Abstract
The strain Pseudomonas sp. strain ADP is able to degrade atrazine as a sole nitrogen source and therefore needs a single source for both carbon and energy for growth. In addition to the typical C source for Pseudomonas, Na2-succinate, the strain can also grow with phenol as a carbon source. Phenol is oxidized to catechol by a multicomponent phenol hydroxylase. Catechol is degraded via the ortho pathway using catechol 1,2-dioxygenase. It was possible to stimulate the strain in order to degrade very high concentrations of phenol (1,000 mg/liter) and atrazine (150 mg/liter) simultaneously. With cyanuric acid, the major intermediate of atrazine degradation, as an N source, both the growth rate and the phenol degradation rate were similar to those measured with ammonia as an N source. With atrazine as an N source, the growth rate and the phenol degradation rate were reduced to ∼35% of those obtained for cyanuric acid. This presents clear evidence that although the first three enzymes of the atrazine degradation pathway are constitutively present, either these enzymes or the uptake of atrazine is the bottleneck that diminishes the growth rate of Pseudomonas sp. strain ADP with atrazine as an N source. Whereas atrazine and cyanuric acid showed no significant toxic effect on the cells, phenol reduces growth and activates or induces typical membrane-adaptive responses known for the genus Pseudomonas. Therefore Pseudomonas sp. strain ADP is an ideal bacterium for the investigation of the regulatory interactions among several catabolic genes and stress response mechanisms during the simultaneous degradation of toxic phenolic compounds and a xenobiotic N source such as atrazine.
Atrazine [2-chloro-4-(ethylamino)-6-(isopropylamino)-1,3,5-triazine] is a herbicide used for controlling broadleaf and grassy weeds and is relatively persistent in soils (14). Atrazine and its metabolites have been detected in ground and surface waters at levels exceeding the Environmental Protection Agency's maximum contaminant level of 3 ppb (14).
Pseudomonas sp. strain ADP was the first isolated bacterium capable of degrading the herbicide atrazine (22, 36). Since then, most of our understanding of the genes and enzymes involved in atrazine degradation has been derived from studies using this strain, in which the first three enzymatic steps of atrazine degradation have been defined (8). The genes atzA, atzB, and atzC, which encode the enzymes atrazine chlorohydrolase (AtzA), hydroxyatrazine ethylaminohydrolase (AtzB), and N-isopropylammelide isopropylaminohydrolase (AtzC), convert atrazine sequentially to cyanuric acid (8). Cyanuric acid is catabolized by Pseudomonas sp. strain ADP to carbon dioxide and ammonia (8). These first three genes have been localized on an ∼100-kb plasmid, pADP-1 (8). Recently, pADP-1 was completely sequenced and was shown to contain the genes for the complete catabolism of cyanuric acid to CO2 and NH3 as well, namely, atzD, atzE, and atzF (25). Structural and functional studies showed that the genes encoding the initial reactions of atrazine catabolism are not organized in an operon but are dispersed and flanked by transposase copies.
However, the strain is able to use atrazine and cyanuric acid as sole nitrogen sources and therefore needs an additional carbon and energy source for growth.
In this study, we investigated the degradation of phenol in the presence of different N sources, including the two s-triazines atrazine and cyanuric acid. Thus, we could demonstrate that the direct effects of the adaptation of the cells to phenol, as well as the degradation of atrazine to cyanuric acid, were the major bottleneck that limits effective degradation of atrazine. We also demonstrated that in the ADP strain, genes encoding phenol degradation are located on the chromosome. A multicomponent phenol hydroxylase converts phenol to catechol, which is further metabolized via the catechol degradation ortho pathway.
MATERIALS AND METHODS
Strains and chemicals.
Pseudomonas sp. strain ADP was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM 11735) and has been described previously (22). The bacterial strains and plasmids used in this study are listed in Table 1. All chemicals were reagent grade and were obtained from commercial sources.
TABLE 1.
Strains and plasmid used in this study
| Strain or plasmid | Genetic characteristics | Reference |
|---|---|---|
| Strains | ||
| Pseudomonas fluorescens PC18 | Phe+; multicomponent phenol hydroxylase genes; meta pathway of catechol-degrading genes on chromosome | 16 |
| P. putida PaW85 | Phe−; ortho pathway of catechol degradation genes on chromosome | 3 |
| P. putida PaW340 | Phe− Trp− Sm+; ortho pathway genes on chromosome | 11 |
| Pseudomonas sp. strain ADP | Atr+ Phe+ | 23 |
| Plasmid | ||
| pVI261 | Ap+; dmpKLMNOP genes | 2 |
Culture conditions.
Pseudomonas sp. strain ADP was cultivated in a mineral medium as described by Hartmans et al. (13). The N and C sources in the medium were modified for the specific purposes of the experiments. Adaptation of the cells to phenol was reached by growing the cells semicontinuously in batch cultures with increasing concentrations of phenol up to 1,000 mg/liter. The adapted cells were maintained on agar plates with 500 mg of phenol/liter as the sole C and energy source. Cells were grown in 50-ml shake cultures in a horizontally shaking water bath at 30°C. Growth was monitored by measuring the turbidity (optical density) at 560 nm (OD560). Atrazine was applied in amounts up to 150 mg/liter by using a small reservoir made of an Eppendorf tube with a semipermeable membrane. This guaranteed the constant presence of the maximum atrazine concentration, which corresponds to the maximum solubility of the compound in water (30 mg/liter). This application hindered the precipitation of atrazine in the medium, which would have affected measurement of the OD.
A 1-ml inoculum from an overnight culture was transferred to 50 ml of fresh medium, and cells were grown exponentially for 3 to 4 h (until an OD560 of 0.6 was reached). For molecular biological experiments, all Pseudomonas strains were grown at 30°C on Luria-Bertani (LB) medium (26) or M9 minimal medium (1). If required, phenol was added to a final concentration of 250 mg/liter. Pseudomonas putida PaW340 was incubated in the presence of streptomycin at a final concentration of 1,000 μg/ml and in the presence of tryptophan at a final concentration of 25 μg/ml.
Incubation with toxins.
For measurements of the toxic effects of the investigated compounds, they were added to exponentially growing nonadapted cultures as described by Heipieper et al. (17). The cultures were incubated in the presence of the toxins for 2 h in a shaking water bath at 30°C. Then, the cells were harvested, washed two times with potassium phosphate buffer (50 mM; pH 7.0), and stored at −20°C prior to use for lipid extraction.
Lipid extraction, transesterification, and fatty acid analysis.
The lipids were extracted with chloroform-methanol-water as described by Bligh and Dyer (5). Fatty acid methyl esters (FAME) were prepared by incubation for 15 min at 95°C in boron trifluoride-methanol, applying the method of Morrison and Smith (28). FAME were extracted with hexane.
Analysis of fatty acid composition by GC-MS.
Analysis of FAME in hexane was performed using a quadruple gas chromatography-mass spectrometry (GC-MS) system (HP6890 and HP5973; Hewlett-Packard, Palo Alto, Calif.) equipped with a split-splitless injector. A CP-Sil 88 capillary column (inside diameter, 0.32 m; length, 30 m; 0.25-μm-thick film) (Chrompack, Middelburg, The Netherlands) was used for the separation of the FAME. The GC conditions were as follows. The injector temperature was held at 250°C. The split flow was 1:10, and the carrier gas was He. The temperature program was 80°C, 1-min isotherm, 15°C/min to 140°C and 4°C/min to 280°C. The MS conditions were ionization mode electron ionization and ionization energy of 70 eV. The peak areas of the carboxylic acids in total ion chromatograms were used to determine their relative amounts. The fatty acids were identified by GC-MS and coinjection of authentic reference compounds obtained from Supelco (Bellefonte, Pa.).
Phenol analysis.
The quantification of phenol was done using the colorimetric method of Martin (24). Phenol reacts with 4-aminoantipyrin (4-amino-2,3-dimethyl-1-phenyl-3-pyrazolin-5-on) and forms a red indophenol dye under alkaline conditions. This dye was measured spectrophotometrically at a wavelength of 460 nm.
Enzyme assays.
Cells of strain ADP were grown overnight in M9 minimal medium containing 250 mg of phenol/liter. Then, the culture was diluted in fresh LB medium, LB medium plus 250 mg of phenol/liter, or M9 minimal medium containing 250 mg of phenol/liter. To avoid any stationary-phase-induced enzyme activity, the bacterial culture was diluted with fresh medium to OD560s of 0.005 and 0.002, those of a starting culture in minimal medium and in LB or LB plus phenol, respectively. The cell samples for enzyme assays were taken from cultures grown for 6, 8, 10, 12, and 24 h.
For the preparation of crude extracts, the collected cells were washed with Na2HPO4-KH2PO4 buffer (100 mM; pH 7.5), resuspended in the same buffer, and sonically disrupted. The crude extracts were purified from cell debris and unbroken cells by centrifugation at 12,000 × g and 4°C for 30 min. The activities of the soluble phenol monooxygenase PheA (20) and TbuD (21) in crude extracts were spectrophotometrically monitored by examining the disappearance of NADPH at 340 nm as described previously (4). For the measurement of endogenous oxidation of NADPH, the phenol substrate was omitted from the reaction mixture.
To detect catechol 1,2-dioxygenase (C12O) and catechol 2,3-dioxygenase (C23O) activities, the formation of the reaction products cis-cis-muconic acid and 2-hydroxymuconic semialdehyde was measured spectrophotometrically at 260 and 375 nm, respectively (10, 15). The protein concentration was measured according to the Bradford method (6).
DNA techniques.
The method of Hansen and Olsen (12) was used for plasmid DNA preparation from Pseudomonas sp. strain ADP. The plasmid DNA was fractionated from the total DNA by a CsCl-ethidium bromide density gradient. Separation of genomic DNA in agarose gels and Southern blotting were carried out by the standard procedures described elsewhere (32). Digestion of DNA with restriction enzymes was performed according to the manufacturers' guidelines.
Two primer pairs were used to amplify the DNA region encoding the largest and most conserved subunit of the multicomponent phenol hydroxylase. The primers dmpN1 (5′ GCTGTTCCTCACCGCCG 3′) and dmpN537 (5′ CCGCGCCAGAACCACTTA 3′) were designed to amplify the dmpN gene of Pseudomonas sp. strain CF600, and the second pair of primers used, phe149 (5′ CGATCGACGAGCTGCGCCA 3′) and phe212 (5′ GTTGGTCAGCACGTACTCGAAGGACAA 3′), was the same as that designed by Watanabe et al. (37) based on the analysis of conserved sequences of the large subunit of the prevalent phenol hydroxylase in the environment.
Conjugation.
Pseudomonas sp. strain ADP as a donor and P. putida PaW340 as a recipient were grown overnight in LB medium, and streptomycin was added to the growth medium of PaW340. Then, the cultures were diluted 1:20 (vol/vol) with fresh LB medium and cultivated for 3 h. The cultures were mixed in equal volumes, and 100 μl of the mixture was spotted onto LB medium plates overnight. The next morning, the bacteria were resuspended and washed with Na2HPO4-KH2PO4 buffer (100 mM; pH 7.5) and plated onto M9 minimal medium plates supplemented with 250 mg of phenol/liter as the only carbon source and with tryptophan. Streptomycin was added to selective plates for contraselection for growth of the donor cells.
RESULTS AND DISCUSSION
Growth of Pseudomonas sp. strain ADP with phenol as a sole C source and atrazine or cyanuric acid as a sole N source.
Pseudomonas sp. strain ADP is able to degrade atrazine as a sole nitrogen source and therefore needs an additional carbon and energy source for growth. Besides the typical C source for Pseudomonas, Na2-succinate, the strain can also grow with phenol as a carbon source. Although phenol showed toxic effects on the strain, it was possible to adapt strain ADP to high concentrations of the substance. For maintenance of the strain and for all further degradation experiments, phenol concentrations of 500 mg/liter were chosen, which caused ∼50 percent growth inhibition and the strongest adaptive reactions.
Using the adapted cells, it was possible to degrade phenol as a carbon and energy source and atrazine as an N source simultaneously. To quantify the simultaneous degradation, it was decided to vary the phenol concentrations, whereas the initial concentrations of the tested N sources were kept stable. For this reason, we applied a nominal atrazine concentration of 150 mg/liter.
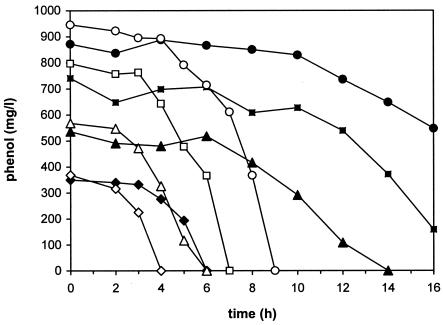
With atrazine as an N source, the strain was able to degrade phenol in amounts of up to 1,000 mg/liter. At higher concentrations, even completely adapted cells were no longer able to grow. Figure 1 shows the degradation of different phenol concentrations; atrazine degradation showed the same depletion curves (data not shown). The highest concentration of phenol was completely degraded after ∼30 h. Thus, the growth rates were up to 0.12 h−1, which corresponds to a doubling time of ∼5.75 h.
FIG. 1.
Comparison of phenol degradation by adapted cells of Pseudomonas sp. strain ADP with 15 mM cyanuric acid (open symbols) and with 150 mg of atrazine/liter (closed symbols) as a sole N source at initial phenol concentrations of 300 (diamonds), 500 (triangles), 750 (squares), and 1,000 (circles) mg/liter.
To compare these data with those for other s-triazines as N sources, the same series of experiments was conducted with the major intermediate of atrazine degradation, cyanuric acid (Fig. 1). In the presence of this N source, the strain degraded phenol much faster; 1,000 mg/liter was completely degraded within 9 h. The growth rates were up to 0.32 h−1, which corresponds to a doubling time of ∼2 h. The degradation rates for phenol were up to 150 mg/h.
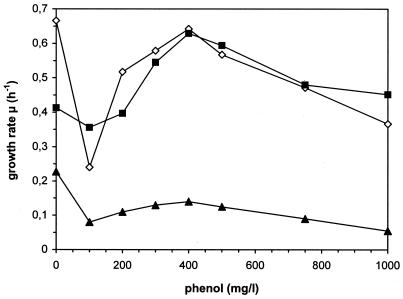
Figure 2 summarizes the measured growth rates, depending on the N source and the initial phenol concentrations, respectively. In the presence of all investigated N sources, the cells showed a typical phenol-dependent course for the growth rates, with a maximum at a phenol concentration of 400 mg/liter. At higher concentrations, the toxic effects of phenol seem to reduce the growth rate of the cells. Comparing the N sources, the growth rates obtained for cyanuric acid were nearly the same as those measured for the N source typically used for this bacterium, ammonia. On the other hand, the growth rates measured for atrazine were only ∼35% of those reached with the other two N sources and the same initial phenol concentrations (Fig. 2).
FIG. 2.
Comparison of growth rates of Pseudomonas sp. strain ADP growing on NH4+ (▪), cyanuric acid (⋄), and atrazine (▴) as N sources. The data given for the concentration of 0 mg of phenol/liter correspond to the control cultures grown on Na2 succinate as a C source.
Contrary to this, cells grown with a constant initial phenol concentration of 500 mg/liter and various atrazine concentrations showed no significant differences in their growth rates. Thus, only a correlation between an increased atrazine concentration and an increasing cell yield could be observed (data not shown).
These data showed that it was possible to cultivate Pseudomonas sp. strain ADP with phenol as a sole C and energy source and simultaneously with atrazine or cyanuric acid as an N source. To our knowledge, this is the first description of the simultaneous degradation of two hazardous compounds used by a single bacterium as the C and N source, respectively.
The differences between growth and phenol degradation rates obtained with cyanuric acid and with atrazine as N sources are difficult to explain. Atrazine had no major toxic effects on the cells, and in previous molecular biological studies, it had been shown that the first three enzymes involved in atrazine degradation to cyanuric acid are constitutively present in the cells (8). However, despite this information the degradation of atrazine to cyanuric acid seems to be the bottleneck that diminishes the growth rate of Pseudomonas sp. strain ADP with atrazine as an N source. One explanation for this obviously reduced efficiency in using atrazine might be the lower bioavailability of this substrate, which shows a much lower solubility than, e.g., cyanuric acid. Therefore, the reduced uptake of atrazine might be the factor causing the diminished growth rates with this compound.
Membrane adaptation of Pseudomonas sp. strain ADP to phenolic compounds and other forms of stress.
The results mentioned above led to the conclusion that in addition to the regulation of the catabolic genes for either atrazine or phenol, toxicity might also directly influence the simultaneous degradation of atrazine and phenol. Therefore, we also investigated the toxic effects of the two compounds on strain ADP. For this reason, nonadapted cells grown in mineral medium with ammonia and succinate as the N and C source, respectively, were incubated with different concentrations of the compounds and growth, as well as membrane-adaptive responses, was observed.
Microorganisms are able to adapt to the presence of toxic organic compounds by using a whole cascade of adaptive mechanisms (for a review, see reference 31). Among the adaptive mechanisms, changes in the fatty acid composition of membrane lipids are the most important reactions of bacteria to membrane-active substances. One adaptive mechanism enabling several Pseudomonas strains to grow in the presence of membrane-disrupting compounds is the isomerization of cis-unsaturated fatty acids to trans-unsaturated fatty acids (17, 35). This mechanism could also be found in Pseudomonas sp. strain ADP. The extent of the isomerization, usually expressed as the trans/cis ratio of unsaturated fatty acids, apparently correlates with the toxicity of organic compounds (18, 19). Additionally, a mutual dependence was found between the activation of this system and the induction or activation of other stress response mechanisms. Therefore, an increase in the trans/cis ratio can be used as an indicator of environmental stress (35).
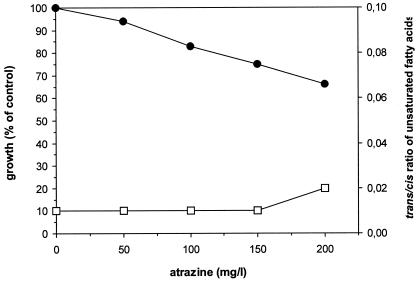
Figure 3 shows the effects of atrazine on the growth and membrane of strain ADP. Atrazine causes only a slight toxic effect on the bacteria. Even at concentrations above the water solubility of the compound (30 mg/liter), the growth of the cells was not completely inhibited. This is also reflected in the trans/cis ratio, where no increase could be observed.
FIG. 3.
Effect of atrazine on growth (•) and trans/cis ratio of unsaturated membrane fatty acids (□) of nonadapted cells of Pseudomonas sp. strain ADP.
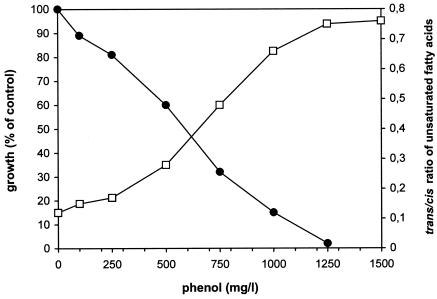
The contrary result can be observed with phenol, as presented in Fig. 4. Although Pseudomonas sp. strain ADP is able to degrade phenol, at the same time, this substrate evokes or causes a strong inhibition of cellular growth. In the presence of phenol, the trans/cis ratio of unsaturated fatty acids also showed a toxicity-dependent increase.
FIG. 4.
Effect of phenol on growth (•) and trans/cis ratio of unsaturated membrane fatty acids (□) of nonadapted cells of Pseudomonas sp. strain ADP.
Increasing concentrations of atrazine had only minor inhibiting effects on the growth rate and cell adaptation of Pseudomonas sp. strain ADP. This can be explained by the very limited water solubility of the compound, which is only ∼30 mg/liter under the experimental conditions. Very surprising was the observation that atrazine showed the slight increase in toxicity even at concentrations which were much higher than the solubility in water. This seems to be an indication of either an accumulation of the compound in the membranes of the cells or the presence of compounds produced by the cells which increase the solubility of atrazine. On the other hand, the simultaneous growth with phenol as a carbon source might provide the explanation for this increased toxicity of atrazine, because phenol also induces or activates the adaptive response mechanisms of the bacteria to this toxic compound. As toxic effects might indeed play a major role in the regulation of the catabolic genes during a simultaneous degradation of the two xenobiotic compounds, it was decided to characterize the phenol degradation in this strain.
Pseudomonas sp. strain ADP uses a multicomponent phenol hydroxylase for phenol oxidation.
The first step of phenol degradation is determined by the phenol hydroxylase, which may contain only one subunit (monocomponent enzyme) or several subunits (multicomponent phenol hydroxylase). The multicomponent phenol hydroxylase of Pseudomonas sp. strain CF600 is well known and has six subunits, although only five seem to be needed for the production of this multicomponent enzyme (30). The genes that encode subunits of the multicomponent phenol hydroxylase of CF600 are located in a plasmid-encoded operon, the dmp operon (29). Many multicomponent phenol hydroxylase-encoding operons have a DNA sequence and organization similar to that of the dmp operon. On the other hand, some bacteria, such as Pseudomonas sp. strain EST1001 (20) and Pseudomonas pickettii strain PKO1 (21), use monocomponent phenol hydroxylases which do not show significant similarities.
Therefore, the first investigations of the molecular biology of phenol degradation in strain ADP were related to the description of the phenol hydroxylase the strain uses for phenol oxidation.
As the monocomponent phenol hydroxylase (PheA) of Pseudomonas sp. strain EST1001 and TbuD of P. pickettii PKO1 are measurable from crude cell extracts and use NADPH to add molecular oxygen to the aromatic ring, we made cell extracts of strain ADP and spectrophotometrically monitored the disappearance of NADPH at 340 nm. No phenol hydroxylase activity was measurable in the cell extracts (data not shown).
The multicomponent phenol hydroxylase is possibly involved in phenol oxidation. In order to prove this possibility, we used the PCR method to try to amplify a DNA segment of the ADP genome by using primers complementary to the gene of the largest and the most conserved subunit of the multicomponent phenol hydroxylase (37). The ∼250-bp PCR product we obtained was sequenced, and the results indicated that ADP contains a sequence for the phenol hydroxylase large subunit gene which is very similar (94% identity [data not shown]) to that of P. putida strain rN1 described by Watanabe et al. (37). The phenol hydroxylase in strain rN1 belongs, together with the Dmp family hydroxylases, to group III of multicomponent phenol hydroxylases (37).
Localization of the phenol degradation genes in Pseudomonas sp. strain ADP.
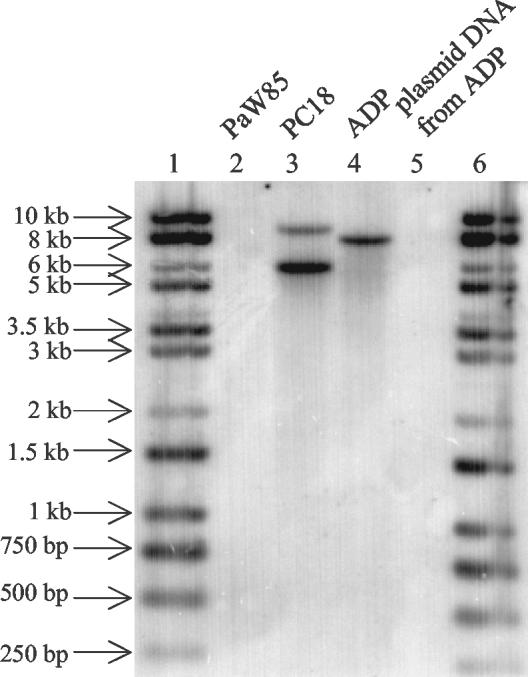
Bacteria can have their phenol-degrading genes located either on plasmids or on the chromosome (27). Pseudomonas sp. strain ADP contains two plasmids, pADP-1 and pADP-2 (8). pADP-1 was sequenced, and no genes exhibiting homology to the genes for phenol degradation could be found on the 109-kb plasmid (25). In order to test whether the genes for phenol hydroxylase could be located on another plasmid, the 53-kb pADP-2, we conjugated ADP as a donor and P. putida strain PaW340 (which contains catechol degradation ortho pathway genes but lacks a phenol hydroxylase gene[s]) as a recipient. No P. putida PaW340 transconjugants were obtained on the selective plates. To specify the location of phenol degradation genes in the ADP strain, Southern blot analysis of a total-DNA preparation of ADP was performed. A radiolabeled 537-bp PCR product, which was amplified from the dmpN gene of plasmid pVI261 by primers dmp1 and dmp537, was used as a specific hybridization probe. The radiolabeled PCR product hybridized with the total DNA of ADP (Fig. 5, lane 4) and with the total DNA of PC18 (lane 3) as a positive control. No hybridization could be detected in the case of the plasmid DNA from ADP (lane 5) and the total DNA of PaW85 as a negative control (lane 2). This indicates that at least one subunit of the phenol hydroxylase gene in strain ADP is localized on the chromosome (Fig. 5).
FIG. 5.
Autoradiograph of Southern analysis of total EcoRI-digested DNA of P. putida PaW85 (lane 2), Pseudomonas fluorescens PC18 (lane 3), and Pseudomonas sp. strain ADP (lane 4) and plasmid DNA of ADP digested with EcoRI (lane 5) hybridized with radiolabeled probe specific for the dmpN gene of multicomponent phenol hydroxylase of Pseudomonas sp. strain CF600. The molecular sizes of the marker DNA (lanes 1 and 6) are given on the left.
Pseudomonas sp. strain ADP utilizes phenol via the ortho pathway of catechol degradation.
Phenol is usually degraded via the catechol degradation pathway. There are two pathways for catechol ring fission, the meta and ortho pathways (34). The majority of bacteria use the meta pathway of catechol degradation, especially if the bacteria have multicomponent phenol hydroxylases, like Pseudomonas sp. strain CF600 (30, 34). However, there is also evidence that the multicomponent phenol hydroxylase and the ortho pathway of catechol degradation can coexist (9). Both meta and ortho pathways are distinguishable by measuring characteristic enzymes, C23O for the meta pathway and C12O for the ortho pathway. The activities of both enzymes were measured in ADP cells grown in phenol minimal media. Activity of C12O, but no evidence of C23O, could be detected in a crude extract of ADP cells. The activity of C12O in the crude extracts of logarithmically growing (6-h) cells was 1.66 ± 0.24 μmol/mg × min. This demonstrates that Pseudomonas sp. strain ADP uses the ortho pathway of catechol degradation on phenol catabolism.
The phenol degradation in ADP is down-regulated in rich medium.
Various reports have demonstrated that promoters of biodegradative operons are down-regulated in response to exponential growth in rich media irrespective of the presence of an effector, a phenomenon referred to as catabolite repression or exponential silencing (reviewed by Cases and de Lorenzo [7]). In order to study whether ADP phenol degradation would also be repressed in rich medium, the strain was grown in LB medium in the presence of phenol and the activity of the ortho pathway enzyme, C12O, was measured (Fig. 6). No C12O activity could be detected when the bacteria were grown in LB medium in the absence of phenol. In the presence of phenol, the C12O activity was measurable only when the cells were entering stationary phase, and it increased during stationary phase, demonstrating that expression of the phenol degradation pathway in Pseudomonas sp. strain ADP is also under physiological control.
FIG. 6.
Growth curve (▪) and measurement of C12O activity (□) from cell extracts of Pseudomonas sp. strain ADP in LB medium supplemented with 250 mg of phenol/liter. Data (means ± standard deviations) from at least three independent experiments are presented.
The genes atzA, atzB, and atzC, which encode the enzymes of atrazine degradation to cyanuric acid, are constitutively expressed and are not regulated, either by induction of atrazine or by repression of other N sources (8, 25). However, the genes for the complete catabolism of cyanuric acid to CO2 and NH3, atzD, atzE, and atzF (25), are regulated by RpoN (σ54) and induced by cyanuric acid and N depletion (E. Santero, personal communication). As RpoN is also known to be involved in the regulation of many promoters that control phenol degradation operons, there might be links between the regulations of both degradation pathways in Pseudomonas sp. strain ADP. Very recently, it was proven that the uptake of atrazine is repressed by more easily consumable N sources, such as ammonia, urea, and serine (E. Santero, personal communication). Thus, the main regulator for nitrogen metabolism, σ54, seems to be responsible for this repression of atrazine uptake and degradation. On the other hand, it has also been shown that several catabolic genes for aromatic compounds are regulated by either σ54 or the main stress regulator, σ38 (σS) (29, 33).
With this background, Pseudomonas sp. strain ADP is an ideal bacterium for the investigation of the regulatory interactions among several catabolic genes and stress response mechanisms during the simultaneous degradation of toxic phenolic compounds and a xenobiotic N source such as atrazine.
Acknowledgments
This work was supported by a bilateral grant given by the German Ministry for Education and Research (WTZ-BMBF), contract EST 02/006. This work was partially supported by contracts QLK3-CT-1999-00041 and QLRT-2001-00435 of the European Commission within its Fifth Framework Programme and by grant 4481 from the Estonia Science Foundation.
REFERENCES
- 1.Adams, M. H. 1959. Bacteriophages, p. 445-447. Interscience Publishers Inc., New York, N.Y.
- 2.Bartilson, M., I. Nordlund, and V. Shingler. 1990. Location and organization of the dimethylphenol catabolic genes of Pseudomonas CF600. Mol. Gen. Genet. 220:294-300. [DOI] [PubMed] [Google Scholar]
- 3.Bayley, S. A., C. J. Duggleby, M. J. Worsey, P. A. Williams, K. G. Hardy, and P. Broda. 1977. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol. Gen. Genet. 154:203-204. [DOI] [PubMed] [Google Scholar]
- 4.Beadle, C. A., and A. R. Smith. 1982. The purification and properties of 2,4-dichlorophenol hydroxylase from a strain of Acinetobacter species. Eur. J. Biochem. 123:323-332. [DOI] [PubMed] [Google Scholar]
- 5.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Cases, I., and V. de Lorenzo. 2001. The black cat/white cat principle of signal integration in bacterial promoters. EMBO J. 20:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza, M. L., L. P. Wackett, and M. J. Sadowsky. 1998. The atzABC genes encoding atrazine catabolism are located on a self-transmissible plasmid in Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 64:2323-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrt, S., F. Schirmer, and W. Hillen. 1995. Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIB8250. Mol. Microbiol. 18:13-20. [DOI] [PubMed] [Google Scholar]
- 10.Feist, C. F., and G. D. Hegeman. 1969. Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J. Bacteriol. 100:869-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin, F. C., and P. A. Williams. 1980. Construction of a partial diploid for the degradative pathway encoded by the TOL plasmid (pWWO) from Pseudomonas putida mt-2: evidence for the positive nature of the regulation by the xyIR gene. Mol. Gen. Genet. 177:321-328. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, J. B., and R. H. Olsen. 1978. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J. Bacteriol. 135:227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartmans, S., J. P. Smits, M. J. van der Werf, F. Volkering, and J. A. M. de Bont. 1989. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl. Environ. Microbiol. 55:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayes, W. J., Jr., and E. R. Laws, Jr. (ed.). 1991. Classes of pesticides, p. 1317-1383. In Handbook of pesticide toxicology, vol. 3. Academic Press, New York, N.Y.
- 15.Hegeman, G. D. 1966. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. 3. Isolation and properties of constitutive mutants. J. Bacteriol. 91:1161-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinaru, E., J. Truu, U. Stottmeister, and A. Heinaru. 2000. Three types of phenol and p-cresol catabolism in phenol- and p-cresol-degrading bacteria isolated from river water continuously polluted with phenolic compounds. FEMS Microbiol. Ecol. 31:195-205. [DOI] [PubMed] [Google Scholar]
- 17.Heipieper, H. J., R. Diefenbach, and H. Keweloh. 1992. Conversion of cis-unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol. 58:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heipieper, H. J., F. J. Weber, J. Sikkema, H. Keweloh, and J. A. M. de Bont. 1994. Mechanisms behind resistance of whole cells to toxic organic solvents. Trends Biotechnol. 12:409-415. [Google Scholar]
- 19.Keweloh, H., and H. J. Heipieper. 1996. trans-unsaturated fatty acids in bacteria. Lipids 31:129-137. [DOI] [PubMed] [Google Scholar]
- 20.Kivisaar, M., R. Horak, L. Kasak, A. Heinaru, and J. Habicht. 1990. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid 24:25-36. [DOI] [PubMed] [Google Scholar]
- 21.Kukor, J. J., and R. H. Olsen. 1992. Complete nucleotide sequence of tbuD, the gene encoding phenol/cresol hydroxylase from Pseudomonas pickettii PKO1, and functional analysis of the encoded enzyme. J. Bacteriol. 174:6518-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandelbaum, R. T., D. L. Allan, and L. P. Wackett. 1995. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol. 61:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandelbaum, R. T., L. P. Wackett, and D. L. Allan. 1993. Mineralization of the s-triazine ring of atrazine by stable bacterial mixed cultures. Appl. Environ. Microbiol. 59:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, R. W. 1949. Rapid colorimetric estimation of phenol. Anal. Chem. 21:1419-1420. [Google Scholar]
- 25.Martinez, B., J. Tomkins, L. P. Wackett, R. Wing, and M. J. Sadowsky. 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 183:5684-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Mishra, V., R. Lal, and Srinivasan. 2001. Enzymes and operons mediating xenobiotic degradation in bacteria. Crit. Rev. Microbiol. 27:133-166. [DOI] [PubMed] [Google Scholar]
- 28.Morrison, W. R., and L. M. Smith. 1964. Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J. Lipid. Res. 5:600-608. [PubMed] [Google Scholar]
- 29.Powlowski, J., and V. Shingler. 1994. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation 5:219-236. [DOI] [PubMed] [Google Scholar]
- 30.Powlowski, J., and V. Shingler. 1990. In vitro analysis of polypeptide requirements of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J. Bacteriol. 172:6834-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramos, J. L., E. Duque, J. J. Rodriguez Herva, P. Godoy, A. Haidour, F. Reyes, and A. Fernandez Barrero. 1997. Mechanisms for solvent tolerance in bacteria. J. Biol. Chem. 272:3887-3890. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sze, C. C., L. M. Bernardo, and V. Shingler. 2002. Integration of global regulation of two aromatic-responsive sigma(54)-dependent systems: a common phenotype by different mechanisms. J. Bacteriol. 184:760-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Schie, H. T., E. M. Bakker, A. M. Jonker, and P. R. van Weeren. 2000. Ultrasonographic tissue characterization of equine superficial digital flexor tendons by means of gray level statistics. Am. J. Vet. Res. 61:210-219. [DOI] [PubMed] [Google Scholar]
- 35.von Wallbrunn, A., H. H. Richnow, G. Neumann, F. Meinhardt, and H. J. Heipieper. 2003. Mechanism of cis-trans isomerization of unsaturated fatty acids in Pseudomonas putida. J. Bacteriol. 185:1730-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wackett, L. P., M. J. Sadowsky, B. Martinez, and N. Shapir. 2002. Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol. 58:39-45. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, K., M. Teramoto, H. Futamata, and S. Harayama. 1998. Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl. Environ. Microbiol. 64:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]