Abstract
Eight different Bacillus subtilis strains and Bacillus atrophaeus were found to produce the bacteriocin subtilosin A. On the basis of the subtilosin gene (sbo) sequences two distinct classes of B. subtilis strains were distinguished, and they fell into the two B. subtilis subspecies (B. subtilis subsp. subtilis and B. subtilis subsp. spizizenii). The entire sequence of the subtilosin gene cluster of a B. subtilis subsp. spizizenii strain, B. subtilis ATCC 6633, was determined. This sequence exhibited a high level of homology to the sequence of the sbo-alb gene locus of B. subtilis 168. By using primer extension analysis the transcriptional start sites of sbo in B. subtilis strains ATCC 6633 and 168 were found to be 47 and 45 bp upstream of the sbo start codon, respectively. Our results provide insight into the incipient evolutionary divergence of the two B. subtilis subspecies.
Almost 4% of the 4.2-Mbp Bacillus subtilis 168 genome codes for proteins similar to the proteins involved in the biosynthesis of antimicrobial metabolites (17). However, B. subtilis 168 produces only a few antibiotics because several of the biosynthetic pathways are not functional, most likely because of the X-ray mutation of the original Marburg strain (6). In contrast, various other B. subtilis wild-type strains produce characteristic cocktails of numerous peptide antibiotics (1, 18). For example, a well-established bioindicator strain for sterilization control, ATCC 6633 (11), was investigated with respect to biosynthesis of the lantibiotic subtilin (4, 8, 16, 27) and its regulation (26, 28). In a series of B. subtilis strains production of the nonribosomally synthesized cyclic lipopeptides surfactin, fengycin, and the iturins, including mycosubtilin, with different compositions has been observed (9, 18, 20, 31).
Subtilosin is a macrocyclic bacteriocin with three intramolecular bridges (14, 19). An acidic isoelectric point differentiates subtilosin from the basic lantibiotics (13, 24). Subtilosin transcription is increased under oxygen-limited and anaerobic conditions (22; T. Stein, S. Düsterhus, A. Stroh, and K.-D. Entian, 10th Int. Conf. Bacilli, abstr. P103, p. 65, 1999). The production of mature subtilosin is based on the expression of the sbo-alb gene cluster encompassing the subtilosin structural gene sbo and genes involved in posttranslational modification and processing of presubtilosin and in immunity (34, 35).
Here we describe subtilosin production by eight different B. subtilis wild-type strains and Bacillus atrophaeus. The sbo genes of these organisms, as well as the entire subtilosin gene cluster of B. subtilis ATCC 6633, were sequenced in order to analyze the genetic variation between B. subtilis wild-type strains.
MATERIALS AND METHODS
Strains and plasmids.
Strains used in this work are listed in Table 1. Recombinant plasmids were amplified in Escherichia coli DH5α or TG1 grown in Luria-Bertani medium (GIBCO, Neu-Isenburg, Germany). B. subtilis was grown either on TY (0.8% tryptone, 0.5% yeast extract [Difco, Detroit, Mich.], 0.5% NaCl) or on Landy medium supplemented with 0.1% yeast extract (33). Antibiotics were used at the following concentrations: 100 μg of ampicillin per ml and 20 μg of chloramphenicol per ml for E. coli and 5 μg of chloramphenicol per ml, 10 μg of kanamycin per ml, and 100 μg of spectinomycin per ml for B. subtilis.
TABLE 1.
Strains used
| Strain | Other designations, description, and/or relevant genotypea | Reference |
|---|---|---|
| Bacillus subtilis strains | ||
| ATCC 6633 | CCM 1999, DSM 347, IAM 1069, NCIB 8054, NCTC 10400 | 6 |
| 168 | Used for genome sequencing; DSM 402, NCIB 10106, B. subtilis subsp. subtilis | 21 |
| 168 Δsbo | sbo::cm (Cmr) | This study |
| DSM 618 | Test strain for detection of antibiotics in meat | |
| DSM 1088 | IFO 13169, Bacillus natto | |
| DSM 2109 | ATCC 11774, NCTC 8236 | |
| DSM 2277 | ATCC 51189, CIP 103406, IAM 1633, NCIB 8649, NCTC 10073, B. atrophaeus | 11 |
| DSM 6405 | Mutant of B. subtilis W23, B. subtilis subsp. spizizenii | 21 |
| 60015 | Marburg strain | 10 |
| 10T | Type strain; ATCC 6051T, CCM 2216T, IAM 12118T, IFO 13719T, NCIB 3610T, NCTC 3610T | |
| ATCC 6633 | 16 | |
| ΔspaS | spaS::cm (Cmr) | |
| ΔspaS | spaS::cm::spec (Specr) | This study |
| Δsbo | sbo::cm (Cmr) | This study |
| ΔspaS Δsbo | spaS::cm::spec (Specr), sbo::cm (Cmr) | This study |
| Micrococcus luteus ATCC 9341 | Test strain for antimicrobial activity | |
| Escherichia coli DH5α | supE44 lacU169 [Φ80lacZM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 23 |
ATCC, American Type Culture Collection; CCM, Czech Collection of Microorganisms; CIP, Collection de l'Institut Pasteur; DSM, German Collection of Microorganisms (http://www.dsmz.de); IAM, Institute of Applied Microbiology; IFO, Institute for Fermentation; NCIB, National Collection of Industrial Bacteria; NCTC, National Collection of Type Cultures.
Plasmid isolation and PCR.
Established protocols were used for molecular biology techniques (25), and E. coli plasmids were isolated by the rapid alkaline extraction procedure (5). DNA amplification with Taq DNA polymerase was performed according to the instructions of the commercial supplier (Boehringer GmbH, Mannheim, Germany) by using a Hybaid R2 Combi-thermal reactor. DNA was cleaved and isolated with a QIAquick purification kit (Qiagen GmbH, Hilden, Germany). Oligonucleotides were purchased from ARK Scientific GmbH Biosystems, Darmstadt, Germany. Sequencing by primer walking was carried out by Scientific Research and Development, Oberursel/Frankfurt, Germany; nucleotide sequences were determined at least two times for each DNA strand.
sbo deletion in B. subtilis 168 and ATCC 6633.
Primers TS13C (GAATTGACACTATCTAGAGAAATGCCG) and TS14 (ATCCGGTGGTGCGGAATTCGATGA) (restriction sites are underlined) were designed by using the genome sequence of B. subtilis 168 (15). A 1,375-bp DNA fragment including the sbo gene was PCR amplified, cleaved with EcoRI and XbaI, and cloned into pUC19. To remove an NdeI restriction site, the resulting plasmid, pTSsbo, was cleaved with NarI and AatII, and blunt ends were generated after exonuclease treatment. The self-ligation product (pSD13) was cleaved with NdeI and the Klenow frag-ment, and the sbo gene was removed by BglII cleavage. Into the resulting blunt and BglII sites, a Sau3A/HincII site-containing cat gene obtained from pCE26 (16) was cloned. The plasmid constructed, pSD15, was linearized and used for transformation of competent B. subtilis 168 cells as described previously (2), with slight modifications as described by Klein et al. (16). An sbo deletion in B. subtilis ATCC 6633 was obtained after transformation with chromosomal DNA obtained from the corresponding B. subtilis 168 strain. For construction of the B. subtilis ΔspaS/Δsbo strain ATCC 6633, the chloramphenicol resistance cassette of the ΔspaS strain (16) was replaced by a gene conferring resistance to spectinomycin (pJL62) by in vivo recombination (30) prior to deletion of sbo.
Primer extension.
Total RNA was prepared from overnight cultures grown in Landy medium by using an RNeasy mini-kit (Qiagen) and was treated with 20 U of high-performance liquid chromatography (HPLC)-grade DNase. (Amersham Biosciences, Freiburg, Germany) in 40 mM Tris-HCl (pH 7.5)-60 mM MgCl2 in a 50-μl (final volume) mixture for 30 min at 37°C. RNA was isolated, precipitated with ethanol, and dissolved in 20 μl of H2O. Primer extension analyses with primer AS26 (CCCCATAGACCGAATAGACCTG) were performed as previously described (26).
Reversed-phase HPLC and mass spectrometry.
Culture supernatants of B. subtilis strains were separated by reversed-phase HPLC by using C18-Hypersil (particle size, 5 μm; precolumn dimensions, 4 by 10 mm; main column dimensions, 2 by 100 mm; Maisch, Ammerbuch, Germany). Eluents A and B were composed of 0.1% (vol/vol) trifluoroacetic acid and 20% (vol/vol) acetonitrile in water and 0.1% (vol/vol) trifluoroacetic acid in acetonitrile, respectively. A sample was applied with 100% eluent A and eluted with segmented gradients of acetonitrile (20% eluent B for 5 min, 20 to 40% eluent B for 20 min, and 40 to 100% eluent B for 5 min) (29). Delayed-extraction matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectra were recorded with a Voyager-RP-DE instrument (PerSeptive, Framingham, Mass.) by using a 337-nm nitrogen laser for desorption and ionization (19). The total acceleration voltage was 20 kV; 11.6 kV was used for the first grid. Reversed-phase HPLC fractions (0.7 μl) were mixed with 0.7 μl of matrix solution (20 μg of α-cyano-β-hydroxycinnamic acid [Sigma] μl−1 in eluent A) on the sample target and dried in the ambient air. Between 128 and 256 single scans were accumulated for each mass spectrum. The delay time was 375 ns.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been deposited in the EMBL nucleotide sequence database under accession number AJ430547.
RESULTS AND DISCUSSION
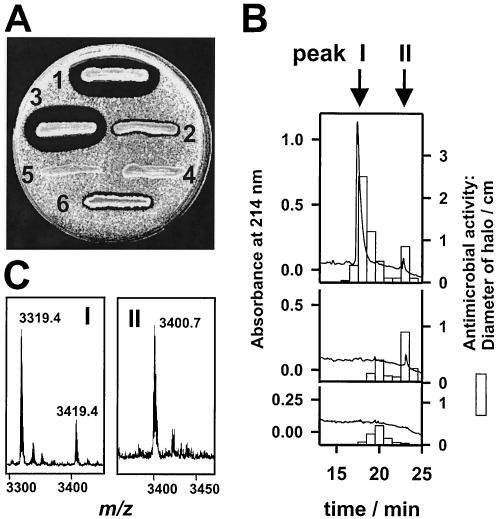
Antibiotic production by different B. subtilis strains was investigated by performing antimicrobial activity tests, reversed-phase HPLC separation of culture supernatants, and MALDI-TOF mass spectrometry (Fig. 1). For strain ATCC 6633, a representative strain, subtilin and its isoform [N-alpha-succinyl-Trp1]-subtilin are responsible for the main anti-Micrococcus activity. Consequently, the ΔspaS subtilin deletion strain exhibited no subtilin production. For the previously unidentified active antimicrobial compound in peak II (Fig. 1B) an m/z value of 3400.7 was determined (Fig. 1C). Both the m/z value and the elution position are consistent with the properties of authentic subtilosin produced by B. subtilis 168 (3, 19). In addition, peak II (subtilosin) was not observed in the supernatant of the ΔspaS/Δsbo double mutant (Fig. 1B), clearly demonstrating that the newly identified ATCC 6633 bacteriocin is based on sbo gene expression and demonstrating that it is identical to subtilosin. In fractions eluting around 20 min (Fig. 1B) the lipopeptides surfactin and mycosubtilin were observed. The production of these compounds was not affected in the gene deletion mutants (data not shown). Surprisingly, all eight B. subtilis wild-type strains investigated, including B. atrophaeus (black-pigmented B. subtilis; formerly B. subtilis DSM 2207 or ATCC 51189) (11), that are listed in Table 1 have been found to produce subtilosin. This finding was unprecedented because most of the known B. subtilis wild-type strains produce individual antibiotic cocktails. For example, subtilin production has been described only for the ATCC 6633 strain, and distinct lipopeptides are produced only by a few individual strains. The widespread occurrence of subtilosin might reflect an important physiological role. As subtilosin is produced at the end of exponential growth, particularly under stress conditions, a specific function of subtilosin as an antibiotic, killing factor (12) or as a pheromone during anaerobic or biofilm growth of B. subtilis (15) has to be considered.
FIG. 1.
Analysis of B. subtilis peptide antibiotics. (A) Micrococcus luteus growth inhibition assay. Streak 1, B. subtilis wild-type strain ATCC 6633; streak 2, strain ATCC 6633 ΔspaS deletion mutant; streak 3, strain ATCC 6633 Δsbo deletion mutant; streak 4, strain ATCC 6633 ΔspaS/Δsbo deletion mutant; streak 5, strain 168 Δsbo deletion mutant; streak 6, B. subtilis wild-type strain 168. (B) Reversed-phase HPLC separation of B. subtilis ATCC 6633 culture supernatants. The flow rate was 0.4 ml/min, and 400-μl fractions were collected. Aliquots (20 μl) of the wild type (top panel), the ΔspaS mutant (middle panel), and the ΔspaS/Δsbo double mutant (bottom panel) were used to inhibit the growth of M. luteus. (C) Detail of MALDI-TOF mass spectra of peak I (m/z 3319.4 and 3419.4 correspond to subtilin and succinylated subtilin, respectively) and peak II (m/z 3400.7 corresponds to subtilosin A). The variance of the m/z measurements was ±0.2 Da.
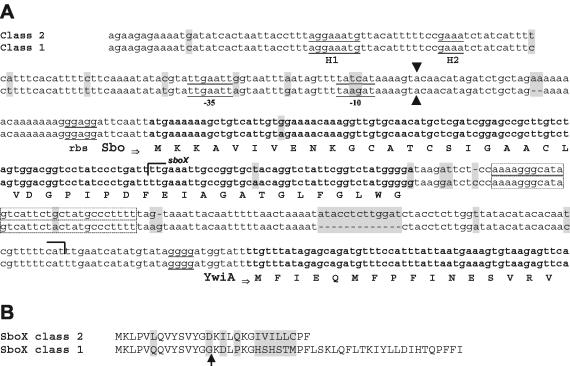
During cloning of B. subtilis ATCC 6633 DNA we observed restriction sites not present in the genome of strain 168 (15), while proposed sites were absent. We sequenced the sbo genes and flanking regions of all investigated B. subtilis wild-type strains in order to analyze the structural basis of these observations and to evaluate possible evolutionary relationships among the subtilosin producers. The main result of a comparison of the sbo alleles was identification of two distinct B. subtilis classes (Fig. 2A). Class 1 (168-like) includes strains 60015 (Marburg strain), 168, and 10T (type strain), as well as DSM 1088 and DSM 2109. Class 2 (W23-like) comprises strains ATCC 6633 and DSM 618, as well as DSM 6405, a mutant of the W23 strain. This observation is in good agreement with the recent classification of strain 168 as B. subtilis subsp. subtilis and the recent classification of W23-related strains as B. subtilis subsp. spizizenii based on DNA reassociation studies (21).
FIG. 2.
Comparison of sbo alleles of two B. subtilis classes. (A) Subtilosin A-encoding gene sequence sbo and flanking regions and the derived amino acid sequence. rbs, standard prokaryotic ribosome binding site. Transcriptional start sites (see Fig. 3) are indicated by arrows, and the positions of derived −10 and −35 regions are indicated. H1 and H2 indicate a putative sigma factor H region located 80 to 100 bp upstream of the transcriptional start site of sbo. A putative termination loop is enclosed in a box. The putative open reading frame sboX (150 nucleotides) is indicated by brackets. (B) Alignment of the amino acid sequences of the putative SboX gene product. The arrow indicates the putative processing site (double Gly motif). Differences between the two alleles are indicated by shading.
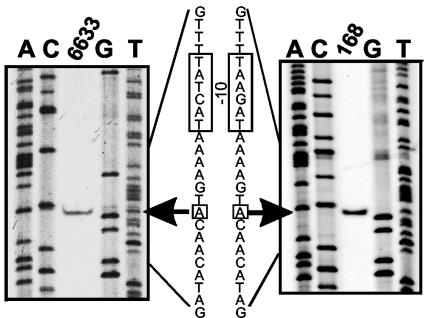
Remarkably, the nucleotide sequences of the sbo genes and flanking regions are identical in strains belonging to the same subspecies, and the sequences differ by three nucleotides in the two subspecies (Fig. 2A). However, the encoded Sbo prepeptides are identical in all cases. Primer extension analyses of sbo transcripts in representatives of both B. subtilis classes revealed transcriptional start sites that are 47 nucleotides (class 2 strain ATCC 6633) and 45 nucleotides (class 1 strain 168) upstream of the start ATG codon of sbo (Fig. 3). Similar 5′ transcriptional start sites are utilized by both B. subtilis classes. Due to a two-nucleotide insertion into class 2 sequences, the sizes of the transcripts differ by two nucleotides. The −10 and −35 regions derived from the transcriptional start sites resemble promoter regions utilized by sigma factor A (Fig. 2A). Remarkably, within the −10 region two nucleotide substitutions in both B. subtilis classes were observed, which suggests that there is an effect on sbo expression. However, a region upstream of the −35 region (positions −70 to −110) is perfectly conserved. This region represents a perfect sigma factor H binding site; however, involvement of this region in regulation of subtilosin biosynthesis has not been shown yet.
FIG. 3.
Primer extension analysis of sbo: mapping of the transcriptional start site of sbo by primer extension analysis with RNA from B. subtilis ATCC 6633 and 168 (middle lanes). The outside lanes show the results of autoradiography of dideoxynucleotide sequencing reactions with primer AS26 complementary to the 5′ region of sbo. The transcriptional start sites are indicated by arrows, and the −10 regions are enclosed by boxes in the derived nucleotide sequence.
Downstream of sbo a gene cluster with seven open reading frames (ywiA and ywhRQPOMN) has been identified and sequenced in B. subtilis ATCC 6633, a representative of the B. subtilis subsp. spizizenii strains (accession number AJ430547). The identified gene cluster exhibits a high level of homology to the sbo-alb gene cluster of B. subtilis 168 involved in the biosynthesis of subtilosin, including the structural gene, as well as genes encoding the posttranslational modification machinery and subtilosin immunity proteins. BLAST alignments (Table 2) revealed that the first four genes are highly conserved with those of B. subtilis subsp. subtilis (96 to 100% amino acid identity), while the remaining four genes are less conserved (83 to 88% identity). These differences reveal incipient evolutionary divergence of the B. subtilis subspecies. This low level of conservation is unprecedented; for example, thymidylate synthases A (thyA) in B. subtilis subsp. spizizenii ATCC 6633 and W23 and B. subtilis subsp. subtilis (168) exhibit more than 95% amino acid identity (32). Even the average level of amino acid identity for the DNA gyrases (gyrA) in seven Bacillus type strains was 95.1% (7).
TABLE 2.
Proteins derived from the subtilosin gene cluster
The nucleotide sequences of the closely related species and subspecies can be used for identification of genes and highly conserved regions in the gene products putatively corresponding to functional domains. For example, in strain 168 a new gene with an unknown function, sboX, encoding a bacteriocin-like product, was hypothesized (Fig. 2A) (34), which resides in an open reading frame overlapping the coding region of sbo. Notably, the expression of sboX would result in a 22-amino-acid truncated peptide in W23-like strains compared to the peptide produced by 168-like strains (Fig. 2B), which makes it unlikely that SboX is produced by W23-like strains.
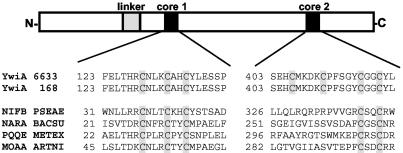
YwiA (AlbA) is involved in subtilosin biosynthesis, most likely in the posttranslational modification of presubtilosin (34, 35), although its molecular function is unknown. The amino acid sequences of YwiA in the two B. subtilis subspecies (Fig. 4) were compared, and two highly conserved regions (amino acids 1 to 90 and 109 to 450) separated by a less conserved linker region (amino acids 91 to 105) were identified. The large conserved domain from amino acid 109 to amino acid 450 exhibits homology to proteins belonging to the MoaA-NifB-PqqE family, which carry Fe-S centers in their active sites. Also, this cysteine-rich cofactor binding region is conserved in YwiA (core 1), as is a second CXXC motif near the C terminus (core 2). A pattern search for core 2 of YwiA proteins (Fig. 4) revealed homology to arylsulfatases and metallothioneins. Upstream of this signature sequence an unusual sulfur-rich motif (CMXXXC) with an unknown function has been found in YwiA proteins.
FIG. 4.
Domain structure of YwiA (AlbA): schematic representation of the putative domain structure of YwiA resulting from amino acid alignment of the sequences of B. subtilis ATCC 6633 and 168. Highly conserved regions (cores) are indicated by solid boxes; a gray box indicates a less conserved region. The first cysteine cluster (core 1) is highly homologous to active sites of proteins belonging to the MoaA-NifB-PqqE family carrying Fe-S centers, like NifB from Pseudomonas aeruginosa (NIFB PSEAE), NarA from B. subtilis (NARA BACSU), PqqE from Methylobacterium extorquens (PQQE METEX), and MoaA from Arthrobacter nicotinovorans (MOAA ARTNI).
As this study revealed, two distinct B. subtilis subspecies (B. subtilis subsp. subtilis and B. subtilis subsp. spizizenii) are distinguishable only on the basis of their sbo genes. Comparisons between the subtilosin gene clusters of the two subspecies led to identification of highly conserved protein domains and also provided insight into incipient evolutionary divergence.
Acknowledgments
We greatly acknowledge Michael Karas, University Frankfurt, for the opportunity to use his MALDI-TOF mass spectrometric equipment and J. Hofemeister for valuable discussions and for the kind gift of various B. subtilis wild-type strains.
REFERENCES
- 1.Ahimou, F., P. Jacques, and M. Deleu. 2000. Surfactin and iturin A effects on Bacillus subtilis surface hydrophobicity. Enzyme Microb. Technol. 27:749-754. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babasaki, K., T. Takao, Y. Shimonishi, and K. Kurahashi. 1985. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. (Tokyo) 98:585-603. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, S., and J. N. Hansen. 1988. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J. Biol. Chem. 263:9508-9514. [PubMed] [Google Scholar]
- 5.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkholder, P. R., and N. H. Giles. 1947. Induced biochemical mutants in Bacillus subtilis. Am. J. Bot. 34:345-348. [PubMed] [Google Scholar]
- 7.Chun, J., and K. S. Bae. 2000. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Leeuwenhoek 78:123-127. [DOI] [PubMed] [Google Scholar]
- 8.Corvey, C., T. Stein, S. Düsterhus, M. Karas, and K.-D. Entian. 2003. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem. Biophys. Res. Commun. 304:48-54. [DOI] [PubMed] [Google Scholar]
- 9.Duitman, E. H., L. W. Hamoen, M. Rembold, G. Venema, H. Seitz, W. Saenger, F. Bernhard, R. Reinhardt, M. Schmidt, C. Ullrich, T. Stein, F. Leenders, and J. Vater. 1999. The mycosubtilin synthetase of Bacillus subtilis ATCC 6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. USA 96:13294-13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freese, E., and P. Fortnagel. 1967. Analysis of sporulation mutants. I. Response of uracil incorporation to carbon sources, and other mutant properties. J. Bacteriol. 94:1957-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritze, D., and R. Pukall. 2001. Reclassification of bioindicator strains Bacillus subtilis DSM 675 and Bacillus subtilis DSM 2277 as Bacillus atrophaeus. Int. J. Syst. Evol. Microbiol. 51:35-37. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Pastor, J. E., E. C. Hobbs, and R. Losick. 2003. Cannibalism by sporulating bacteria. Science 301:510-513. [DOI] [PubMed] [Google Scholar]
- 13.Jack, R. W., and G. Jung. 2000. Lantibiotics and microcins: polypeptides with unusual chemical diversity. Curr. Opin. Chem. Biol. 4:310-317. [DOI] [PubMed] [Google Scholar]
- 14.Kawulka, K., T. Sprules, R. T. McKay, P. Mercier, C. M. Diaper, P. Zuber, and J. C. Vederas. 2003. Structure of subtilosin A, an antimicrobial peptide from Bacillus subtilis with unusual posttranslational modifications linking cysteine sulfurs to alpha-carbons of phenylalanine and threonine. J. Am. Chem. Soc. 125:4726-4767. [DOI] [PubMed] [Google Scholar]
- 15.Kearns, D. B., and R. Losick. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol. Microbiol. 49:581-590. [DOI] [PubMed] [Google Scholar]
- 16.Klein, C., C. Kaletta, N. Schnell, and K.-D. Entian. 1992. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl. Environ. Microbiol. 58:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 18.Leenders, F., T. H. Stein, B. Kablitz, P. Franke, and J. Vater. 1999. Rapid typing of Bacillus subtilis strains by their secondary metabolites using matrix-assisted laser desorption/ionization mass spectrometry of intact cells. Rapid Commun. Mass Spectrom. 13:943-949. [Google Scholar]
- 19.Marx, R., T. Stein, K.-D. Entian, and S. J. Glaser. 2001. Structure of the Bacillus subtilis peptide antibiotic subtilosin A determined by 1H-NMR and matrix assisted laser desorption/ionization time-of-flight mass spectrometry. J. Protein Chem. 6:501-506. [DOI] [PubMed] [Google Scholar]
- 20.Menkhaus, M., C. Ullrich, B. Kluge, J. Vater, D. Vollenbroich, and R. M. Kamp. 1993. Structural and functional organization of the surfactin synthetase multienzyme system. J. Biol. Chem. 268:7678-7684. [PubMed] [Google Scholar]
- 21.Nakamura, L. K., M. S. Roberts, and F. M. Cohan. 1999. Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int. J. Syst. Bacteriol. 49:1211-1215. [DOI] [PubMed] [Google Scholar]
- 22.Nakano, M. M., G. Zheng, and P. Zuber. 2000. Dual control of sbo-alb operon expression by the Spo0 and ResDE systems of signal transduction under anaerobic conditions in Bacillus subtilis. J. Bacteriol. 182:3274-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raleigh, E. A., R. Trimarchi, and H. Revel. 1989. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics 122:279-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Stein, T., S. Borchert, P. Kiesau, S. Heinzmann, S. Klöss, C. Klein, M. Helfrich, and K.-D. Entian. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44:403-416. [DOI] [PubMed] [Google Scholar]
- 27.Stein, T., and K.-D. Entian. 2002. Maturation of the lantibiotic subtilin: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to monitor precursors and their proteolytic processing in crude bacterial cultures. Rapid Commun. Mass Spectrom. 16:103-110. [DOI] [PubMed] [Google Scholar]
- 28.Stein, T., S. Heinzmann, P. Kiesau, B. Himmel, and K.-D. Entian. 2003. The spa-box for transcriptional activation of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 47:1627-1636. [DOI] [PubMed] [Google Scholar]
- 29.Stein, T., S. Heinzmann, I. Solovieva, and K.-D. Entian. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89-94. [DOI] [PubMed] [Google Scholar]
- 30.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 31.Steller, S., D. Vollenbroich, F. Leenders, T. Stein, B. Conrad, J. Hofemeister, P. Jacques, P. Thonart, and J. Vater. 1999. Structural and functional organization of the fengycin synthetase multienzyme system from Bacillus subtilis b213 and A1/3. Chem. Biol. 6:31-41. [DOI] [PubMed] [Google Scholar]
- 32.Tam, N. H., and R. Borriss. 1998. Genes encoding thymidylate synthases A and B in the genus Bacillus are members of two distinct families. Mol. Gen. Genet. 258:427-430. [DOI] [PubMed] [Google Scholar]
- 33.Ullrich, C., B. Kluge, Z. Palacz, J. Vater, F. Baumgart, B. Kluge, C. Ullrich, J. Vater, and D. Ziessow. 1991. Cell-free biosynthesis of surfactin, a cyclic lipopeptide produced by Bacillus subtilis. Identification of amino acid substitutions in the lipopeptide surfactin using 2D NMR spectroscopy. Biochemistry 30:6503-6508. [DOI] [PubMed] [Google Scholar]
- 34.Zheng, G., R. Hehn, and P. Zuber. 2000. Mutational analysis of the sbo-alb locus of Bacillus subtilis: identification of genes required for subtilosin production and immunity. J. Bacteriol. 182:3266-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng, G., L. Z. Yan, J. C. Vederas, and P. Zuber. 1999. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J. Bacteriol. 181:7346-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]