmg.b.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proceedings of the National Academy of Sciences. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao SY, Lin SH, Liu CM, Hsieh MH, Hwang TJ, Liu SK, Guo SC, Hwu HG, Chen WJ. Genetic variants in COMT and neurocognitive impairment in families of patients with schizophrenia. Genes Brain Behav. 2009;8:228–237. doi: 10.1111/j.1601-183X.2008.00467.x. [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Bjork C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. The American Journal of Human Genetics. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nature Genetics. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Paterson AD, Szatmari P. Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biological Psychiatry. 2008;64:561–570. doi: 10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, He Z, Fang W, Tang W, Huang K, Li Z, He G, Xu Y, Feng G, Zheng T, Zhou J, He L, Shi Y. The Ser9Gly polymorphism of the dopamine D3 receptor gene and risk of schizophrenia: an association study and a large meta-analysis. Schizophrenia Research. 2008;101:26–35. doi: 10.1016/j.schres.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Mansour HA, Talkowski ME, Wood J, Pless L, Bamne M, Chowdari KV, Allen M, Bowden CL, Calabrese J, El-Mallakh RS, Fagiolini A, Faraone SV, Fossey MD, Friedman ES, Gyulai L, Hauser P, Ketter TA, Loftis JM, Marangell LB, Miklowitz DJ, Nierenberg AA, Patel J, Sachs GS, Sklar P, Smoller JW, Thase ME, Frank E, Kupfer DJ, Nimgaonkar VL. Serotonin gene polymorphisms and bipolar I disorder: focus on the serotonin transporter. Ann Med. 2005;37:590–602. doi: 10.1080/07853890500357428. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Gao P, Goldstein JL, Wilson AF, Pugh EW, Furbert-Harris P, Dunston GM, Malveaux FJ, Togias A, Barnes KC, Beaty TH, Huang SK. A graphical assessment of p-values from sliding window haplotype tests of association to identify asthma susceptibility loci on chromosome 11q. BMC Genet. 2006;7:38. doi: 10.1186/1471-2156-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell E, Family interview for genetic studies (FIGS) Intramural Research Program – clinical neurogenetics branch. National Institute of Mental Health; Rockville: 1992. [Google Scholar]
- Mowry BJ, Gratten J. The emerging spectrum of allelic variation in schizophrenia: current evidence and strategies for the identification and functional characterization of common and rare variants. Mol Psychiatry. 2013;18(1):38–52. doi: 10.1038/mp.2012.34. [DOI] [PubMed] [Google Scholar]
- Ng MY, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T, Riley B, Paunio T, Pulver AE, Irmansyah, Holmans PA, Escamilla M, Wildenauer DB, Williams NM, Laurent C, Mowry BJ, Brzustowicz LM, Maziade M, Sklar P, Garver DL, Abecasis GR, Lerer B, Fallin MD, Gurling HM, Gejman PV, Lindholm E, Moises HW, Byerley W, Wijsman EM, Forabosco P, Tsuang MT, Hwu HG, Okazaki Y, Kendler KS, Wormley B, Fanous A, Walsh D, O’Neill FA, Peltonen L, Nestadt G, Lasseter VK, Liang KY, Papadimitriou GM, Dikeos DG, Schwab SG, Owen MJ, O’Donovan MC, Norton N, Hare E, Raventos H, Nicolini H, Albus M, Maier W, Nimgaonkar VL, Terenius L, Mallet J, Jay M, Godard S, Nertney D, Alexander M, Crowe RR, Silverman JM, Bassett AS, Roy MA, Merette C, Pato CN, Pato MT, Roos JL, Kohn Y, Amann-Zalcenstein D, Kalsi G, McQuillin A, Curtis D, Brynjolfson J, Sigmundsson T, Petursson H, Sanders AR, Duan J, Jazin E, Myles-Worsley M, Karayiorgou M, Lewis CM. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Molecular Psychiatry. 2009;14:774–785. doi: 10.1038/mp.2008.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, Stephens M, Bustamante CD. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunokawa A, Watanabe Y, Kaneko N, Sugai T, Yazaki S, Arinami T, Ujike H, Inada T, Iwata N, Kunugi H, Sasaki T, Itokawa M, Ozaki N, Hashimoto R, Someya T. The dopamine D3 receptor (DRD3) gene and risk of schizophrenia: case-control studies and an updated meta-analysis. Schizophrenia Research. 2010;116:61–67. doi: 10.1016/j.schres.2009.10.016. [DOI] [PubMed] [Google Scholar]
- O’Donovan MC, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Giegling I, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Adolfsson R, Osby U, Terenius L, Jonsson EG, Cichon S, Nothen MM, Gill M, Corvin AP, Rujescu D, Gejman PV, Kirov G, Craddock N, Williams NM, Owen MJ. Analysis of 10 independent samples provides evidence for association between schizophrenia and a SNP flanking fibroblast growth factor receptor 2. Molecular Psychiatry. 2009;14:30–36. doi: 10.1038/mp.2008.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okochi T, Ikeda M, Kishi T, Kawashima K, Kinoshita Y, Kitajima T, Yamanouchi Y, Tomita M, Inada T, Ozaki N, Iwata N. Meta-analysis of association between genetic variants in COMT and schizophrenia: an update. Schizophrenia Research. 2009;110:140–148. doi: 10.1016/j.schres.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Owen MJ, O’Donovan MC, Gottesman II. editor Schizophrenia. Oxford UniversityPress; Oxford U.K.: 2002. [Google Scholar]
- Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N, Brun M, Szelinger S, Coon KD, Zismann VL, Webster JA, Beach T, Sando SB, Aasly JO, Heun R, Jessen F, Kolsch H, Tsolaki M, Daniilidou M, Reiman EM, Papassotiropoulos A, Hutton ML, Stephan DA, Craig DW. Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. The American Journal of Human Genetics. 2007;80:126–139. doi: 10.1086/510686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Jakobsson M, Conrad DF, Coop G, Wall JD, Pritchard JK, Patel PI, Rosenberg NA. Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India. Ann Human Genetics. 2008;72:535–546. doi: 10.1111/j.1469-1809.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones RM, Calderin YC, Dominguez M, Bravo TM, Berazain AR, Garcia A, Caballero A, Reyes MM. Heritability of Trail Making Test performance in multiplex schizophrenia families: implications for the search for an endophenotype. Eur Arch Psychiatry Clin Neurosci. 2009;259:475–481. doi: 10.1007/s00406-009-0012-6. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa EC, Dickinson D, Apud J, Weinberger DR, Elvevag B. COMT Val158Met polymorphism, cognitive stability and cognitive flexibility: an experimental examination. Behavioral and Brain Functions. 2010;6:53. doi: 10.1186/1744-9081-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmond DA, Weickert CS, Webster MJ. Developmental changes in human dopamine neurotransmission: cortical receptors and terminators. BMC Neurosci. 2012;13:18. doi: 10.1186/1471-2202-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Chant D, Welham J, McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2:e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P. Atypical antipsychotics: mechanism of action. Can J Psychiatry. 2002;47:27–38. [PubMed] [Google Scholar]
- Seeman P, Kapur S. Schizophrenia: more dopamine, more D2 receptors. Proceedings of the National Academy of Sciences. 2000;97:7673–7675. doi: 10.1073/pnas.97.14.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltman H, Roeder K, Devlin B. Transmission/disequilibrium test meets measured haplotype analysis: family-based association analysis guided by evolution of haplotypes. The American Journal of Human Genetics. 2001;68:1250–1263. doi: 10.1086/320110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh JP, Volavka J, Czobor P, Van Dorn RA. A meta-analysis of the Val158Met COMT polymorphism and violent behavior in schizophrenia. PLoS One. 2012;7:e43423. doi: 10.1371/journal.pone.0043423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Smoller JW, Fan J, Ferreira MA, Perlis RH, Chambert K, Nimgaonkar VL, McQueen MB, Faraone SV, Kirby A, de Bakker PI, Ogdie MN, Thase ME, Sachs GS, Todd-Brown K, Gabriel SB, Sougnez C, Gates C, Blumenstiel B, Defelice M, Ardlie KG, Franklin J, Muir WJ, McGhee KA, MacIntyre DJ, McLean A, VanBeck M, McQuillin A, Bass NJ, Robinson M, Lawrence J, Anjorin A, Curtis D, Scolnick EM, Daly MJ, Blackwood DH, Gurling HM, Purcell SM. Whole-genome association study of bipolar disorder. Molecular Psychiatry. 2008;13:558–569. doi: 10.1038/sj.mp.4002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Ortiz S, Perez-Luque E, Morado-Crespo L, Gutierrez-Munoz M. Executive functions and selective attention are favored in middle-aged healthy women carriers of the Val/Val genotype of the catechol-o-methyltransferase gene: a behavioral genetic study. Behavioral and Brain Functions. 2010;6:67. doi: 10.1186/1744-9081-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Deshpande SN, Thelma BK. Dopaminergic pathway gene polymorphisms and genetic susceptibility to schizophrenia among north Indians. Neuropsychobiology. 2010;61:64–70. doi: 10.1159/000265131. [DOI] [PubMed] [Google Scholar]
- Staddon S, Arranz MJ, Mancama D, Perez-Nievas F, Arrizabalaga I, Anney R, Buckland P, Elkin A, Osborne S, Munro J, Mata I, Kerwin RW. Association between dopamine D3 receptor gene polymorphisms and schizophrenia in an isolate population. Schizophrenia Research. 2005;73:49–54. doi: 10.1016/j.schres.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2:e212. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of General Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Lin D, Tzeng JY, van den Oord E, Perkins D, Stroup TS, Wagner M, Lee S, Wright FA, Zou F, Liu W, Downing AM, Lieberman J, Close SL. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Molecular Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kato M, Takano H, Arakawa R, Okumura M, Otsuka T, Kodaka F, Hayashi M, Okubo Y, Ito H, Suhara T. Differential contributions of prefrontal and hippocampal dopamine D(1) and D(2) receptors in human cognitive functions. Journal of Neuroscience. 2008;28:12032–12038. doi: 10.1523/JNEUROSCI.3446-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Kirov G, Bamne M, Georgieva L, Torres G, Mansour H, Chowdari KV, Milanova V, Wood J, McClain L, Prasad K, Shirts B, Zhang J, O’Donovan MC, Owen MJ, Devlin B, Nimgaonkar VL. A network of dopaminergic gene variations implicated as risk factors for schizophrenia. Human Molecular Genetics. 2008;17:747–758. doi: 10.1093/hmg/ddm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Mansour H, Chowdari KV, Wood J, Butler A, Varma PG, Prasad S, Semwal P, Bhatia T, Deshpande S, Devlin B, Thelma BK, Nimgaonkar VL. Novel, replicated associations between dopamine D3 receptor gene polymorphisms and schizophrenia in two independent samples. Biological Psychiatry. 2006;60:570–577. doi: 10.1016/j.biopsych.2006.04.012. [DOI] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, Wu PP, Wang Y, Spoonde AY, Koehler RT, Peyret N, Chen C, Broomer AJ, Ridzon DA, Zhou H, Hoo BS, Hayashibara KC, Leong LN, Ma CN, Rosenblum BB, Day JP, Ziegle JS, De La Vega FM, Rhodes MD, Hennessy KM, Wenz HM. The SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. Journal of Biomolecular Techniques. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya K, Shinkai T, Sakata S, Yamada K, Chen HI, De Luca V, Hwang R, Ohmori O, Nakamura J. Genetic association between the dopamine D3 receptor genepolymorphism (Ser9Gly) and tardive dyskinesia in patients with schizophrenia: a reevaluation in East Asian populations. Neurosci Lett. 2012;17(507):52–56. doi: 10.1016/j.neulet.2011.11.050. [DOI] [PubMed] [Google Scholar]
- Watson DJ, Marsden CA, Millan MJ, Fone KC. Blockade of dopamine D(3) but not D(2) receptors reverses the novel object discrimination impairment produced by post-weaning social isolation: implications for schizophrenia and its treatment. International Journal of Neuropsychopharmacology. 2011;15:471–484. doi: 10.1017/S1461145711000435. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.