Abstract
The fluoropyrimidines are the mainstay chemotherapeutic agents for the treatment of many types of cancers. Detoxifying metabolism of fluoropyrimidines requires dihydropyrimidine dehydrogenase (DPD, encoded by the DPYD gene), and reduced or absent activity of this enzyme can result in severe, and sometimes fatal, toxicity. We summarize evidence from the published literature supporting this association and provide dosing recommendations for fluoropyrimidines based on DPYD genotype (updates at http://www.pharmgkb.org).
The purpose of this guideline is to provide information to allow the interpretation of clinical dihydropyrimidine dehydrogenase (DPYD) genotype tests so that the results can be used to guide dosing of fluoropyrimidines (5-fluorouracil, capecitabine, and tegafur). Detailed guidelines for use of fluoropyrimidines, their clinical pharmacology (see ref. 1 for review), and analyses of the cost-effectiveness are beyond the scope of this article. The Clinical Pharmacogenetics Implementation Consortium guidelines consider the situation of patients for whom genotype data are already available.2
Focused Literature Review
A systematic literature review focused on the DPYD genotype and the use of 5-fluorouracil, capecitabine, and tegafur (details in Supplementary Material online) was conducted, with reviews used as summaries of earlier literature.
Gene: DPYD
Background
Dihydropyrimidine dehydrogenase (DPD) is the rate-limiting enzyme for fluoropyrimidine catabolism and eliminates >80% of administered 5-fluorouracil.3 DPD levels show high inter- and intraindividual variation, and this variability is likely to influence response of patients to 5-fluorouracil with respect to toxicity, resistance, and efficacy.4 In patients who are deficient in DPD, 5-fluorouracil can cause profound toxicity, such as myelosuppression, mucositis, neurotoxicity, hand–foot syndrome, and diarrhea. Familial studies have demonstrated that this is an autosomal codominantly inherited trait.5
DPYD, the gene encoding DPD, is a large gene with 4,399 nucleotides in 23 coding exons spanning 950 kb on chromosome 1p22.6 The most well-studied variant is DPYD*2A (also known as DPYD:IVS14 + 1G>A, c.1905+1G>A, or rs3918290).7 It is a single-nucleotide polymorphism at the intron boundary of exon 14 that results in a splicing defect, skipping of the entire exon, and a nonfunctional protein.8 Recently, Offer et al. measured the relative sensitivity to 5-fluorouracil of cells expressing DPD variations and confirmed that DPYD*2A is catalytically inactive.9 This allele is considered relatively rare, although it is more common than most other known inactivating variants in DPYD. Estimates of the frequency of the *2A allele range from <0.005 (in the HapMap CEU, YRI, JPT, and HCB populations and several other studies) to 3.5% in a Swedish population.10
The most frequently observed variants are *5 (rs1801159 T>C), *6 (rs1801160 C>T), and *9A (rs1801265 A>G) at frequencies of 11.5–30, 0.7–9, and 2.9–13.7%, respectively, and data regarding their effects on DPD activity are contradictory.6,11,12,13,14 However, the Dutch Pharmacogenetics Working Group has designated these alleles as “functional” on the basis of the lack of an association with toxicity reported in studies and/or decreased clearance or activity.15 DPYD*3 (rs72549303 C>del), *13 (rs55886062 A>C), and rs67376798T>A are also relatively rare but result in low DPD activity and/or 5-fluorouracil toxicity (see Supplementary Tables S1 and S2 online).5,16,17 Moreover, most variants of phenotypic consequence in DPYD are of very low frequency, and several studies did not observe any individuals with these variants.11,13,14,18,19 Recently, a novel DPYD variant (Y186C) was identified only in the African-American population (found in 26% of African-American patients with reduced DPD activity). Individuals carrying this allele had a 46% reduction in DPD activity as compared with noncarriers.20
Patients with <70% of the mean observed leukocyte DPD protein activity in the normal population are considered at risk for the development of severe toxicity after administration of 5-fluorouracil (or its prodrugs).21 The relationship between DPYD genotype and phenotype is complicated; although several variants have been associated with low DPD activity (*2A, *13, and rs67376798) and fluoropyrimidine toxicity (*2A, *13, and rs67376798) or have been observed in other cases of toxicity (*2A, *4, *6, *9A, *13, and rs67376798), the presence of these variants does not always result in toxicity, and associations have not been consistently replicated (discussed in refs. 9,11,19,22 and other publications).
The inconsistency in study results may be explained by the substantial variation in treatment regimens across studies. In a study by Schwab et al.11 including 683 cancer patients, DPYD*2A was found to play a limited role in 5-fluorouracil-related toxicity; of note, only patients receiving 5-fluorouracil monotherapy were included. By contrast, Morel et al. found strong associations in a cohort of 487 patients between both DPYD*2A and rs67376798 and severe 5-fluorouracil toxicity in patients receiving combination therapy. Studies linking DPYD*2A to toxicity generally include patients on combination therapies, suggesting that concomitant drugs may enhance the effect of DPYD risk alleles. Furthermore, Schwab et al. also observed a higher rate of severe toxicities in patients receiving bolus-based 5-fluorouracil than in patients receiving continuous infusion, suggesting a dose-dependent effect of 5-fluorouracil. Moreover, several studies have shown that only ~50% of heterozygote carriers of a low-activity allele develop severe 5-fluorouracil toxicity.11,18,19 This may indicate allelic regulation of DPYD or compensation by another DPYD variant on the second allele, resulting in greater DPD activity.22 Recently, DPYD haplotypes (e.g., haplotype B3) have been considered to be more predictive in identifying patients at risk for severe 5-fluorouracil-related19 and capecitabine-related toxicities (grade ≥3).23 However, data on functional consequences of these haplotypes are so far incomplete. Promoter methylation altered expression in cell lines;24 however, methylation was not associated with toxicity in patients.11 MicroRNAs have also been implicated in the regulation of DPYD, but their relevance for the modulation of DPD phenotypes with respect to drug response has not been tested.25 Nevertheless, >20% (23.3–38%) of 5-fluorouracil toxicities can be explained by combining multiple DPYD variants, suggesting a significant importance of DPYD variation for the risk of 5-fluorouracil-related toxicities.18,19,26,27
Genetic test interpretation
Each named * allele is defined by the genotype at one or more specific single-nucleotide polymorphisms (Supplementary Table S1 online). DPD function associated with the most common allelic variants is summarized in Supplementary Table S2 online. Table 1 summarizes the assignment of the probable DPD phenotype on the basis of the * allele diplotypes, and these assignments are used to link genotypes with fluoropyrimidine dosing. Briefly, homozygotes of *2A, *13, and rs67376798 are considered deficient in DPD; heterozygotes for any combination of *2A, *13, and rs67376798 have intermediate or partial DPD activity; and those with none of these alleles are likely to have normal, high activity. DPYD alleles have been extensively studied in multiple geographically, racially, and ethnically diverse groups and are summarized in Supplementary Tables S3 and S4 online. Because of conflicting data or weak evidence for alleles other than *2A, *13, and rs67376798, this guideline does not currently report dosing recommendations for other variants of DPYD. Reports of other variants and phenotypes are discussed in the Supplementary Material online.
Table 1. Assignment of likely DPD phenotype based on genotype.

Available genetic test options
There are several testing options for DPYD genotype, although, at present most test only for the DPYD*2A variant. A list of testing services is provided in an online linked format at PharmGKB (http://www.pharmgkb.org/gene/PA145) and the National Institutes of Health Genetic Testing Registry (http://www.ncbi.nlm.nih.gov/gtr/conditions/C2720286/ or http://www.ncbi.nlm.nih.gov/gtr/conditions/CN077983/).
Incidental findings
Individuals who harbor one copy of variant DPYD can be considered to have carrier status for an inborn error of metabolism, and consideration should be given to its potential effects on offspring. Patients homozygous for inactivating variants of the DPYD gene have DPD deficiency, a disease that shows large phenotypic variability, ranging from no symptoms to severe convulsive disorders with motor and mental retardation.28,29
Other considerations
Several other genes may influence responses to 5-fluorouracil3,11 (see Supplementary Material online). The well-studied genes among these are ABCB1, MTHFR, and TYMS, although results have been inconsistent to date, and predictive dosing strategies have yet to be successfully applied. Some of the testing options for 5-fluorouracil toxicity and DPYD also include testing for other gene variants in TYMS and MTHFR. For a summary of pharmacogenomic studies of 5-fluorouracil, see the PGx Research tab at http://www.pharmgkb.org/drug/PA128406956.
There are alternatives to genotyping of DPYD that assess DPD activity directly, including dihydrouracil/uracil ratio determination in plasma, the uracil breath test method, measurement of DPD activity in peripheral mononuclear cells, and pharmacokinetically guided strategies such as the 5-fluorouracil test dose method (see ref. 30 for further information). Studies using dose reduction of 5-fluorouracil in patients with DPD deficiency, as evidenced by the use of one of these functional tests, have shown a reduction in drug-related toxicities while maintaining efficacy in these patients.31,32
Drugs: Fluoropyrimidines
Background
Fluoropyrimidines such as 5-fluorouracil, capecitabine, and tegafur are widely used in the treatment of solid tumors, including colorectal and breast cancer and cancers of the aerodigestive tract. More than 2 million patients receive these types of drugs annually.19 Approximately 10–40% of 5-fluorouracil patients develop severe, and sometimes life-threatening, toxicity (neutropenia, nausea, vomiting, severe diarrhea, stomatitis, mucositis, hand–foot syndrome, and neuropathy).19
Only 1–3% of the administered 5-fluorouracil dose has been found to be metabolized to cytotoxic metabolites, with ~80% of the administered dose being degraded or excreted in the urine. DPD is the first and rate-limiting step in the catabolic pathway converting 5-fluorouracil to dihydrofluorouracil.3 Dihydrofluorouracil is then converted to fluoro-β-ureidopropionate and fluoro-β-alanine, which are then excreted in the urine (http://www.pharmgkb.org/pathway/PA150653776).3 Capecitabine and tegafur are prodrugs of 5-fluorouracil that are converted to 5-fluorouracil and then metabolized by DPD as described above.
Fluoropyrimidines are mostly used in combination with various other antineoplastic drugs. Disease and treatment regimens (which are also related to disease background—for example, breast cancer patients tend to receive bolus 5-fluorouracil, whereas colorectal cancer patients tend to receive infusion 5-fluorouracil) may also influence the importance of DPD activity to risk for toxicity.
Linking genetic variability to variability in drug-related phenotypes
There is substantial evidence linking DPYD genotype with phenotypic variability in DPD enzyme activity, 5-fluorouracil clearance, and subsequently 5-fluorouracil toxicity (Supplementary Table S5 online). Evidence providing the basis for the dosing recommendations (Table 2) is from two large prospective studies,11,18 small studies with retrospective genotyping of patients with severe toxicity, and case studies (see Supplementary Table S5 online).
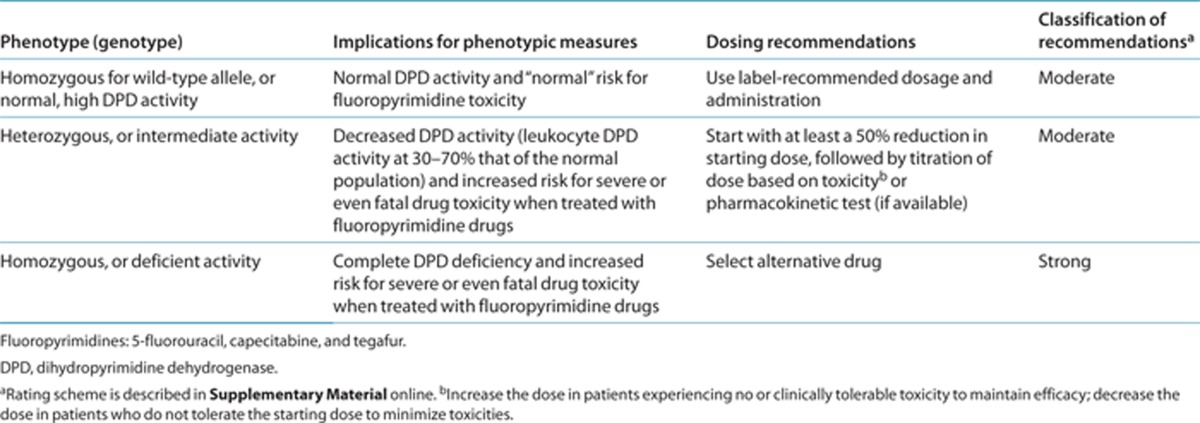
Table 2. Recommended dosing of fluoropyrimidines by DPD phenotype.
5-Fluorouracil has a relatively narrow therapeutic window, resulting in a small difference between an efficacious dose and the maximum tolerable dose. Reduced activity of DPD, resulting in reduced clearance and increased half-life of 5-fluorouracil, results in increased risk of dose-dependent severe toxicities.33,34,35 Morel et al.18 compared the 5-fluorouracil clearances of patients with DPYD*2A (n = 8 heterozygotes; n = 1 homozygote), *13 (n = 1 heterozygote), and rs67376798 (n = 10 heterozygotes) with those of patients without a known variant or with DPYD*9A or c.1590T>C. Mean clearances were 74.9 ± 38.3 l/h·m2(range: 21.2–183.5 l/h·m2) and 132.3 ± 46.6 l/h·m2 (range: 36.8–369.7 l/h·m2), respectively. They were statistically different (P < 0.001). Boisdron-Celle et al.35 compared the 5-fluorouracil clearances in patients with DPYD*2A (n = 2 heterozygotes) and rs67376798 (n = 7 heterozygotes) with the clearances in those with no relevant variant (n = 163). 5-Fluorouracil clearances in patients heterozygous for either DPYD*2A or rs67376798 were 80 and 40–58% (depending on treatment regimen), respectively, less than the clearances in the group with no variant. One patient had three heterozygote variants: DPYD*2A, rs67376798, and 85 T > C, resulting in a 5-fluorouracil plasma clearance close to zero. Both studies found a significant difference in grade 3–4 toxicities in patients with these variants as compared with patients lacking variants. These data indicate that patients heterozygous for these variants have significantly reduced 5-fluorouracil clearances, ranging from 40 to 80% less than the clearances in patients without these variants.
Dosage recommendations
Table 2 summarizes the genetics-based dosing recommendations for DPYD genotype and fluoropyrimidines. The strength of the dosing recommendations is based on the facts that some variants (DPYD*2A, *13, and rs67376798) clearly affect DPD activity, DPD activity is clearly related to 5-fluorouracil clearance, and 5-fluorouracil exposure is associated with its toxic effects. Therefore, reduction of fluoropyrimidine dosage in patients with these variants may prevent severe and possibly life-threatening toxicities. However, available evidence does not clearly indicate a degree of dose reduction needed to prevent fluoropyrimidine-related toxicities. Supplementary Table S6 online summarizes the effects of these variants on 5-fluorouracil clearance and DPD activity. Although the data suggest that patients with the DPYD*2A variant may need a greater dose reduction than a patient with the rs67376798 variant, it is unclear to what extent the dose should be reduced. Furthermore, patients who are heterozygous for the nonfunctional DPYD variants mostly demonstrate partial DPD deficiency (leukocyte DPD activity at 30–70% that of the normal population).16,21,26,34,35 Thus, our recommendation is to start with at least a 50% reduction of the starting dose; followed by an increase in dose in patients experiencing no or clinically tolerable toxicity, to maintain efficacy; and a decrease in dose in patients who do not tolerate the starting dose, to minimize toxicities. An alternative is pharmacokinetic-guided dose adjustment (if available). Patients who are homozygous for DPYD*2A, *13, or rs67376798 may demonstrate complete DPD deficiency,17,21,23 and the use of 5-fluorouracil or capecitabine is not recommended in these patients. Because capecitabine and tegafur are converted to 5-fluorouracil and then metabolized by DPD, the clearance of and exposure to 5-fluorouracil, in addition to its toxic effects, are similar in patients with these variants.23,36
The US Food and Drug Administration (FDA) has added statements to the drug labels for 5-fluorouracil (topical only) and capecitabine that contraindicate use in patients with DPD enzyme deficiency. The FDA drug label also warns to use precaution with intravenous 5-fluorouracil in these patients. The Dutch Pharmacogenetics Working Group has evaluated therapeutic dose recommendations for 5-fluorouracil, capecitabine, and tegafur (5-fluorouracil prodrug combined with uracil; not available in United States).15 The Working Group recommends the use of an alternative drug for homozygous carriers of a decreased-activity allele and a reduced dose or alternative drug to capecitabine or 5-fluorouracil for heterozygous carriers of a decreased-activity allele.15
At the time of this writing, there are no data available on the possible role of DPYD*2A, *13, or rs67376798 in 5-fluorouracil toxicities in pediatric patient populations; however, there is no reason to suspect that variant DPYD alleles would affect 5-fluorouracil metabolism differently in children as compared with adults.
Recommendations for incidental findings
DPD deficiency is a clinically heterogeneous autosomal recessive disorder of pyrimidine metabolism resulting in wide variability of clinical presentations.28 These symptoms generally present in childhood, with the majority of patients showing symptoms within the first year of life. Currently, there is no correlation between symptom severity and DPD function and/or genetics. However, early diagnosis is crucial because of the potential of life-threatening defects. Therefore, early phenotypic (e.g., urine screening of uracil and its degradation products) and/or genetic testing (pre- or postnatal) of offspring of DPYD-variant carriers could aid in early diagnosis and prevent unnecessary and costly diagnostic testing.29,37
Other considerations
Some studies have suggested that the patient's gender may influence the likelihood of 5-fluorouracil toxicity, although the results have been contradictory and the mechanism is unknown. Several studies showed increased numbers of women, as compared with men, among patients with 5-fluorouracil toxicity,11,26 although in one study, this was not significant when excluding patients with breast cancer, suggesting in that instance that the effect may have been an artifact of different treatment regimens.26 Early studies showed lower DPD activity and lower clearance of 5-fluorouracil in women as compared with men.38,39 In one of the largest studies of DPYD variants and 5-fluorouracil toxicity, the association of DPYD*2A with increased risk for toxicity was more significant in men than in women, even though there was no difference between DPD activity and protein content in histologically normal liver tissues of male and female donors.11 The use of folinic acid and the mode of 5-fluorouracil infusion have also been associated with 5-fluorouracil toxicity and should be considered when estimating a patient's individual risk of toxicity with 5-fluorouracil (see ref. 11 for further information).
There is some evidence from the work of Gamelin et al.40 that individual pharmacokinetically guided dosing of 5-fluorouracil therapy in patients with metastatic colorectal cancer improves treatment outcome with a reduced number of 5-fluorouracil-related adverse drug reactions. Of note, in a recent paper by Deenen et al., data are provided indicating that the cumulative dose of capecitabine per course was significantly decreased in patients heterozygous for DPYD*2A (~50%) or rs67376798 (~25%) as compared with that in patients with the wild-type allele, and therapy could be continued safely in variant cases. Unfortunately, no data on capecitabine pharmacokinetics are included. Indirect determination of the endogenous uracil/dihydrouracil plasma ratio as a surrogate for DPD activity could be used to titrate 5-fluorouracil-based chemotherapy with reduction of severe 5-fluorouracil-related toxicities.31
Potential Benefits and Risks for the Patient
The main benefit to the patient would be the potential to avoid toxicity by using either alternative therapy or lower fluoropyrimidine doses. The aim is to prevent the most severe and fatal instances of toxicity, but some patients who would not have experienced this degree of toxicity and who would have benefited from fluoropyrimidine therapy may be advised against it. Moreover, heterozygous patients who receive a lower dose of a fluoropyrimidine and who would not have experienced this degree of toxicity may not experience the full benefit of fluoropyrimidine therapy; therefore, it is important to increase the dose in patients experiencing no or clinically tolerable toxicity to maintain efficacy. Patients who proceed with 5-fluorouracil therapy may still experience lower-grade toxicity that may be acceptable and even necessary in order to achieve efficacy. Some patients without a variant DPYD allele may still experience severe toxicity due to other genetic, environmental, or other factors.
A possible risk is the misreporting or misinterpretation of genotype test results. This mistake could be recorded in the patient record and could also influence further treatments.
Caveats: Appropriate use and/or Potential Misuse of Genetic Tests
The positive predictive value and negative predictive value of DPYD*2A genotyping to predict development of severe toxicity (grade 3) are ~50 and ~95%, respectively;11 however, taking into account other variant alleles, such as rs67376798 and DPYD*13, increases the positive predictive value to 62% (the negative predictive value remains unchanged).18 Furthermore, the sensitivity calculated in this study for this genotype test was only 31%; therefore, the absence of these variants does not rule out DPD defects. Although many additional variants of DPYD are known (see Supplementary Tables S1, S3, and S4 online), the frequencies are often very low, and evidence for their functionality is limited.
Disclaimer
Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written and are intended only to assist clinicians in decision making and to identify questions for further research. New evidence may have emerged since the time a guideline was submitted for publication. Guidelines are limited in scope and are not applicable to interventions or diseases that are not specifically identified. Guidelines do not account for individual variations among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the health-care provider to determine the best course of treatment for a patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be made solely by the clinician and the patient. CPIC assumes no responsibility for any injury or damage to persons or property arising out of or related to any use of CPIC's guidelines or for any errors or omissions.
Acknowledgments
We acknowledge the critical input of members of CPIC of the Pharmacogenomics Research Network, particularly Mary V. Relling (St Jude Children's Research Hospital), funded by the National Institutes of Health (NIH). This work was funded by NIH grants R24 GM61374, U01 GM092666, and U01 HL0105918. This work was also supported by the German Federal Ministry of Education and Research (BMBF grant 03 IS 2061C) and the Robert Bosch Foundation, Stuttgart, Germany.
The authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Supplementary Material
References
- Diasio R.B., Harris B.E. Clinical pharmacology of 5-fluorouracil. Clin. Pharmacokinet. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- Relling M.V., Klein T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn C.F., Marsh S., Carrillo M.W., McLeod H.L., Klein T.E., Altman R.B. PharmGKB summary: fluoropyrimidine pathways. Pharmacogenet. Genomics. 2011;21:237–242. doi: 10.1097/FPC.0b013e32833c6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming R.A., et al. Correlation between dihydropyrimidine dehydrogenase activity in peripheral mononuclear cells and systemic clearance of fluorouracil in cancer patients. Cancer Res. 1992;52:2899–2902. [PubMed] [Google Scholar]
- Johnson M.R., Wang K., Diasio R.B. Profound dihydropyrimidine dehydrogenase deficiency resulting from a novel compound heterozygote genotype. Clin. Cancer Res. 2002;8:768–774. [PubMed] [Google Scholar]
- Wei X., et al. Characterization of the human dihydropyrimidine dehydrogenase gene. Genomics. 1998;51:391–400. doi: 10.1006/geno.1998.5379. [DOI] [PubMed] [Google Scholar]
- Wei X., McLeod H.L., McMurrough J., Gonzalez F.J., Fernandez-Salguero P. Molecular basis of the human dihydropyrimidine dehydrogenase deficiency and 5-fluorouracil toxicity. J. Clin. Invest. 1996;98:610–615. doi: 10.1172/JCI118830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kuilenburg A.B., et al. Heterozygosity for a point mutation in an invariant splice donor site of dihydropyrimidine dehydrogenase and severe 5-fluorouracil related toxicity. Eur. J. Cancer. 1997;33:2258–2264. doi: 10.1016/s0959-8049(97)00261-x. [DOI] [PubMed] [Google Scholar]
- Offer S.M., Wegner N.J., Fossum C., Wang K., Diasio R.B. Phenotypic profiling of DPYD variations relevant to 5-fluorouracil sensitivity using real-time cellular analysis and in vitro measurement of enzyme activity. Cancer Res. 2013;73:1958–1968. doi: 10.1158/0008-5472.CAN-12-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofverholm A., Arkblad E., Skrtic S., Albertsson P., Shubbar E., Enerbäck C. Two cases of 5-fluorouracil toxicity linked with gene variants in the DPYD gene. Clin. Biochem. 2010;43:331–334. doi: 10.1016/j.clinbiochem.2009.09.024. [DOI] [PubMed] [Google Scholar]
- Schwab M., et al. German 5-FU Toxicity Study Group Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J. Clin. Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- He Y.F., et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in Chinese cancer patients. J. Clin. Pharm. Ther. 2008;33:307–314. doi: 10.1111/j.1365-2710.2008.00898.x. [DOI] [PubMed] [Google Scholar]
- Ben Fredj R., et al. Mutational spectrum of dihydropyrimidine dehydrogenase gene (DPYD) in the Tunisian population. C. R. Biol. 2007;330:764–769. doi: 10.1016/j.crvi.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Maekawa K., et al. Genetic variations and haplotype structures of the DPYD gene encoding dihydropyrimidine dehydrogenase in Japanese and their ethnic differences. J. Hum. Genet. 2007;52:804–819. doi: 10.1007/s10038-007-0186-6. [DOI] [PubMed] [Google Scholar]
- Swen J.J., et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- van Kuilenburg A.B., et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: identification of new mutations in the DPD gene. Clin. Cancer Res. 2000;6:4705–4712. [PubMed] [Google Scholar]
- Vreken P., Van Kuilenburg A.B., Meinsma R., van Gennip A.H. Dihydropyrimidine dehydrogenase (DPD) deficiency: identification and expression of missense mutations C29R, R886H and R235W. Hum. Genet. 1997;101:333–338. doi: 10.1007/s004390050637. [DOI] [PubMed] [Google Scholar]
- Morel A., et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol. Cancer Ther. 2006;5:2895–2904. doi: 10.1158/1535-7163.MCT-06-0327. [DOI] [PubMed] [Google Scholar]
- Amstutz U., Farese S., Aebi S., Largiadèr C.R. Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics. 2009;10:931–944. doi: 10.2217/pgs.09.28. [DOI] [PubMed] [Google Scholar]
- Offer S.M., Lee A.M., Mattison L.K., Fossum C., Wegner N.J., Diasio R.B. A DPYD variant (Y186C) in individuals of african ancestry is associated with reduced DPD enzyme activity. Clin. Pharmacol. Ther. 2013;94:158–166. doi: 10.1038/clpt.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kuilenburg A.B., Meinsma R., Zoetekouw L., Van Gennip A.H. Increased risk of grade IV neutropenia after administration of 5-fluorouracil due to a dihydropyrimidine dehydrogenase deficiency: high prevalence of the IVS14+1g>a mutation. Int. J. Cancer. 2002;101:253–258. doi: 10.1002/ijc.10599. [DOI] [PubMed] [Google Scholar]
- Amstutz U., Froehlich T.K., Largiadèr C.R. Dihydropyrimidine dehydrogenase gene as a major predictor of severe 5-fluorouracil toxicity. Pharmacogenomics. 2011;12:1321–1336. doi: 10.2217/pgs.11.72. [DOI] [PubMed] [Google Scholar]
- Deenen M.J., et al. Relationship between single nucleotide polymorphisms and haplotypes in DPYD and toxicity and efficacy of capecitabine in advanced colorectal cancer. Clin. Cancer Res. 2011;17:3455–3468. doi: 10.1158/1078-0432.CCR-10-2209. [DOI] [PubMed] [Google Scholar]
- Noguchi T., et al. Aberrant methylation of DPYD promoter, DPYD expression, and cellular sensitivity to 5-fluorouracil in cancer cells. Clin. Cancer Res. 2004;10:7100–7107. doi: 10.1158/1078-0432.CCR-04-0337. [DOI] [PubMed] [Google Scholar]
- Hirota T., et al. Dihydropyrimidine dehydrogenase (DPD) expression is negatively regulated by certain microRNAs in human lung tissues. Lung Cancer. 2012;77:16–23. doi: 10.1016/j.lungcan.2011.12.018. [DOI] [PubMed] [Google Scholar]
- Van Kuilenburg A.B., Meinsma R., Zoetekouw L., Van Gennip A.H. High prevalence of the IVS14 + 1G>A mutation in the dihydropyrimidine dehydrogenase gene of patients with severe 5-fluorouracil-associated toxicity. Pharmacogenetics. 2002;12:555–558. doi: 10.1097/00008571-200210000-00007. [DOI] [PubMed] [Google Scholar]
- Gross E., et al. Detailed analysis of five mutations in dihydropyrimidine dehydrogenase detected in cancer patients with 5-fluorouracil-related side effects. Hum. Mutat. 2003;22:498. doi: 10.1002/humu.9201. [DOI] [PubMed] [Google Scholar]
- van Gennip A.H., Abeling N.G., Vreken P., van Kuilenburg A.B. Inborn errors of pyrimidine degradation: clinical, biochemical and molecular aspects. J. Inherit. Metab. Dis. 1997;20:203–213. doi: 10.1023/a:1005356806329. [DOI] [PubMed] [Google Scholar]
- Schmidt C., et al. Comprehensive analysis of pyrimidine metabolism in 450 children with unspecific neurological symptoms using high-pressure liquid chromatography-electrospray ionization tandem mass spectrometry. J. Inherit. Metab. Dis. 2005;28:1109–1122. doi: 10.1007/s10545-005-0133-7. [DOI] [PubMed] [Google Scholar]
- van Staveren M.C., Guchelaar H.J., van Kuilenburg A.B., Gelderblom H., Maring J.G. Pharmacogenomics J. e-pub ahead of print 16 July 2013; 2013. Evaluation of predictive tests for screening for dihydropyrimidine dehydrogenase deficiency. [DOI] [PubMed] [Google Scholar]
- Yang C.G., et al. DPD-based adaptive dosing of 5-FU in patients with head and neck cancer: impact on treatment efficacy and toxicity. Cancer Chemother. Pharmacol. 2011;67:49–56. doi: 10.1007/s00280-010-1282-4. [DOI] [PubMed] [Google Scholar]
- Capitain O., Boisdron-Celle M., Poirier A.L., Abadie-Lacourtoisie S., Morel A., Gamelin E. The influence of fluorouracil outcome parameters on tolerance and efficacy in patients with advanced colorectal cancer. Pharmacogenomics J. 2008;8:256–267. doi: 10.1038/sj.tpj.6500476. [DOI] [PubMed] [Google Scholar]
- Diasio R.B., Beavers T.L., Carpenter J.T. Familial deficiency of dihydropyrimidine dehydrogenase. Biochemical basis for familial pyrimidinemia and severe 5-fluorouracil-induced toxicity. J. Clin. Invest. 1988;81:47–51. doi: 10.1172/JCI113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maring J.G., et al. Reduced 5-FU clearance in a patient with low DPD activity due to heterozygosity for a mutant allele of the DPYD gene. Br. J. Cancer. 2002;86:1028–1033. doi: 10.1038/sj.bjc.6600208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisdron-Celle M., et al. 5-Fluorouracil-related severe toxicity: a comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Lett. 2007;249:271–282. doi: 10.1016/j.canlet.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Cellier P., et al. Phase II study of preoperative radiation plus concurrent daily tegafur-uracil (UFT) with leucovorin for locally advanced rectal cancer. BMC Cancer. 2011;11:98. doi: 10.1186/1471-2407-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurecka A. Inborn errors of purine and pyrimidine metabolism. J. Inherit. Metab. Dis. 2009;32:247–263. doi: 10.1007/s10545-009-1094-z. [DOI] [PubMed] [Google Scholar]
- Etienne M.C., et al. Population study of dihydropyrimidine dehydrogenase in cancer patients. J. Clin. Oncol. 1994;12:2248–2253. doi: 10.1200/JCO.1994.12.11.2248. [DOI] [PubMed] [Google Scholar]
- Milano G., Etienne M.C., Pierrefite V., Barberi-Heyob M., Deporte-Fety R., Renée N. Dihydropyrimidine dehydrogenase deficiency and fluorouracil-related toxicity. Br. J. Cancer. 1999;79:627–630. doi: 10.1038/sj.bjc.6690098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamelin E., et al. Individual fluorouracil dose adjustment based on pharmacokinetic follow-up compared with conventional dosage: results of a multicenter randomized trial of patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:2099–2105. doi: 10.1200/JCO.2007.13.3934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


