Abstract
IL-17 is a proinflammatory cytokine produced by activated Th17 cells and other immune cells. IL-17–producing Th17 cells are major contributors to chronic inflammatory and autoimmune diseases, such as multiple sclerosis, rheumatoid arthritis, and inflammatory bowel disease. Although the transcriptional regulation of Th17 cells is well understood, the posttranscriptional regulation of IL-17 gene expression remains unknown. The RNA-binding protein HuR positively regulates the stability of many target mRNAs via binding the AU-rich elements present in the 3′ untranslated region of many inflammatory cytokines including IL-4, IL-13, and TNF-α. However, the regulation of IL-17 expression by HuR has not been established. CD4+ Th17 cells from HuR knockout mice had decreased IL-17 steady-state mRNA and protein levels compared with wild-type Th17 cells, as well as decreases in frequency of IL-17+ cells. Moreover, we demonstrated that HuR directly binds to the IL-17 mRNA 3′ untranslated region by using RNA immunoprecipitation and biotin pulldown assays. In addition, the knockout of HuR decreased cellular proliferation of CD4+ T cells. Mice with adoptively transferred HuR KO Th17 cells had delayed initiation and reduced disease severity in the onset of experimental autoimmune encephalomyelitis compared with wild-type Th17 cells. Our results reveal a HuR-induced posttranscriptional regulatory mechanism of Th17 differentiation that influences IL-17 expression. These findings may provide novel therapeutic targets for the treatment of Th17-mediated autoimmune neuroinflammation.
Introduction
CD4+ T cells differentiate into distinct subsets with different functions and patterns of cytokine production. Besides Th1 and Th2 cells, a third subset, known as Th17 cells, has received considerable attention because they appear to be principal mediators of pathogenesis in several inflammatory and autoimmune disorders, including asthma, colitis, experimental autoimmune encephalomyelitis (EAE), and rheumatoid arthritis (1, 2).
Th17 cells are characterized by production of IL-17A (in this work, IL-17 represents IL-17A) and the related family of cytokines. Th17 cell differentiation has been extensively studied from a transcriptional but not posttranscriptional framework. High-dose Ag-loaded dendritic cells induce Th17 cell differentiation. TGF-β and IL-6 combinations are very effective to induce naive CD4+ T cells to become Th17 cells (2, 3). Initial studies in mice suggested that IL-23 induces IL-17 expression. However, further studies showed that the IL-23 receptor is only expressed on T cells after activation, and therefore, IL-23 promotes IL-17 production by expansion of Th17 cells, but it cannot act on naive T cells to induce Th17 differentiation (4). IL-6 is sufficient to induce IL-23R expression, and IL-23 further amplifies its receptor expression (5). However, other cytokines, such as, IL-2, IFN-γ, and IL-27, inhibit Th17 differentiation (2, 3, 6).
Several transcription factors have been reported to regulate Th17 cell differentiation. The cytokines IL-23, IL-21, and IL-6 that induce IL-17 production all activate Stat3 (7, 8). Furthermore, deletion of Stat3 in T cells abrogates Th17 cell differentiation (9). Stat3 directly controls expression of many of the other transcription factors that participate in Th17 cell differentiation including retinoic acid–related orphan receptor γt (RORγt) (10). RORγt has been reported to bind to the IL-17 gene promoter, and its overexpression promotes IL-17 expression (10, 11). In contrast, IL-2 and IL-27 inhibit IL-17 production via different mechanisms (2, 6, 12). Recent studies demonstrated that the induction of Stat5 binding by IL-2 was associated with less binding of Stat3 to the IL-17 promoter (12), whereas IL-27 controls IL-17 production via induction of the ligand programmed cell death ligand-1 in a Stat1-dependent manner (6).
Reagents targeting Th17-related molecules have been under clinical investigation for several diseases, but this has not always been effective in controlling disease activity (13). Consistent with this, it has become evident that there are substantial differences in Th17 cell development. Although the cytokines and transcription factors that control Th17 differentiation have been extensively investigated, the posttranscriptional regulation of IL-17 mRNA is not well understood.
The contribution of posttranscriptional regulation in control of gene expression of inflammatory cytokines is increasingly recognized as being an important control point in gene expression (14, 15). Stabilization or degradation of labile mRNAs by AU-rich elements (AREs) in their 3′ untranslated region (UTR) is an important mechanism of posttranscriptional gene regulation of inflammatory cytokines. The best-characterized ARE-binding proteins are the Elav/Hu family of RNA-binding proteins (RBPs; HuR [HuA], HuB, HuC, and HuD), including the ubiquitously expressed family member HuR. HuR protein contains three RNA-recognition motif domains (16). Of these, the two N-terminal domains bind U-rich RNA sequences, and the C-terminal RNA-recognition motif stabilizes Hu–RNA complexes, enhances their translation, or both (17–21). HuR further regulates cell proliferation by stabilization of mRNAs encoding cyclins A and B1 (22).
The RBP HuR positively regulates stability of many target mRNAs via binding AREs present in the 3′ UTR, including inflammatory cytokines such as IL-4, IL-13, and TNF-α (23–26). In this study, we demonstrated that IL-17 mRNA and protein levels in polarized Th17 cells from HuR conditional knockout (KO) mice are reduced compared with wild-type (WT) Th17 cells. We verified that HuR directly binds to IL-17 mRNA 3′ UTR and controls transcript stability. HuR-deficient Th17 cells using an adoptive transfer model have delayed disease onset and less severe pathogenesis in the development of EAE compared with WT Th17 cells. In summary, our results reveal an underappreciated posttranscriptional control mechanism of IL-17 by HuR. These findings may have implications not only for treatment of IL-17–mediated inflammatory diseases such as EAE but in Th17 differentiation as well.
Materials and Methods
Animals
HuRfl/fl mice were generated as previously described (M. M. Gubin, P. Techasintana, J.D. Magee, G.M. Dahm, R. Calaluce, J.L. Martindale, M.S. Whitney, C.L. Franklin, C. Besch-Williford, J.W. Hollingsworth, K. Abdelmohsen, M. Gorospe, and U. Atasoy, et al., submitted for publication). Eight- to 10-wk-old control (HuRfl/fl) female mice and HuR conditional KO mice (OX40-cre HuRfl/fl) were used in which a portion of the promoter region and exons 1 and 2 of the HuR gene were deleted when T cells were activated to express cre. All mice are on the C57BL/6 background and were bred at the animal facility of the University of Missouri. Animal experiments were approved by the Institutional Animal Care and Use Committee and done according to federal and institutional guidelines.
Isolation of naive CD4+ T cells and cell culture
Spleens were isolated from 8-wk-old female HuRf/fmice and OX40-cre HuRf/f mice. Naive CD4+ T cells were purified from splenocytes using CD4 (L3T4) MicroBeads (Miltenyi Biotec) and MACS columns following the manufacturer’s protocol. Cells were cultured in RPMI 1640 media (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine (Invitrogen), penicillin (100 U/ml)/streptomycin (100 μg/ml), sodium pyruvate, and 2-ME.
Th17 cell differentiation In vitro
Purified naive CD4+ T cells were activated with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (3 μg/ml) in the presence of no cytokine (activation) or anti–IFN-γ (10 μg/ml), anti–IL-4 (10 μg/ml), TGF-β (3 ng/ml), IL-6 (20 ng/ml), and IL-23 (20 ng/ml) (Th17 polarization). All cytokines were purchased from R&D Systems and Abs from eBioscience.
ELISA
Cytokine levels in cell-culture supernatants were determined with the ELISA Ready-SET-Go kit (eBioscience).
RNA isolation and quantitative RT-PCR
Cells were collected, and total RNA was extracted using TRIzol (Invitrogen). A total of 500 ng RNA was reverse transcribed into cDNA using the Gene Amp Kit (PerkinElmer) according to the manufacturer’s protocols. The resulting cDNA template was subjected to real-time PCR using Applied Biosystems Step One PCR system with SYBR Green Reagent Kit (Invitrogen) using the following cycling parameters: 95°C, 2 min; 40 cycles of 95°C, 15 s; 60°C, 30 s; and 72°C, 1 min. HuR or IL-17A mRNA levels were normalized to GAPDH levels for each sample run in triplicate. Forward and reverse primers for specific murine target genes are listed in Table I.
Table I. Sequences of mouse quantitative RT-PCR primers and mouse RT-PCR primers in biotin pulldown.
| Sequences of Mouse Quantitative RT-PCR Primers | |
|---|---|
| HuR forward | 5′-ACTGCA GGGATGACATTGGGAGAA-3′ |
| HuR reverse | 5′-AAGCTTTGCAGATTCAACCTCGCC-3′ |
| Stat3 forward | 5′-TAGCCGATTCCTGCAAGAGTCCAA-3′ |
| Stat3 reverse | 5′-CGGGCAATTTCCATTGGCTTCTCA-3′ |
| IFN-γ forward | 5′-ACAGCCACTGCATTCCCAGTTT-3′ |
| HuR forward | 5′-TCTCGGAAGGACTTGCAGACAT-3′ |
| HuR reverse | 5′-TGTGTCTCTGATGCTGTTGCTGCT-3′ |
| IL-17 reverse | 5′-AGGAAGTCCTTGGCCTCAGTGTTT-3′ |
| GM-CSF forward | 5′-TGGAAGCATGTAGAGGCCATCA-3′ |
| GM-CSF reverse | 5′-GCGCCCTTGAGTTTGGTGAAAT-3′ |
| Bcl6 forward | 5′-CATACAGAGATGTGCCTCCATAC-3′ |
| Bcl6 reverse | 5′-CCCATTCTCACAGCTAGA ATC C-3′ |
| Foxp3 forward | 5′-TACACCCAGGAAAGACAGCAACCT-3′ |
| Foxp3 reverse | 5′-TCTGCTTGGCAGTGCTTGAGAA-3′ |
| RORγt forward | 5′-ACAGCCACTGCATTCCCAGTTT-3′ |
| RORγt reverse | 5′-TCTCGGAAGGACTTGCAGACAT-3′ |
| Sequence of Mouse RT-PCR Primers in Biotin Pulldown | |
| IL-17 ORF forward+T7 | 5′-GCT TCT AAT ACG ACT CAC TAT AGG ATG AGT CCA GGG AG-3′ |
| IL-17 ORF reverse | 5′-TTA GGC TGC CTG GCG GAC AA-3′ |
| IL-17 sect1 forward+T7 | 5′-GCT TCT AAT ACG ACT CAC TAT AGG ACA GAG ACC CGC GG-3′ |
| IL-17 sect1 reverse | 5′-GTC TTG AAT TTT GTC TTC TTC AG-3′ |
| IL-17 sect2 forward+T7 | 5′-GCT TCT AAT ACG ACT CAC TAT AGG CCA GAA ATT TTA TT-3′ |
| IL-17 sect2 reverse | 5′-CCA AAA CTT TAT TAT TAC AGT GAT ATT GTT ACA A-3′ |
| IL-17 3′ UTR forward+T7 | 5′-GCT TCT AAT ACG ACT CAC TAT AGG ACA GAG ACC CGC GG-3′ |
| IL-17 3′ UTR reverse | 5′-CCA AAA CTT TAT TAT TAC AGT GAT ATT GTT ACA A-3′ |
| GM-CSF ORF forward+T7 | 5′-GCT TCT AAT ACG ACT CAC TAT AGG ATG TGG CTG CAG AA-3′ |
| GM-CSF ORF reverse | 5′-TCA TTT TTG GCC TGG TTT TTT GCA TTC AAA GGG G-3′ |
| GM-CSF 3′ UTR forward+T7 | 5′-GCT TCT AAT ACG ACT CAC TAT AGG GGA AGC CCA GGC CAG-3′ |
| GM-CSF 3′ UTR reverse | 5′-CTGGTAAGACATTCTCAATAAATAGAGTTGC-3′ |
Western blotting
Cells were pelleted and resuspended in triple-detergent RIPA buffer (1% Nonidet P-40, 50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.5% deoxycholate, 1% SDS, and 1 mM EDTA) and cOmplete Protease Inhibitor Cocktail Tablets (Roche Applied Science). Cells were lysed for 30 min on ice. Protein lysates (25 μg) were loaded on a 10% SDS-PAGE gel and transferred onto nitrocellulose membrane. The membrane was blotted with anti-HuR clone 3A2 (1 μg/ml; Santa Cruz Biotechnology) and anti–β-tubulin (1 μg/ml; Sigma-Aldrich), followed by a sheep anti-mouse Ig conjugated with HRP secondary Ab, and detected by ECL (GE Healthcare). Protein levels were further analyzed using Quantity One software (Bio-Rad).
Immunoprecipitation of endogenous messenger RNP complexes
RNA immunoprecipitation (RIP) was performed according to established protocol (25, 26) with slight modification. Briefly, Th17-polarized cells were lysed using polysomal lysis buffer (25, 26). The lysates were precleared for 30 min at 4°C by adding 30 μg IgG1 (BD Bioscience) and 100 μl Protein-A Sepharose beads (50% slurry; Sigma-Aldrich) swollen in NT2 buffer (150 mM NaCl, 50 mM Tris [pH 7.4], 1 mM MgCl, and 0.05% Nonidet P-40) with 5% BSA. Beads (100 μl) were coated by adding 30 μg either IgG1 (B&D Life Sciences) as control or anti-HuR Ab, 3A2, and incubated overnight at 4°C. After extensive washes of precoated PAS beads, 100 μl precleared lysate was added and incubated for 4 h at 4°C with additives, and then 30 μg proteinase K was added and incubated for 30 min at 55°C to digest protein. RNA was extracted and reverse transcribed to cDNA. Real-time RT-PCR was performed to verify the presence of specific target mRNAs.
In vitro biotin pulldown assay
Biotinylated transcripts were synthesized by using templates and primers. Forward primers that contained the T7 RNA polymerase promoter sequence (5′-CCAAGCTTCTAATACGACTCACTATAGGGAGA-3′ [T7]) and reverse primers used to generate cDNA are listed in Table I. PCR-amplified products were purified and used as templates for the synthesis of biotinylated RNA with T7 RNA polymerase and biotin-conjugated UTP for murine RNAs. Biotinylated transcripts (1 μg) were incubated with NIH/3T3 cell lysate (40 μg) for 15 min at room temperature, and then RNP complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Invitrogen). Bound HuR protein in the pulldown pellet was analyzed by Western blotting analysis.
Induction of EAE
For induction of active EAE, female 8–10-wk-old control (HuRfl/fl) mice and HuR conditional KO mice (OX40-cre HuRfl/fl) were immunized s.c. at two sides on the back with 200 μg MOG35–55 in CFA, and 500 ng pertussis toxin in PBS was administered i.p. on days 0 and 2. EAE was clinically assessed by daily assignment of scores on a scale of 0–5 as follows: partially limp tail, 0.5; completely limp tail, 1; limp tail and waddling gait, 1.5; limp tail and weakness of hind legs, 2; limp tail and paralysis of one hind limb, 2.5; limp tail and complete paralysis of hind legs, 3; limp tail, complete paralysis of hind legs, and weakness of front legs, 3.5; limp tail, complete hind leg and one front leg paralysis, or paralysis of trunk, 4; moribund, 4.5; and death, 5 (27).
For adoptive transfer of EAE, splenocytes and lymph nodes from control and KO mice were harvested 10–14 d later following MOG35–55 immunization. Cells were restimulated with MOG35–55 (20 μg/ml) and recombinant murine IL-23 (20 ng/ml) for 3 d. CD4+ T cells were isolated with a MACS kit (Miltenyi Biotec), and 6 to 7 × 106 cells were injected i.v. into sublethally irradiated (450 rad) syngeneic mice. The recipients were given 500 ng pertussis toxin i.p. on days 0 and 2 posttransfer. Clinical disease was monitored daily as described above.
Isolation of mononuclear cells from spinal cords
Before spinal cord dissection, mice were perfused with cold PBS to remove blood from internal organs. The brain was dissected, and the spinal cord was flushed out by hydrostatic pressure. The brain and spinal cord were cut into small pieces and digested in solution containing 0.2 U/ml Liberase DL (Roche) and 1 mg/ml DNAse I (Roche) in DMEM at 37°C for 45 min. After the end of the digestion, single-cell suspension was prepared by passing through a 70-μm cell strainer. The cells were washed once in PBS, placed in 8 ml 37% Percoll solution, and overlaid with 4 ml 70% solution, then centrifuged at 1800 rpm for 20 min, and the interphase layer (mononuclear cell fraction) was transferred into a fresh tube and used for subsequent experiments.
Intracellular cytokine staining
Cells obtained from in vitro culture or from dissection of brain and spinal cords were incubated for 4 h with 50 ng/ml PMA (Sigma-Aldrich), 500 ng/ml ionomycin (Sigma-Aldrich), and 10 μg/ml brefeldin A (Invitrogen). Cells were stained for surface markers, then fixed in 2% formaldehyde, permeabilized with 0.2% saponin, and stained for intracellular cytokines.
Immunohistochemical staining
Paraffin-embedded sections of spinal cord from mice with EAE were deparaffinized in xylene and dehydrated in graded alcohol. The slides were washed in PBS (0.1 M, pH 7.6). Pretreatment of tissue with heat-induced epitope retrieval is done by using microwave. The slides were blocked for 1 h with 1.5% normal goat serum. Anti-CD3 (Dako North America) was used as primary Ab (1:50–1:100 dilution) and isotype rabbit IgG or BSA as negative control. Biotinylated goat-anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) was used as secondary Ab, followed by incubation with Vectastain Elite avidin-biotin complex (Vector Laboratories, Burlingame, CA). Peroxidase activity was visualized using Nova Red substrate (Vector Laboratories) (28). Cell nuclei were counterstained with hematoxylin (Vector Laboratories).
Isolation of naive CD4+ T lymphocytes from human PBMCs
Buffy coats were obtained from healthy subjects (Applied Biological Materials). PBMCs were separated by Histopaque centrifugation (Sigma-Aldrich). Naive CD4+ T lymphocytes were subsequently isolated using the human CD4+ T cell Isolation Kit II (Miltenyi Biotec).
Human CD4+ T cells differentiation assay
Plates were precoated overnight at 4°C with 5 μg/ml of anti-CD3 mAb (UCHT1; BD Biosciences). Ab solution was removed; plates were rinsed once with PBS. CD4+ T cells were cultured in the complete RPMI 1640 medium (Invitrogen) with 2.5 μg/ml soluble anti-CD28 (clone 28.2; eBioscience), in the presence of recombinant human (rh)IL-1β (20 ng/ml), rhIL-6 (20 ng/ml), rhIL-23 (20 ng/ml), human TGF-β (5 ng/ml), anti–IL-4(10 μg/ml), and anti–IFN-γ (10 μg/ml). The cells were cultured at 37°C for 7 d. For Th17 cell transfection using small interfering RNA (siRNA) experiments, naive CD4+ T cells were cultured and transfected using HuR siRNA or HuR scramble molecules (300 nM; Qiagen) with the Human T Cell Nucleofector Kit (Lonza).
Statistical analysis
The results were analyzed for statistical significance by paired sample t test.
Results
Increased IL-17A mRNA expression correlates with the increased HuR protein expression during Th17 cell differentiation
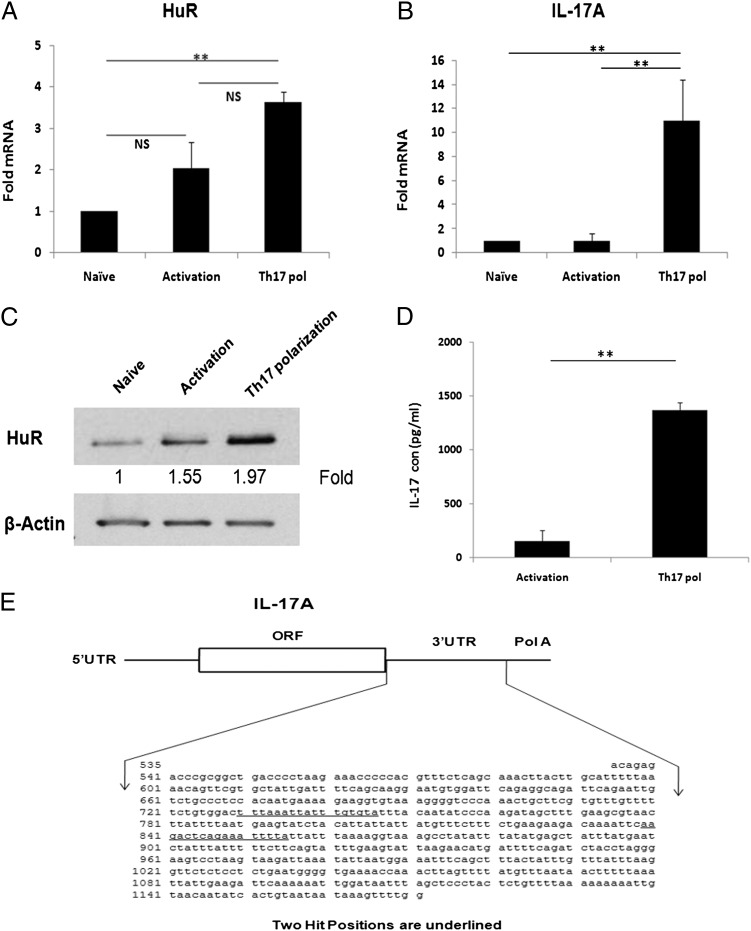
To determine whether HuR would affect Th17 cell differentiation, we first examined HuR mRNA and protein expression levels in naive, activated, and differentiated CD4+ T cells. After 4 d culture, we collected cells and isolated RNA for RT-PCR and cell protein extracts for Western analysis. We analyzed mRNA levels by RT-PCR using primer sequences in Table I. Compared to the HuR mRNA levels in naive CD4+ T cells, the level of HuR mRNA in activated CD4+ T cells was modestly increased but this was not statistically significant (Fig. 1A). However, Th17-polarized cells had significantly higher HuR mRNA steady-state levels compared with naive unactivated cells (Fig. 1A).
FIGURE 1.
Analysis of HuR expression in CD4+ T cells. CD4+ T cells were isolated from spleen of naive WT mice. Cells were stimulated with plate-coated anti-CD3 (10 μg/ml) plus anti-CD28 (3 μg/ml) (activation) or in the presence of Th17 cell–polarizing cytokines for 4 d according to the methods described. (A and B) Total RNA was isolated from activated and Th17-polarized cells and reversely transcribed into cDNA. The mRNA levels of HuR and IL-17 were analyzed by real-time RT-PCR. (C) Cells were lysed with triple-detergent lysis buffer, and HuR protein expression was analyzed by Western blotting (top panel) using β-actin as a loading control (bottom panel). (D) Cell-culture supernatant was collected, and IL-17 cytokine levels were measured by ELISA. (E) Schematic representation of IL-17 3′ UTR. Two predicted HuR binding sites are underlined in 3′ UTR of IL-17 mRNA. Data were presented as mean ± SEM. Data in (A)–(D) were derived from at least three independent experiments. **p < 0.001.
As expected, the IL-17 mRNA levels were significantly increased in Th17 cells compared with naive or activated CD4+ T cells (Fig. 1B). Consistent with HuR mRNA data, Western blot assay showed increased HuR protein expression in both activated and Th17 cells, with slightly greater amounts in the later (Fig. 1C). Therefore, these results suggested that both HuR mRNA and protein expression are increased in Th17 cells, which correlated with increased IL-17 mRNA (Fig. 1A, 1C) and IL-17 cytokine levels (Fig. 1D). Furthermore, by using computational analysis of IL-17 3′ UTR, we found two predicted hits of a HuR murine gene motif (access number NM_010552.3) (Fig. 1E), which are similar to human IL-17 (NM_002190.2) sequences.
HuR genetic ablation decreases IL-17A mRNA and protein expression in Th17 cells
Because HuR protein expression correlated with IL-17 mRNA and protein expression (Fig. 1) in Th17 cells, we asked whether HuR regulates IL-17 mRNA expression. To address this question, we generated HuR conditional KO (OX40-cre HuRfl/fl) mice. OX40 is not expressed in naive, unactivated CD4+ T cells but is upregulated upon T cell activation, thus deleting HuR. A major advantage of using such an approach is that it does not affect T cell development, because HuR levels are lowered following activation.
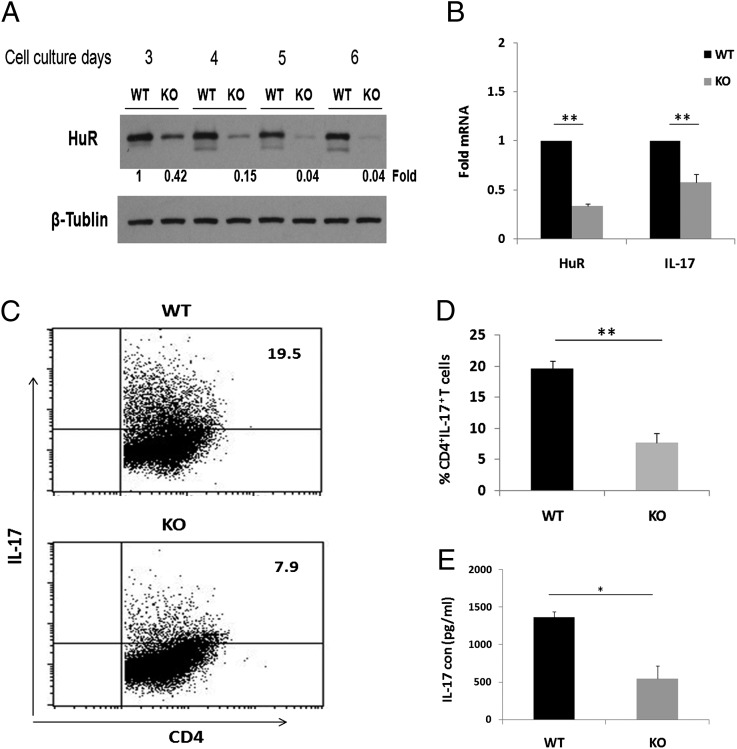
We first examined kinetics of HuR protein expression in polarized Th17 cells from KO mice during culture. Naive CD4+ T cells from WT and KO mice were cultured under Th17-polarizing conditions, and HuR levels were quantified by Western blotting. These results showed that endogenous HuR was gradually diminished following cell activation and differentiation in KO CD4+ T cells beginning at day 3 after culture (Fig. 2A). Compared to WT (100%) HuR levels in CD4+ T cells, HuR levels decreased to 42% on day 3 and 15% by day 4. HuR was barely detected after day 5 (Fig. 2A), which revealed that the HuR gene was genetically ablated in activated T cells. This was further confirmed by measuring HuR mRNA expression using real-time-PCR (Fig. 2B). Taken together, these data argue that HuR genetic ablation does work but it takes several days for the protein levels to maximally decrease.
FIGURE 2.
Reduced IL-17 expression in HuR KO Th17 cells. CD4+ T cells were isolated from spleens of control mice and HuR-deficient mice. Cells were stimulated as described in Fig. 1 (activation or Th17 polarization) for 3, 4, and 5 d. (A) HuR protein levels in Th17 cells were detected by Western blot analysis. HuR protein levels began to diminish starting on day 3 and were hardly detected on days 5 and 6 in KO Th17 cells. (B) Cells were collected at day 4. RNA was extracted, and quantitative RT-PCR was performed following reverse transcription using primer sequences listed in Table I. Decreased IL-17 mRNA was associated with reduced HuR mRNA in KO Th17 cells. IFN-γ mRNA levels were unaffected by HuR knockdown (data not shown). (C) CD4+ T cells were isolated from spleens of control mice and HuR-deficient mice. Cells were stimulated with plate-coated anti-CD3 (10 μg/ml) plus anti-CD28 (3 μg/ml) (activation) or Th17 cell–polarizing cytokines (Th17 polarization) as described in Materials and Methods. Cells were collected at day 5, and intercellular staining was performed for flow cytometric analysis. (D) HuR KO Th17 cells produced lower percentage of IL-17+ than WT Th17 cells. (E) Cell supernatant was collected at day 4, and IL-17 cytokine levels were measured by ELISA. KO Th17 cells produced lower levels of IL-17 protein than WT Th17 cells. Error bars represent ± SEM. Data were derived from at least three independent experiments. *p < 0.005, **p < 0.001.
Because there were reductions in IL-17 expression in KO CD4+ T cells, we asked whether this resulted in disturbances in Th17 differentiation. Fig. 2C demonstrates significant reductions in IL-17+ cells in KO versus control CD4+ T cells (7.9 versus 19.5%). We also measured expression of other relevant genes in Th17 differentiation and found no differences in RORγt and IL-17F and only modest increases in IL-22 (Supplemental Fig. 1).
Next, we examined IL-17A mRNA and protein levels following HuR ablation in KO and WT Th17 cells. Real-time PCR analysis showed that KO Th17 cells had significantly less IL-17 mRNA as compared with WT Th17 cells on day 4 after culture (Fig. 2B). In all of our experiments, IFN-γ mRNA and protein levels were unaffected (data not shown). In addition, we measured IL-17 protein level in culture supernatants by ELISA. Consistent with reduction in IL-17 steady-state mRNA levels, KO Th17 cells produced notably less IL-17 (64%) compared with WT Th17 cells (Fig. 2E). These results indicated that HuR ablation reduced steady-state levels of IL-17 mRNA and protein in Th17 cells and, furthermore, decreased frequency of IL-17+ cells in Th17 differentiation.
HuR regulates IL-17 expression by directly binding to the 3′ UTR
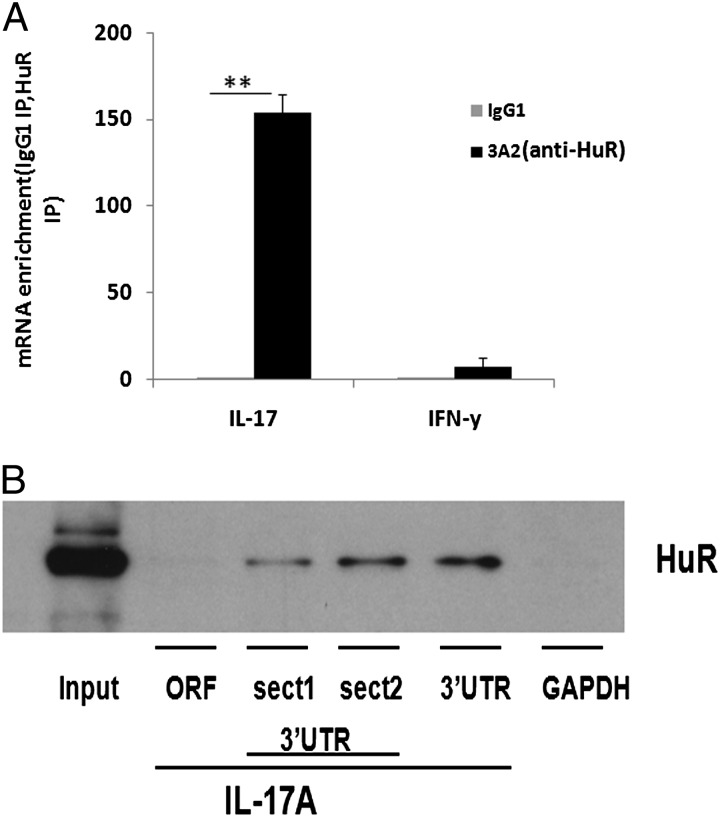
The regulation of IL-17 expression by HuR suggested that IL-17 mRNA might be a direct target of HuR. To evaluate the potential Th17 cell–extrinsic function of HuR in vitro, we first performed RIP assay to test whether HuR protein directly binds IL-17 mRNA. The association of HuR with IL-17 mRNA was monitored by isolating RNA from the preserved RNP complexes using anti-HuR Ab. These data showed a significant IL-17 mRNA enrichment (>100-fold) in the anti-HuR immunoprecipitation (IP) sample comparing with isotype-matched IgG control. IFN-γ mRNA was not appreciably enriched in the HuR pulldown (Fig. 3A). To validate HuR interactions with IL-17 mRNA, we used a second independent method, biotin pulldown.
FIGURE 3.
IL-17 mRNA is a direct target of HuR. Naive CD4+ T cells were isolated from spleen of WT mice and stimulated as described in Fig. 1 in the presence of Th17 cell–polarizing cytokines for 5 d. (A) IP of RNP complexes assayed for detection of IL-17 mRNA in Th17 cell cytoplasmic extracts immunoprecipitated with anti-HuR or isotype-matched Ab. Real-time PCR showed IL-17 mRNA enrichment in the anti-HuR IP samples compared with isotype-matched IgG1. IFN-γ mRNA was hardly detected in any IP samples. (B) Biotin pulldown was used to determine whether HuR directly binds to 3′ UTR of IL-17 mRNA. HuR protein could be detected by Western blotting in biotinylated transcripts spanning IL-17 3′ UTR, but not in IL-17 mRNA ORF biotinylated transcripts. GAPDH biotinylated transcripts were unreactive. Error bars represent ± SEM. Data in (A) were derived from at least three independent experiments. Data in (B) were derived from two independent experiments. **p < 0.001.
We generated four biotinylated transcripts that are complementary to sequences in IL-17 mRNA 3′ UTR and open reading frames (ORF) (Figs. 1D, 3B). The biotinylated transcripts were incubated with cytoplasmic protein lysates from WT Th17 cells. As expected, HuR was detected by Western blot in the samples precipitated by the biotinylated transcripts corresponding to putative binding sites section 1 and section 2 of IL-17 mRNA 3′ UTR, but not to IL-17 mRNA ORF (Fig. 3B). As a negative control, HuR protein did not associate with GAPDH 3′ UTR biotinylated probes and GAPDH mRNA complex, because this transcript lacks HuR binding domains (Fig. 3B). Taken together, these results indicated that HuR directly binds to IL-17 3′ UTR mRNA, suggesting a regulatory relationship.
HuR posttranscriptionally regulates IL-17 by stabilizing its mRNA
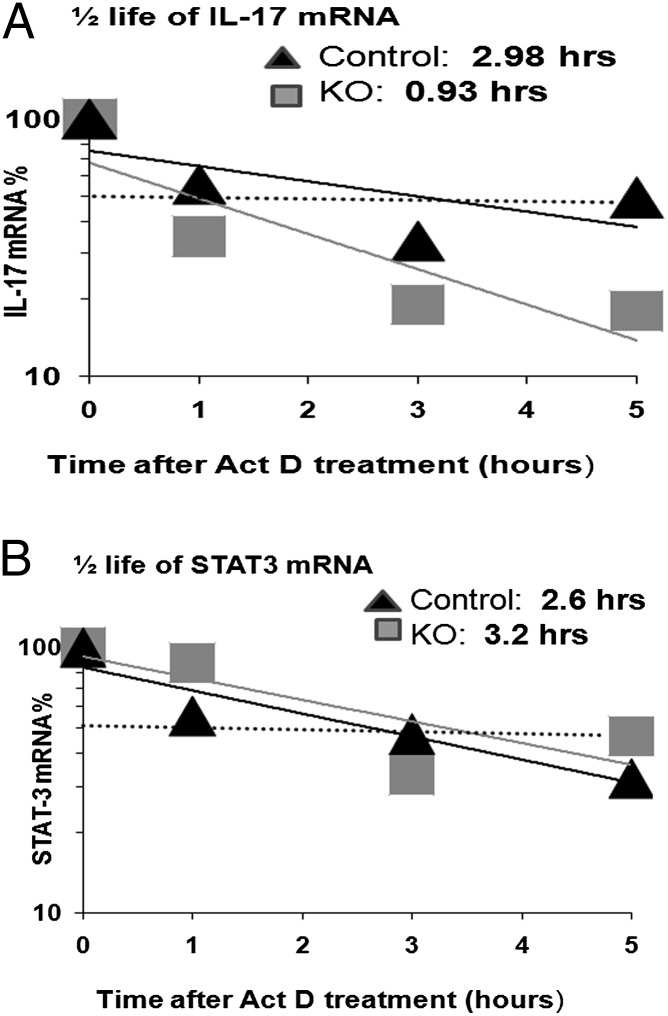
HuR has previously been described as regulating its mRNA targets in part by increasing mRNA stability, translation, or both (19–22). To identify the function of HuR for protecting IL-17 mRNA from degradation, IL-17 mRNA decay rates were measured using actinomycin D treatment in WT and KO Th17 cells.
As seen in Fig. 4A, there is a 3-fold reduction in IL-17 mRNA stability (3 to 0.9 h) in HuR KO Th17 cells. In comparison, STAT3 mRNA stability is not altered under these conditions (Fig. 4B). These results suggested that HuR directly binds and stabilizes IL-17 mRNA, which in turn leads to increased mRNA accumulation and protein production in Th17 cells. Therefore, HuR modulates IL-17 mRNA by transcript stabilization and thus promotes IL-17 expression.
FIGURE 4.
HuR deficiency in Th17 cells impaired IL-17 mRNA stability. CD4+ T cells were isolated from spleen of WT control mice and HuR KO mice. Cells were stimulated as described in Fig. 1 in the presence of Th17 cell–polarizing cytokines for 5 d. Th17-polarized cells were untreated or treated with actinomycin D (Act D; 3 μg/ml) and harvested at 1, 3, and 5 h. Levels of IL-17 (A) and Stat-3 (B) mRNA were determined by real-time PCR. Data in (A) were derived from at least three independent experiments. Data in (B) were derived from two independent experiments.
HuR deficiency impairs cell proliferation
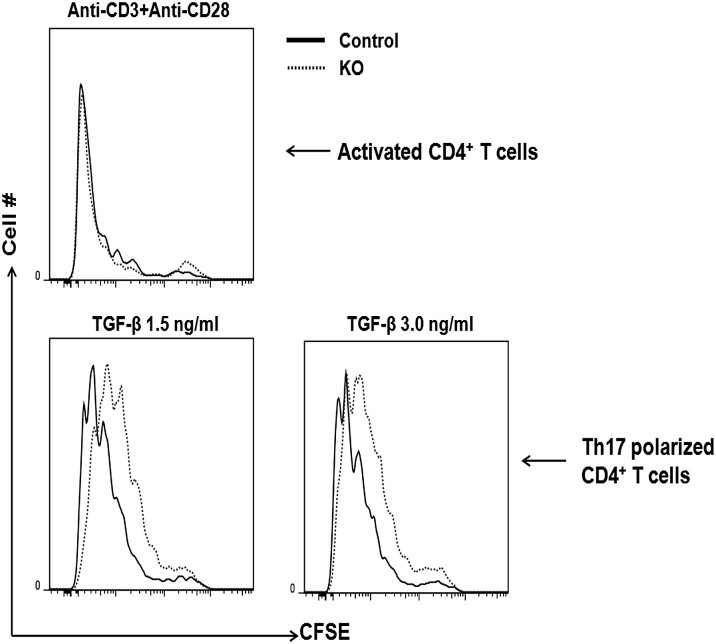
The levels of HuR are increased in many cancers (29, 30), and it promotes colorectal carcinoma proliferation by regulating cyclin A and cyclin B1 mRNA stability (22). To our knowledge, so far there are no reports about HuR regulating Th17 cell proliferation and differentiation. To address this question, we isolated KO and WT CD4+ T cells and labeled them with CFSE to measure proliferation under Th17-polarization conditions. The results showed that HuR KO CD4+ T cells had slightly decreased cell proliferation under Th17-polarization conditions as compared with WT CD4+ T cells (Fig. 5). Because IL-2 plays an important role in T cell proliferation, we measured IL-2 levels but did not find any differences between KO and WT CD4+ T cells, even when supplemented with exogenous TGF-β. (Supplemental Fig. 2). These data suggested that HuR also functions to promote activated T cell proliferation. In contrast, there were minimal differences in cellular differentiation when CD4+ T were merely activated.
FIGURE 5.
HuR deletion in Th17 cells impairs cellular proliferation. CD4+ T cells were isolated from spleens of control and HuR KO mice, labeled with CFSE, and cultured under Th17-polarizing conditions for 5 d. Cell proliferation was measured by flow cytometry assay. HuR KO Th17 cells had reduced proliferation as compared with WT Th17 cells.
We further investigated whether HuR ablation affected T follicular helper (Tfh) and regulatory T cell (Treg) differentiation. We polarized naive CD4+ T cells under either Tfh or Treg conditions and assayed for expression of the relevant transcription factors for these lineages and asked whether there were any alterations in HuR expression. We did not find any appreciable differences in transcriptional factor or HuR expression in Tfh or Treg cells (Supplemental Fig. 3).
HuR deficiency delays onset and reduces severity in EAE development
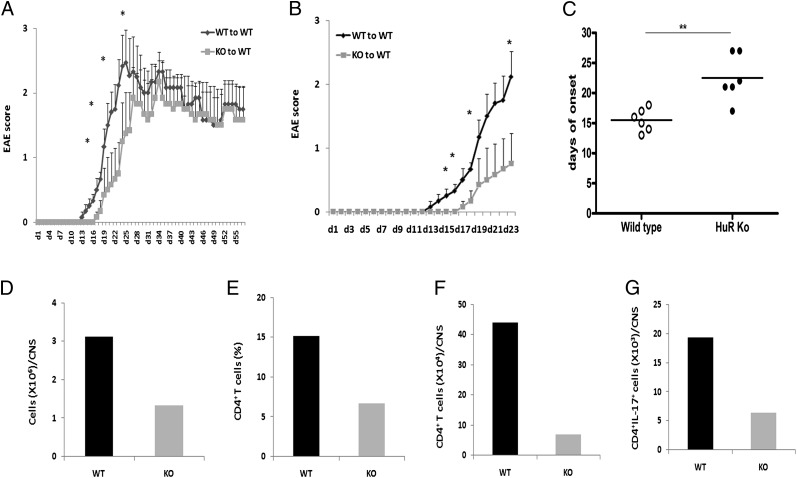
Previously published reports have demonstrated that Th17 cells play a pathogenic role in EAE and multiple sclerosis (13, 31–34). To investigate the function of HuR in vivo, we studied the effect of HuR deficiency on the development of EAE by adoptive transfer of MOG-specific CD4+ T cells into naive mice. WT and KO mice were immunized with MOG and CFA according to methods previously described (34). We monitored EAE disease score severity in recipients daily according to disease criteria (see Materials and Methods). We first performed adoptive transfer experiments and observed the mice for up to 55 d to ascertain the entire course of disease progression. These results suggested that there may be significant differences in disease onset and severity up to day 23; however, these differences were not apparent later on (Fig. 6A). We repeated the adoptive transfer experiments and sacrificed the animals at earlier time points to discover whether there were any differences in trafficking of cells to the CNS.
FIGURE 6.
HuR is required for initiation of EAE. Splenocytes and lymph node cells from immunized control and HuR KO mice were cultured in the presence of MOG35–55 (20 μg/ml) and murine IL-23 (20 ng/ml) for 3 d. CD4+ T cells were isolated, and 6 to 7 × 106 cells were injected i.v. into sublethally irradiated WT mice following two dosed pertussis toxin injections. (A) Disease onset in mice receiving adoptively transferred WT or HuR KO CD4+ T cells and observed until day 55 shows delay in disease initiation. Data in (A) and (B) were summarized for at least six mice per group and are representative of two independent experiments. (B) Development of clinical disease in early time points in recipients after adoptive transfer of WT control or HuR KO CD4+ Th17 polarized T cells. These data represent an independent experiment from (A). (C) Disease onset in mice. Data in (B) were summarized for at least six mice per group and representative of two independent experiments. Error bars in (A) and (B) represent ± SEM. (D–G) Proinflammatory cells in spinal cord of recipients that received WT and HuR KO Th17 cells at day 21 after transfer were isolated and analyzed by flow cytometry. These data are representative of four pairs of mice from two independent experiments. *p < 0.05, **p < 0.01.
The recipients that received WT CD4+ Th17 cells began to show clinical signs of EAE ∼11 d after transfer and reached maximum severity score around day 23 (Fig. 6B). However, in contrast, onset of clinical EAE signs in mice that received KO CD4+ Th17 cells was remarkably delayed (Fig. 6B, 6C), and the development of EAE disease severity was also significantly reduced (Fig. 6C) compared with mice receiving WT CD4+ T cells.
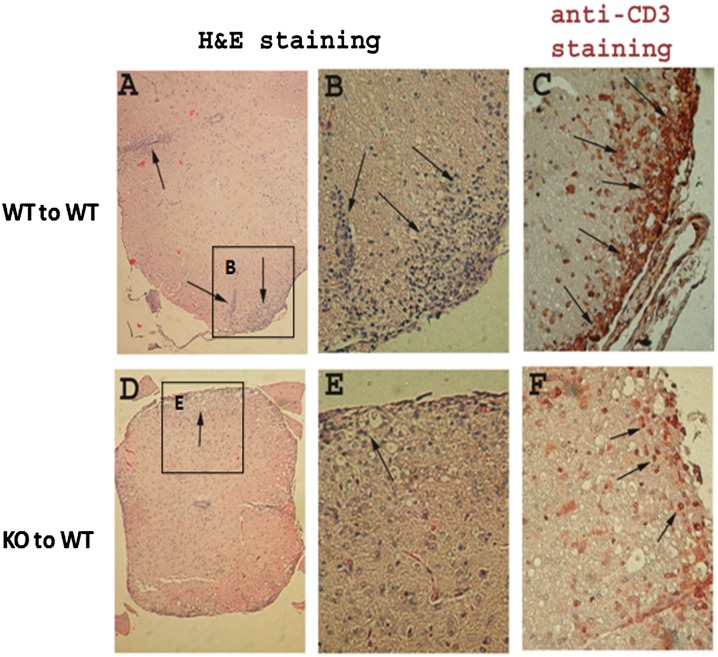
We further isolated monocytes from spinal cords to quantitate and identify CNS inflammatory cells and frequency of IL-17–producing cells by flow cytometry. The results showed that there were reduced numbers of CD4+ T cells and IL-17–positive cells in spinal cords of mice who received KO CD4+ T cells, as compared with cells in recipients that received WT CD4+ T cells (Fig. 6D–G). Consistent with the clinical scores, histological analysis showed that the proinflammatory cell infiltration of the spinal cord was much reduced in recipients of KO CD4+ T cells compared with that of WT CD4+ T cells (Fig. 7A, 7B versus 7D, 7E). Anti-CD3 immunohistochemical staining demonstrated that there was increased inflammatory T cell infiltration of spinal cords of recipients with WT CD4+ T cells, but fewer numbers in recipients of KO CD4+ T cells (Fig. 7C versus 7F).
FIGURE 7.
Histopathology of spinal cords of mice with EAE. Spinal cords were obtained from recipients that received WT (A–C) or HuR KO CD4+ T cells (D–F) at ∼21 d after cell transfer. Representative H&E staining of spinal cord sections (A, B, D, and E) and anti-CD3 immunohistochemical staining of spinal cord sections (C, F) are shown. There are higher numbers of inflammatory cellular infiltrates of spinal cords in mice that received WT CD4+ T cells (A–C) than those that received HuR KO CD4+ T cells (D–F). Staining is representative of four pairs of mice. Original magnification in (A) and (D) is ×100; in (B), (C), (E), and (F), ×400. Arrows point to areas of cellular infiltration. Insets in (A) and (D) are enlarged to full size in (B) and (E), respectively.
These observations revealed that MOG35–55-specific CD4+ T cells from KO mice are less efficient in initiating pathogenesis of EAE. Taken together, these experiments reveal HuR as an important initiating factor in Th17 cell–mediated neuroinflammatory autoimmune disease.
The cytokine GM-CSF has also been previously described as playing an important role in the EAE model of multiple sclerosis (27, 35, 36). Therefore, we asked if HuR ablation altered GM-CSF expression in ex vivo experiments. HuR genetic ablation results in significant decreases in GM-CSF steady-state mRNA levels in activated CD4+ T cells that are commensurate with HuR mRNA reductions but not in protein (data not shown).
Knockdown of HuR in human cells affects IL-17 expression
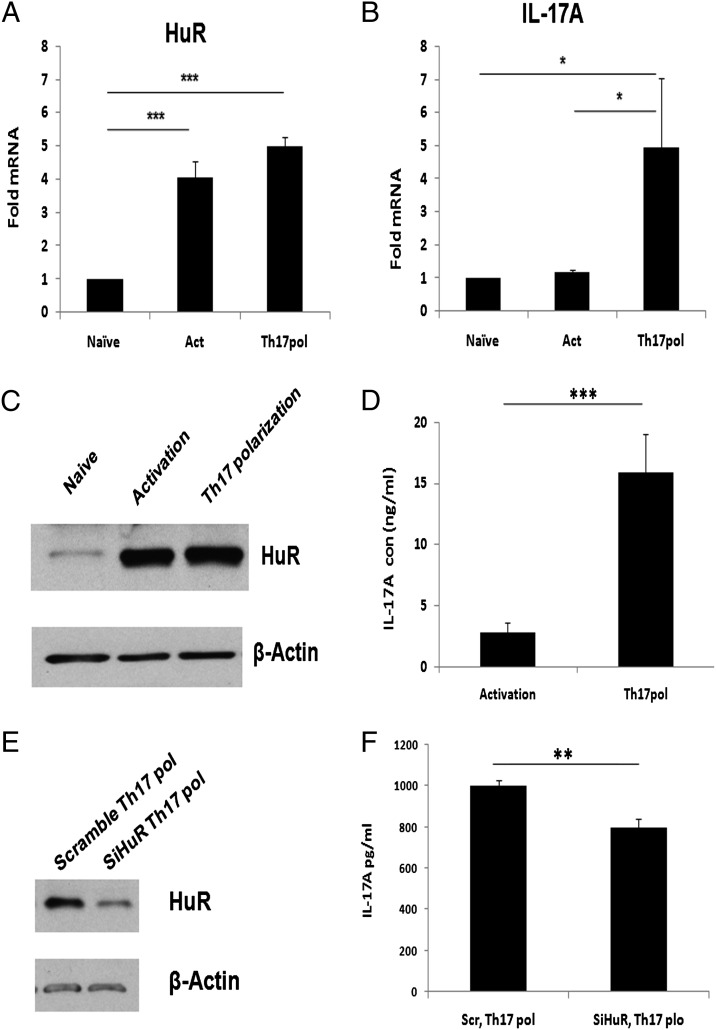
The functional properties of IL-17 in the neuropathology of multiple sclerosis have begun to be elucidated. Studies have demonstrated increased IL-17 levels in the cerebrospinal fluid of the patients with the more severe, opticospinal form of the disease (1, 37, 38). Another study showed that human Th17-differentiated cells could more effectively traverse the blood–brain barrier endothelial cells than Th1-differentiated cells (39). We asked whether HuR-mediated IL-17 expression obtained with murine models could be extended to the human system, because mouse and human cytokines may be regulated differently. To address this question, we isolated naive CD4+ T cells from human PBL and polarized them under Th17 cell conditions. Interestingly, the data showed that increased IL-17 and mRNA and protein levels correlated with increased HuR mRNA level (Fig. 8A, 8B) and protein levels (Fig. 8C, 8D). In order to determine if HuR functionally regulated human IL-17 expression, we knocked down HuR using siRNA in human CD4+ T cells. The results confirmed that knocking down HuR decreased IL-17 expression (Fig. 8E, 8F). These data suggest that HuR also contributes to regulation of IL-17 expression in human CD4+ T cells and thus may also play a role in human disease.
FIGURE 8.
Knockdown of HuR in human CD4+ T cells affected IL-17 expression. (A and B) Naive CD4+ T lymphocytes were isolated from human PBL cells and cultured in the plate precoated with 5 μg/ml of anti-CD3 and 2.5 μg/ml of anti-CD28 (activated) or in the presence of Th17 cell–polarizing cytokines according to the methods described. Cells were collected. (A and B) HuR and IL-17 mRNA expression was measured by real-time PCR. (C and D) Protein levels of HuR and IL-17 were detected by immunoblotting and ELISA, respectively. (E and F) Naive CD4+ T cells were transfected using HuR siRNA or HuR scramble molecules (300 nM) by the Human T Cell Nucleofector Kit and cultured under Th17-differentiation conditions. (E) Western blot assay confirmed that HuR siRNA transfection reduced HuR protein in CD4+ Th17 cells. (F) ELISA assay showed that knocking down HuR in CD4+ T cells decreased IL-17 expression. Data in (A)–(D) were derived from three independent experiments. Data in (E) and (F) were derived from two independent experiments. Error bars represent ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001.
Discussion
HuR has been reported to stabilize transcripts of multiple proinflammatory cytokines including, but not limited to, cyclooxygenase 2, TNF-α, vascular endothelial growth factor, and IL-13 (23–25). In this study, we report that HuR directly binds to IL-17 3′ UTR mRNA and stabilizes its transcript, which in turn results in increases in both IL-17 mRNA and protein levels. Additionally, HuR ablation in CD4+ Th17 cells interferes with Th17 differentiation and decreased IL-17 expression in an adoptive transfer model of EAE. This in turn results in reductions in disease initiation and early severity.
Studies on the regulation of IL-17 expression in T cells have thus far focused on transcriptional activation and associated signaling cascades, which play important roles in its regulation (2–5, 8–12). However, a growing body of evidence indicates that posttranscriptional gene regulation also plays a key but poorly understood role in T cell differentiation following activation (24). HuR plays critical roles in ARE-mediated mRNA stability and translation (40–42). HuR performs its functions closely related with its translocation from the nucleus into cytoplasm by modulating mRNA stability and translation. The function of HuR has been reported in regulating gene expression in many cellular processes, such as cell-cycle control, tumorigenesis, inflammation, apoptosis, and cell stress response (40–44). We previously reported that the rapid decay of mRNA in activated T cells could be changed by the activation of HuR, which binds IL-13 mRNA 3′ UTR in Th2 cells (24). To determine if HuR was expressed in Th17 cells, we assayed both mRNA and protein expression. The results showed that HuR protein is robustly expressed in Th17 cells and HuR KO reduced frequency of IL-17+ cells, as well as IL-17 mRNA and protein levels, as compared with WT Th17 cells. Thus, HuR may play an important posttranscriptional role during Th17 differentiation.
We computationally detected two hits of the putative HuR signature motif in IL-17 mRNA 3′ UTR, suggesting that IL-17 mRNA might be a direct target of HuR. We demonstrated that HuR interacts with IL-17 mRNA in its 3′ UTR region using RIP and biotin pulldown techniques. These results indicated that HuR directly binds to IL-17 mRNA 3′ UTR, but not IL-17 mRNA ORF region.
In T cells, HuR functions by increasing mRNA stabilization, cytoplasmic accumulation, or translation of mRNA transcripts (24–26). In our current study, the significant changes in IL-17 mRNA and protein levels correlated well with HuR levels. This provided additional evidence that HuR acts as a positive regulator for IL-17 mRNA stability similar to what has previously been reported with other proinflammatory cytokine mRNAs, such as IL-13, TNF-α, and GM-CSF (24, 45). Consistent with previous reports, we observed that HuR KO significantly reduced IL-17 mRNA t1/2 compared with that of WT Th17 cells. Therefore, HuR associated with IL-17 mRNA, which makes the IL-17 mRNA more stable. Indeed, HuR has been reported to stabilize IL-4 and IL-13 mRNA in activated T cells (24).
Th17 cells play a pathogenic role in EAE and multiple sclerosis (1, 27, 33–35, 37–39). Our current study showed that mice that received HuR KO MOG-specific Th17 cells exhibited delayed onset, reduced severity scores, and ameliorated histological changes in the initiation of EAE. Flow cytometry analysis showed that the IL-17–producing cells are much reduced in the spinal cord and brain compared with that of WT CD4+ T cells. These results are consistent with earlier reports that in vivo neutralization of IL-17 or genetically mutated IL-17 mice have significantly reduced EAE severity (33). However, we observed these results only in disease initiation and not in overall disease severity at later time points. We hypothesize this may be due to the timing of HuR genetic ablation, because in our system, it takes about 4 d to achieve profound HuR KO. We speculate that earlier HuR genetic ablation, prior to T cell activation and thus Th17 polarization, may perhaps result in more significant phenotypic changes, though this remains to be experimentally determined. Another interesting point is that IFN-γ plays an important role later on in EAE production. Our evidence to date suggests that HuR regulates Th2 and Th17 cytokines but not Th1, such as IFN-γ. Thus, our observations may be due to a lag in IL-17 expression early on in EAE pathogenesis, but IFN-γ production later results in similar disease progression.
It seems plausible that RBPs such as HuR cooperate not only with each other, but also with miRNAs to ultimately affect transcript fate, as has been suggested by Srikantan and colleagues (46). Thus, it will be important in future studies to interrogate IL-17 mRNA milieus to discover trans-acting factors that control its expression and therefore may be novel targets in EAE disease pathogenesis.
Taken together, our study indicates the importance of HuR-controlled posttranscriptional regulatory events in regulation of IL-17 expression in activated CD4+ T cells and extends evidence that HuR functions as an important initiator of autoimmune neuroinflammation. These results also elucidate the important role posttranscriptional events play in Th17 differentiation. Further understanding of molecular pathways of HuR function may identify novel therapeutic targets for intervention in EAE and multiple sclerosis.
Acknowledgments
We thank Habib Zaghouani for assistance with the EAE model and Myriam Gorospe and Yongping Yue for technical assistance.
This work was supported by National Institutes of Health Grants R01AI080870-01 and R21AI079341-01 (to U.A.).
The online version of this article contains supplemental material.
- ARE
- AU-rich element
- EAE
- experimental autoimmune encephalomyelitis
- IP
- immunoprecipitation
- KO
- knockout
- ORF
- open reading frame
- RBP
- RNA-binding protein
- rh
- recombinant human
- RIP
- RNA immunoprecipitation
- RORγt
- retinoic acid–related orphan receptor γt
- siRNA
- small interfering RNA
- Tfh
- T follicular helper
- Treg
- regulatory T cell
- UTR
- untranslated region
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Tesmer L. A., Lundy S. K., Sarkar S., Fox D. A. 2008. Th17 cells in human disease. Immunol. Rev. 223: 87–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. 2005. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6: 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y. K., Turner H., Maynard C. L., Oliver J. R., Chen D., Elson C. O., Weaver C. T. 2009. Late developmental plasticity in the T helper 17 lineage. Immunity 30: 92–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGeachy M. J., Bak-Jensen K. S., Chen Y., Tato C. M., Blumenschein W., McClanahan T., Cua D. J. 2007. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat. Immunol. 8: 1390–1397 [DOI] [PubMed] [Google Scholar]
- 5.Zhou L., Ivanov I. I., Spolski R., Min R., Shenderov K., Egawa T., Levy D. E., Leonard W. J., Littman D. R. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8: 967–974 [DOI] [PubMed] [Google Scholar]
- 6.Hirahara K., Ghoreschi K., Yang X. P., Takahashi H., Laurence A., Vahedi G., Sciumè G., Hall A. O., Dupont C. D., Francisco L. M., et al. 2012. Interleukin-27 priming of T cells controls IL-17 production in trans via induction of the ligand PD-L1. Immunity 36: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Laurence A., Kanno Y., Pacher-Zavisin M., Zhu B. M., Tato C., Yoshimura A., Hennighausen L., O’Shea J. J. 2006. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl. Acad. Sci. USA 103: 8137–8142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei L. A., Laurence A., Elias K. M., O’Shea J. J. 2007. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 282: 34605–34610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathur A. N., Chang H. C., Zisoulis D. G., Stritesky G. L., Yu Q., O’Malley J. T., Kapur R., Levy D. E., Kansas G. S., Kaplan M. H. 2007. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 178: 4901–4907 [DOI] [PubMed] [Google Scholar]
- 10.Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126: 1121–1133 [DOI] [PubMed] [Google Scholar]
- 11.Ichiyama K., Yoshida H., Wakabayashi Y., Chinen T., Saeki K., Nakaya M., Takaesu G., Hori S., Yoshimura A., Kobayashi T. 2008. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J. Biol. Chem. 283: 17003–17008 [DOI] [PubMed] [Google Scholar]
- 12.Yang X. P., Ghoreschi K., Steward-Tharp S. M., Rodriguez-Canales J., Zhu J. F., Grainger J. R., Hirahara K., Sun H. W., Wei L., Vahedi G., et al. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12: 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada H. 2010. Current perspective on the role of IL-17 in autoimmune disease. J. Inflamm. Res. 3: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMullen M. R., Cocuzzi E., Hatzoglou M., Nagy L. E. 2003. Chronic ethanol exposure increases the binding of HuR to the TNF-alpha 3′-untranslated region in macrophages. J. Biol. Chem. 278:38333–38341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H., Jarujaron S., Gurley E. C., Chen L., Ding H., Studer E., Pandak W. M., Jr., Hu W., Zou T., Wang J. Y., Hylemon P. B. 2007. HIV protease inhibitors increase TNF-alpha and IL-6 expression in macrophages: involvement of the RNA-binding protein HuR. Atherosclerosis 195: e134–e143 [DOI] [PubMed] [Google Scholar]
- 16.Abdelmohsen K., Gorospe M. 2010. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA. 1:214–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinman M. N., Lou H. 2008. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 65: 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan X. C., Steitz J. A. 1998. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 17: 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng S. S., Chen C. Y., Xu N., Shyu A. B. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 17: 3461–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford L. P., Watson J., Keene J. D., Wilusz J. 1999. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 13: 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atasoy U., Watson J., Patel D., Keene J. D. 1998. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J. Cell Sci. 111: 3145–3156 [DOI] [PubMed] [Google Scholar]
- 22.Wang W., Caldwell M. C., Lin S., Furneaux H., Gorospe M. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 19: 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vavassori S., Covey L. R. 2009. Post-transcriptional regulation in lymphocytes: the case of CD154. RNA Biol. 6: 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casolaro V., Fang X., Tancowny B., Fan J., Wu F., Srikantan S., Asaki S. Y., De Fanis U., Huang S. K., Gorospe M., et al. 2008. Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR. J. Allergy Clin. Immunol. 121: 853–859, e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stellato C., Gubin M. M., Magee J. D., Fang X., Fan J., Tartar D. M., Chen J., Dahm G. M., Calaluce R., Mori F., et al. 2011. Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. J. Immunol. 187: 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atasoy U., Curry S. L., López de Silanes I., Shyu A. B., Casolaro V., Gorospe M., Stellato C. 2003. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 171: 4369–4378 [DOI] [PubMed] [Google Scholar]
- 27.El-Behi M., Ciric B., Dai H., Yan Y., Cullimore M., Safavi F., Zhang G. X., Dittel B. N., Rostami A. 2011. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12: 568–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu S., Sharp G. C., Braley-Mullen H. 2008. TGF-β promotes thyroid epithelial cell hyperplasia and fibrosis in IFN-γ-deficient NOD.H-2h4 mice. J. Immunol. 181: 2238–2245 [DOI] [PubMed] [Google Scholar]
- 29.Denkert C., Weichert W., Winzer K. J., Müller B. M., Noske A., Niesporek S., Kristiansen G., Guski H., Dietel M., Hauptmann S. 2004. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin. Cancer Res. 10: 5580–5586 [DOI] [PubMed] [Google Scholar]
- 30.López de Silanes I., Fan J., Yang X., Zonderman A. B., Potapova O., Pizer E. S., Gorospe M. 2003. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 22: 7146–7154 [DOI] [PubMed] [Google Scholar]
- 31.Kang Z., Altuntas C. Z., Gulen M. F., Liu C., Giltiay N., Qin H., Liu L., Qian W., Ransohoff R. M., Bergmann C., et al. 2010. Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity 32: 414–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siffrin V., Radbruch H., Glumm R., Niesner R., Paterka M., Herz J., Leuenberger T., Lehmann S. M., Luenstedt S., Rinnenthal J. L., et al. 2010. In vivo imaging of partially reversible th17 cell-induced neuronal dysfunction in the course of encephalomyelitis. Immunity 33: 424–436 [DOI] [PubMed] [Google Scholar]
- 33.Komiyama Y., Nakae S., Matsuki T., Nambu A., Ishigame H., Kakuta S., Sudo K., Iwakura Y. 2006. IL-17 plays an important role in the development of experimental autoimmune encephalomyelitis. J. Immunol. 177: 566–573 [DOI] [PubMed] [Google Scholar]
- 34.Kroenke M. A., Carlson T. J., Andjelkovic A. V., Segal B. M. 2008. IL-12- and IL-23-modulated T cells induce distinct types of EAE based on histology, CNS chemokine profile, and response to cytokine inhibition. J. Exp. Med. 205: 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Codarri L., Gyülvészi G., Tosevski V., Hesske L., Fontana A., Magnenat L., Suter T., Becher B. 2011. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 12: 560–567 [DOI] [PubMed] [Google Scholar]
- 36.Ponomarev E. D., Shriver L. P., Maresz K., Pedras-Vasconcelos J., Verthelyi D., Dittel B. N. 2007. GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J. Immunol. 178: 39–48 [DOI] [PubMed] [Google Scholar]
- 37.Matusevicius D., Kivisäkk P., He B., Kostulas N., Ozenci V., Fredrikson S., Link H. 1999. Interleukin-17 mRNA expression in blood and CSF mononuclear cells is augmented in multiple sclerosis. Mult. Scler. 5: 101–104 [DOI] [PubMed] [Google Scholar]
- 38.Ishizu T. M., Osoegawa M., Mei F. J., Kikuchi H., Tanaka M., Takakura Y., Minohara M., Murai H., Mihara F., Taniwaki T., Kira J. 2005. Intrathecal activation of the IL-17/IL-8 axis in opticospinal multiple sclerosis. Brain 128: 988–1002 [DOI] [PubMed] [Google Scholar]
- 39.Kebir H. K., Kreymborg K., Ifergan I., Dodelet-Devillers A., Cayrol R., Bernard M., Giuliani F., Arbour N., Becher B., Prat A. 2007. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 13: 1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelmohsen K., Lal A., Kim H. H., Gorospe M. 2007. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle 6: 1288–1292 [DOI] [PubMed] [Google Scholar]
- 41.Brennan C. M., Steitz J. A. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58: 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gorospe M. 2003. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle 2: 412–414 [PubMed] [Google Scholar]
- 43.Ghosh M. G., Aguila H. L., Michaud J., Ai Y., Wu M. T., Hemmes A., Ristimaki A., Guo C., Furneaux H., Hla T. 2009. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J. Clin. Invest. 119: 3530–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gubin M. M., Calaluce R., Davis J. W., Magee J. D., Strouse C. S., Shaw D. P., Ma L., Brown A., Hoffman T., Rold T. L., Atasoy U. 2010 Overexpression of the RNA binding protein HuR impairs tumor growth in triple negative breast cancer associated with deficient angiogenesis. Cell Cycle 9: 3337–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esnault S., Malter J. S. 2003. Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J. Immunol. 171: 6780–6787 [DOI] [PubMed] [Google Scholar]
- 46.Srikantan S., Tominaga K., Gorospe M. 2012. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 13: 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]