Abstract
Nitrogenase converts N2 to NH3, at one face of an Fe-Mo-S cluster (FeMo-co) buried in the protein. Through exploration of cavities in the structures of nitrogenase proteins, a pathway for the egress of ammonia from its generation site to the external medium is proposed. This pathway is conserved in the three species Azotobacter vinelandii, Klebsiella pneumoniae and Clostridium pasteurianum. A molecular mechanism for the translocation of NH3 by skipping through a sequence of hydrogen bonds involving eleven water molecules and surrounding aminoacids has been developed. The putative mechanism requires movement aside of some water molecules by up to ~ 1Å. Consistent with this, the surrounding protein is comprised of different chains and has little secondary structure: protein fluctuations are part of the mechanism. This NH3 pathway is well separated from the water chain and embedded proton wire that have been proposed for serial supply of protons to FeMo-co. Verification procedures are suggested.
The enzyme nitrogenase converts dinitrogen to ammonia N2 to NH3, with some reduction of protons to dihydrogen: N2 + 6H+ + 6e− → 2NH3; 2H+ + 2e− → H2. There is considerable understanding of the biochemical processes of this enzyme. The two proteins that comprise nitrogenase are the MoFe protein in which the chemical catalysis occurs at the iron-molybdenum cofactor (FeMo-co), and the Fe protein where the reducing potential is generated. The catalytic site, FeMo-co, is an unprecedented CFe7MoS9 cluster (Fig. 1(a)) containing obligatory homocitrate (HCA, 2-hydroxybutane-1,2,4-tricarboxylate). Also in the MoFe protein is a redox-variable Fe8S7 cluster called the P-cluster, approximately 12Å from FeMo-co, and understood to mediate electron transfer to FeMo-co. The Fe protein contains an Fe4S4 cluster and the nucleotide (MgATP or MgADP) binding sites. The biochemical cycle involves docking of the Fe protein with the MoFe protein, hydrolysis of 2MgATP, and transfer of one electron via the Fe4S4 cluster at the docking interface to the P-cluster and then to FeMo-co1,2,3,4,5,6,7,8,9. This biochemical cycle of association, hydrolysis, electron transfer, dissociation is repeated eight times during reduction of N2 to 2NH3 with concomitant formation of H2.
Figure 1.
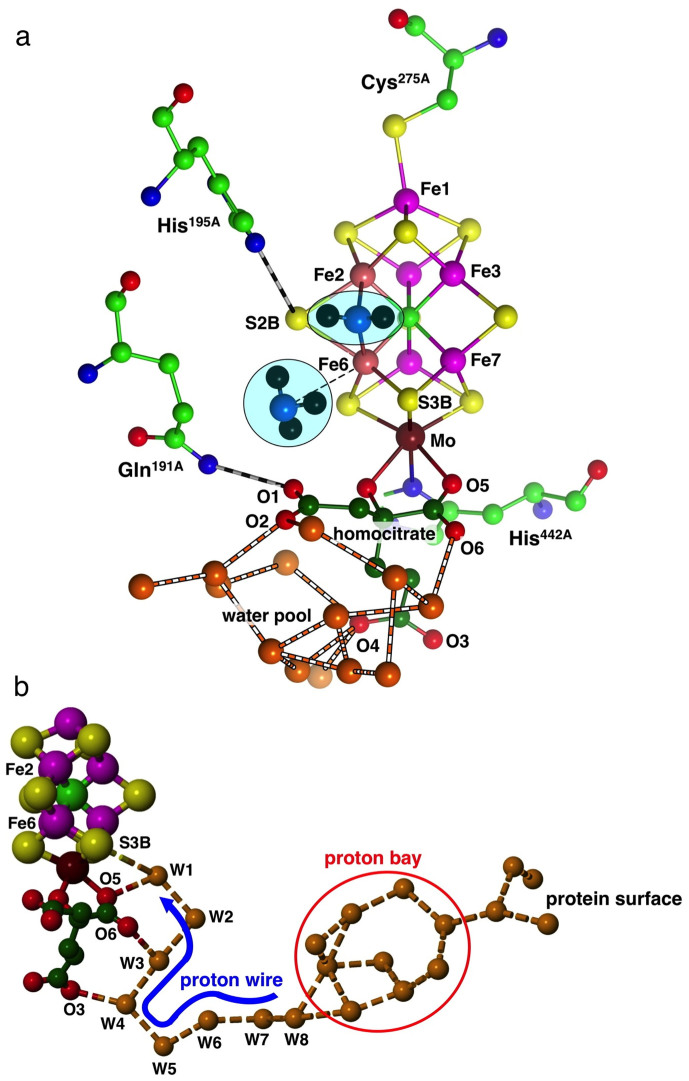
(a) The structure of FeMo-co including homocitrate (carbon dark green) and ligating residues α-275Cys at Fe1 and α-442His at Mo: atom labels are for the Azotobacter vinelandii protein in PDB 3U7Q. Also shown are key hydrogen bonded residues Gln191A and His195A, and water molecules (orange) of the water pool around homocitrate, The first product NH3 molecule is dissociating from Fe6, the second is to be formed from NH2 bridging Fe2 and Fe6 (in this and some following figures Fe2 and Fe6 are coloured differently). (b) The chain of hydrogen bonded water molecules from the protein surface to S3B of FeMo-co. The proton wire section W8 to W1 is fully conserved, while the branched proton bay section and the path to the surface are variable.
The understanding of mechanism at the chemical level is poorer, in part due to experimental difficulties in trapping and characterising intermediates. However, investigations of the reactivities of proteins with modified residues have revealed the locus of chemical catalysis, the Fe2,Fe3,Fe6,Fe7, face of FeMo-co, and particularly the Fe2-S2B-Fe6 region directly underneath the side chain of α-70Val 10,11,12. The surrounding residues α-195His, α-191Gln, α-69Gly and α-96Arg when modified also influence the reactivity of nitrogenase6,8,13,14,15,16,17,18,19,20,21,22.
A series of density functional investigations23,24,25,26,27 has led to a detailed 21-step mechanism for the conversion of N2 to NH328,29. A key aspect of this mechanism is the serial formation, migration, and accumulation of multiple H atoms on FeMo-co, and their sequential transfer to bound substrates and intermediates to generate hydrogenated products25,30. Coupled electron transfer and proton transfer to FeMo-co are proposed to generate the H atoms at S3B. A requirement of this general intramolecular hydrogenation mechanism for nitrogenase is a controlled pathway for serial provision of external protons to S3B, and in a recent paper I described the chain of water molecules that leads from the surface of the protein to S3B, Fig. 1(b). A Grotthuss mechanism for serial translocation of protons along the fully conserved single chain of hydrogen bonded water molecules W8 to W1 (see Fig. 1(b)) to S3B was developed31. This inner section is a tight proton wire: the outer section is variable and is postulated to function as a proton bay.
In this paper I report explorations for the pathway used for diffusion of product NH3/NH4+ from the reaction site to the surface of the protein. This product egress aspect of the activity of nitrogenase has been largely ignored. Durrant32 recognized the water chain that transports protons (Fig. 1(b)) and suggested that it could be used for transport of ammonia, but ammonia and protons cannot travel in opposite directions along the same water channel. The eight water molecules comprising the inner section of the proton wire are ideally hydrogen bonded to each other and to the enclosing homocitrate and key residues, in order to facilitate and control the Grotthuss proton transfer. Intrusion or substitution of NH3 here, and creation of NH4+, would block proton transfer to FeMo-co (by the formation of too many hydrogen bonds to allow the Grotthuss proton hopping) and in the same manner would freeze translocation of ammonia. It is clear that separate pathways are needed for the proton supply to FeMo-co and the efflux of NH3. In the structures of MoFe proteins there is a collection of water molecules around homocitrate2, and it has been suggested that this water pool could absorb product ammonia31. However this does not account for transport of product ammonia to the protein surface, which is the question addressed here.
The translocation of ammonia through other proteins has been investigated in detail, principally for the ammonium transporter (Amt) proteins which control movement of NH3/NH4+ across cell membranes in bacteria, archaea, fungi, plants and animals33,34,35,36, and for a collection of glutamine amidotransferase (GAT) enzymes. These latter enzymes utilise NH3, derived from the hydrolysis of glutamine, in a synthase reaction. They transport the NH3 from the generation site to a distant synthase domain through intramolecular tunnels protected from the environment37,38,39,40,41,42. The consensus interpretation, based on structural information, targeted mutations, molecular dynamics simulations, and calculation of intra-protein pKa, is that NH3 rather than NH4+ is the moving entity in these glutamine amidotransferases37,43,44,45,46,47. However, the Amt proteins (and related human Rhesus proteins48,49) have different functions, including transport of NH4+ against a concentration gradient and discrimination against K+. Three mechanisms are under consideration: transport of NH3, transport of NH4+, and co-transport of NH3 and H+ 33,34,35,50,51,52,53,54,55,56,57,58,59.
Results
This investigation is based on detailed exploration of the structures of crystals containing the MoFe protein, from different species and in various trapped states, and including the independent occurrences of FeMo-co and surrounds in each protein. Unless stated otherwise, amino-acid numbering is that of the Azotobacter vinelandii (Av) MoFe protein, and water molecules are labeled as in the highest resolution structure, PDB 3U7Q60. Residue labels contain letters A, B. to denote the chain in the PDB file. In the following the term ‘water path' is reserved for the collection shown in Fig. 1 (b), and the term ‘water pool' refers to the different collection of water molecules around homocitrate (Fig. 1(a)).
Generation of NH3
Product NH3 molecules are expected to be generated close to the reaction face of FeMo-co. In the complete mechanism proposed for conversion of N2 to NH328, the NH3 molecules are formed (late in the catalytic cycle) at atoms Fe6 and Fe2 of FeMo-co. The first NH3 is generated at the exo position of Fe6, from which it dissociates in the exo direction. The second NH3 is generated at the endo position of Fe2, and dissociates in an endo direction, looping around the endo side of Fe6. Residue Val70A and its neighbours in the α-helix that covers the face of FeMo-co constrain both product NH3 molecules to leave in the exo direction from Fe6. This is illustrated in Fig. 1(a) that shows the first NH3 leaving Fe6, and the nascent second NH3 as an NH2 group bridging Fe2 and Fe6. The formation of the second NH3 is completed by H atom transfer from S3B, and after dissociation it is proposed to follow the path of the first NH3. Fig. 1(a) also locates the hydrogen bonded water molecules that comprise the water pool, clustering around the uncoordinated carboxylate atoms O1, O2 and O4 of homocitrate. The separate water chain that supports the proton wire to S3B of FeMo-co uses different homocitrate atoms O5, O6 and O3 (Fig. 1(b)). Residue Gln191A, hydrogen bonded to O1 of homocitrate, lies approximately in the path of the departing NH3 molecules, whereas His195A is out of the way.
Strategy
The search for possible channels and pathways was undertaken first by a search for cavities in the MoFe protein in the vicinity of FeMo-co and the water pool. The species generality of the cavities so located was checked, together with the generality of their occurrence in the best available crystal structures. The most promising cavities near the NH3 generation position were examined for size and extent, water content, and immediate surroundings. Then, in the one feasible cavity, possible locations for a moving NH3 molecule were modeled, with hydrogen bonding to water and protein surrounds. Finally, the most probable pathway was assessed in terms of protein tertiary structure and likely dynamic fluctuations during NH3 egress.
Calculations of pockets in the protein
The initial search for cavities involved calculations of pockets in the protein, using the program fpocket61. Pockets are clusters of intersecting alpha spheres. An alpha sphere contacts four atoms on its boundary and contains no internal atom: the four atoms are equally distant from the sphere centre, defining the radius of the sphere. The results reported used alpha spheres with a minimum radius of 2.8Å, and required at least 35 intersecting alpha spheres to constitute a pocket. Water molecules are ignored in the calculation of pockets, but were included in the results, in order to examine the presence of water in or near the pockets. Thus the pocket calculations yielded domains in the protein sufficiently large to contain NH3 or NH4+, and possibly containing water.
Calculations were made for all of the high-resolution crystal structures (PDB 3U7Q60, 1M1N62, 2MIN63, 3MIN63, 2AFH64, 2AFI64, 1MIO65, 1QGU66, 1QH166, 1QH866, 1H1L67) that include the MoFe protein and a high proportion of resolved water molecules, and encompassed the species Azotobacter vinelandii (Av), Klebsiella pneumoniae (Kp) and Clostridium pasteurianum (Cp).
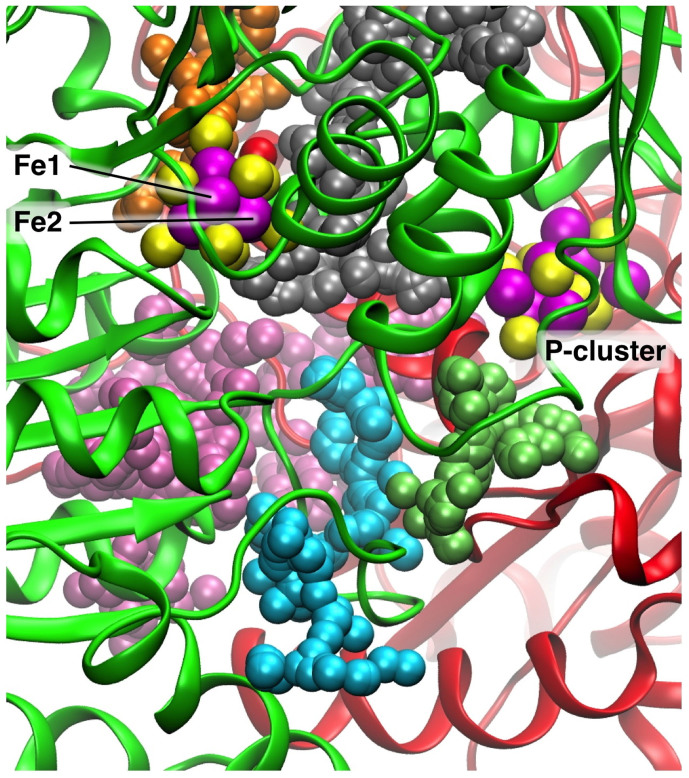
Figure 2 shows the significant pockets (drawn as aggregates of alpha spheres) found in the vicinity of FeMo-co for the Av protein (PBD 3U7Q). The view direction in this figure is directly along the pseudo-threefold axis of FeMo-co, from the top. Two protein chains that envelop the pockets and cofactors are coloured green and red. In this and the following descriptions the pockets are labeled by their colours. The silver pocket contains the water molecules of the water pool, while the water path that supports the proton wire appears as the orange pocket extending upwards from FeMo-co. The key pocket is that coloured cyan, which extends from an edge of the silver pocket, twisting towards the surface of the protein. The inner section of the cyan pocket is very similar for the Av and Kp proteins, but variation occurs as the pocket reaches the protein surface. The gap between the inside edge of the cyan pocket and the silver pocket is devoid of protein main chain, and the point of approach of the cyan and silver pockets is essentially unchanged for all of the protein crystals investigated. The lime pocket extends from the P-cluster, and has variable shape and size. In some protein structures it is larger than shown in Fig. 2, extending to the protein surface near where the cyan pocket meets the surface. The lime pocket is not near the water pool, and is separated from it by a section of polypeptide chain, and therefore the lime pocket is not believed to play any role in the movement of NH3, and it will not be considered further. The only other pocket near the silver pocket is that coloured mauve: this pocket has variable size and shape in different proteins, and is separated from the silver pocket by a gap of 6.4Å that is blocked by the sidechain of Lys426 (Av, Lys424 in Kp).
Figure 2. Relevant pockets calculated for Av protein, PDB 3U7Q. The pockets are represented as assemblies of alpha spheres, differentiated by colour.
The view direction is directly along the pseudo-threefold axis of FeMo-co, from the top: Mo and homocitrate (HCA) are on the distant side of FeMo-co. The rotational orientation of FeMo-co is signified by the marked position of Fe2, and by the location of the P-cluster. The two protein chains that envelope the pockets are coloured green and red.
These pocket calculations focus attention on the cyan pocket as the most likely domain for NH3 efflux. Before proceeding with more detailed investigation of this silver-plus-cyan domain of the protein, two further checks were made. The first involved the location of the region where the Fe protein docks with the MoFe protein during the electron transfer cycle, in relation to the postulated exit point for NH3 on the surface of the MoFe protein. This was to determine whether diffusion of NH3 from the surface of the MoFe protein might be blocked when the Fe protein is docked. Crystal structures 2AFH and 2AFI (Av) contain the Fe protein docked with the MoFe protein, using two different but adjacent docking domains, dependent on the state of the nucleotide in the Fe protein64. In both structures, the surface terminus of the cyan pocket is not part of the docking domain, and does not encounter the polypeptide chains of the Fe protein (supplementary Fig. S1).
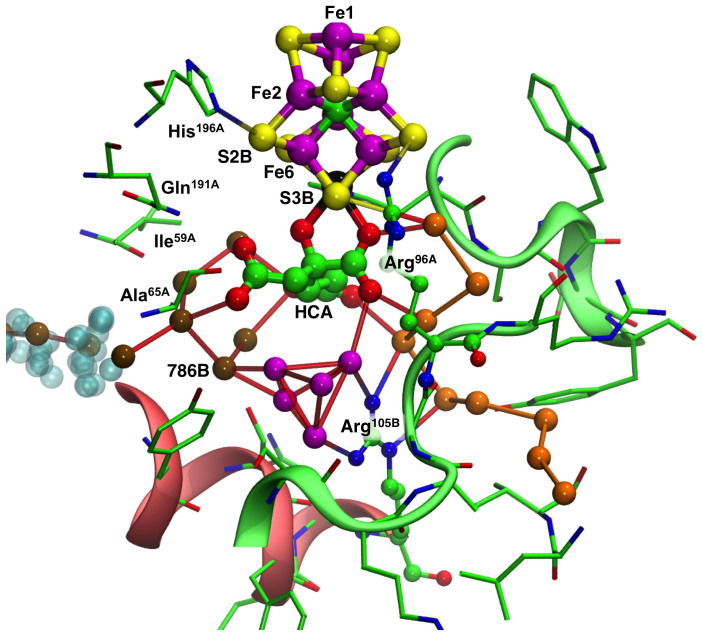
The second check involved the relationship between the water pool postulated for NH3 movement away from FeMo-co and the chain of water molecules that constitutes the proton wire. Figure 3 shows the relevant part of the protein, around and ‘underneath' homocitrate. The conserved chain of hydrogen-bonded water molecules comprising the proton wire (terminating with a hydrogen bond to S3B) is depicted with orange spheres, while water molecules extending from the other side of homocitrate towards the alpha spheres of the cyan pocket are coloured brown. There is an additional cluster of water molecules, coloured magenta, directly under homocitrate. The composition and structure of this water cluster varies in the different crystal structures, and the geometry is such that not all of the hydrogen bond distances within this cluster could be hydrogen bonds. This cluster is hydrogen bonded to one ‘brown' water molecule (786B) and is not hydrogen bonded directly to the proton wire. The side-chain of arginine105B is a barrier between the magenta cluster and the proton wire, and, as previously described, is a crucial part of the proton wire31.
Figure 3. The water pool around homocitrate in the MoFe protein (Av, PDB 3U7Q).
The hydrogen bonded water molecules of the proton wire are coloured orange. The water molecules extending from homocitrate towards the cyan pocket are coloured brown. Between these two sets of water molecules is a compact cluster of water molecules coloured magenta. Connecting lines from the water molecules signify contacts at hydrogen bond distances. Two significant amino acids, Arg105B and Arg96A, are drawn with atom spheres. Cartoon representations of significant parts of chain A (green) and B (red) are included.
It is clear from Fig. 3 that the putative ammonia path into the cyan pocket and the proton path occur in opposite directions through the protein, as required chemically. The water pool near homocitrate is comprised of the ‘brown' and ‘magenta' water molecules, not directly linked to the proton wire. The magenta water cluster does not appear to have a functional role other than as a small compact reservoir for water molecules.
Contents and surrounds of the postulated NH3 exit pathway
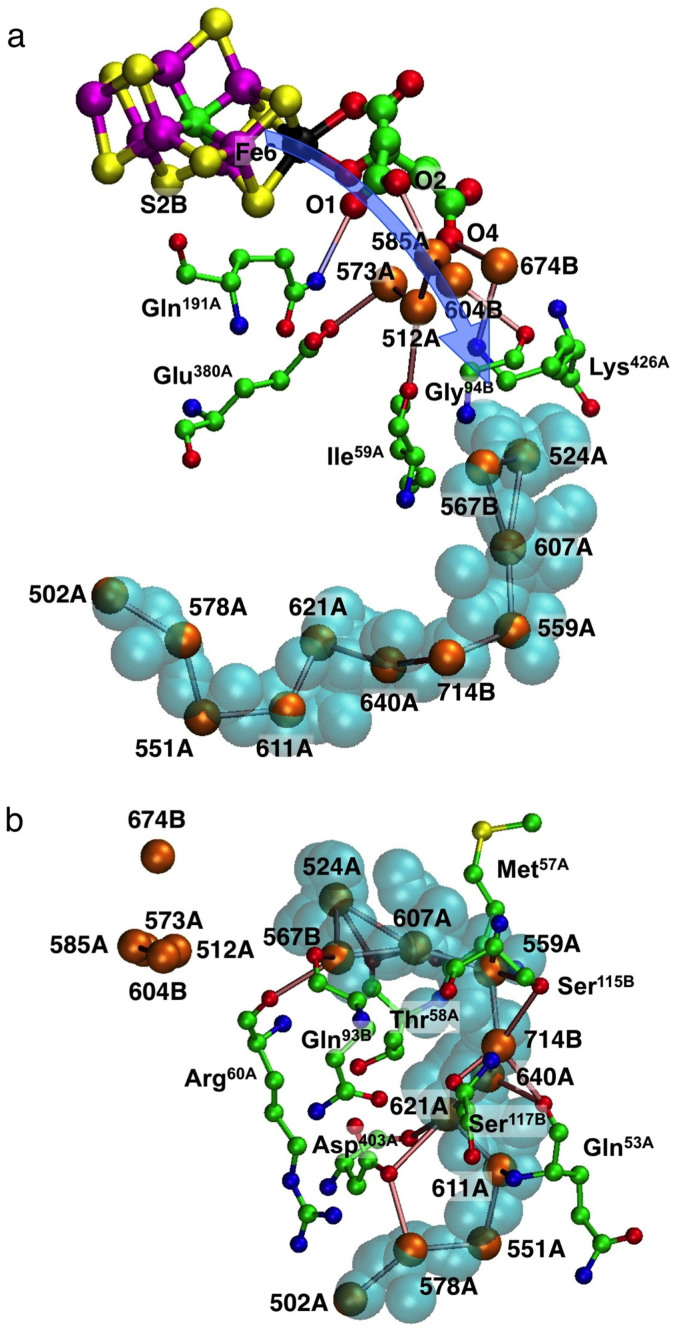
The next step was to examine closely the contents and immediate surroundings of the indicated ammonia pathway towards and through the cyan pocket. The blue arrow in Fig. 4(a) shows the proposed general direction of movement from the vicinity of Fe6 (cf Fig. 1(a)), over homocitrate and associated water molecules in the water pool, and towards the water molecules at the entrance of the cyan pocket. The relevant water molecules close to O1, O2 and O4 of homocitrate are 573A, 512A, 585A, 604B and 674B, with hydrogen bond connections to amino acids, Gln191A, Glu380A, Lys426A, Ile59A and Gly94B (all labels here and following are those of the Av protein, PDB 3U7Q). The gap between these water molecules and the entrance to the cyan pocket (the gap between silver and cyan pockets in Fig. 2) is surrounded by but not blocked by Lys426A, Ile59A and Gly94B.
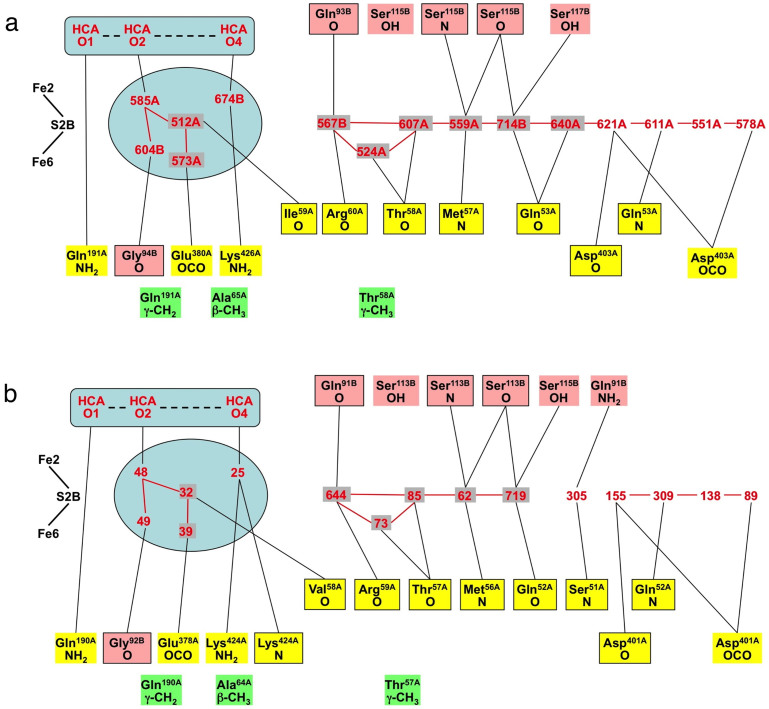
Figure 4. The contents and immediate surroundings of the approach to the cyan pocket, and the cyan pocket, labeled for the Av protein (3U7Q).
Atoms O1, O2, O4 of homocitrate are evident in (a), and the blue arrow depicts the general direction of NH3 movement from the reaction face of FeMo-co. Part (b) shows the relevant residues surrounding the cyan pocket and hydrogen bonding to the chain of water molecules. Hydrogen bonds between water molecules are marked.
The cyan pocket (of the Av protein) contains a sequence of eleven hydrogen bonded water molecules, extending roughly in an arc. Around these there are interactions with Met57A, Thr58A, Arg60A, Gln53A, Asp403A, Ser115B and Ser117B, marked in Fig. 4(b).
It is postulated that NH3 egress starts from Fe2/Fe6 on FeMo-co, following the direction of the blue arrow (Fig. 4(a)) and probably engaging with some of the hydrogen bonding components O1, O2, O4 of homocitrate, the five associated water molecules, and side-chains of Gln191A, Glu380A, Lys426A. At this point the NH3 molecule would be positioned at the entrance of the cyan cavity, ready to move to the end of this cavity, engaging with the contained water and surrounding residues. If this hypothesis is correct, these features should be conserved in the various protein structures, and therefore details of these components and surroundings have been compared for the highest resolution crystal structures for Av (3U7Q) and Kp (1QH1). Fig. 5, a comparison of the hydrogen bonding connectivities, demonstrates that the two proteins are very similar. The domains in this depiction, left to right, are first the water pool and its homocitrate components, then a gap to the sequence of water molecules and protein surrounds in the vicinity of the cyan cavity. Functions from the A and B chains are differentiated by colour.
Figure 5. Schematic representations of the atoms involved in the postulated ammonia efflux channel in (a) Av protein structure 3U7Q and (b) Kp protein structure 1QH1.
The numbering of O1/O2 and O3/O4 of HCA is reversed in PDB 1QH1, and has been changed to the 3U7Q numbering in this presentation. Water molecules are labeled red; protein atoms are differentiated as chain A yellow and chain B salmon, with main chain atoms boxed and side-chain atoms unboxed; connecting lines are possible hydrogen bonds. The groups coloured green are hydrophobic side-chain components that are potential steric barriers. The aqua enclosures signify the relevant components of the water pool.
The similarity of the two proteins is evident. The principal difference is the absence in Kp of a water molecule in the same position as water 640A in Av, breaking the chain of hydrogen bonded water molecules, and changing some hydrogen bonding interactions with surrounding protein. This water is halfway through the cyan pocket (and more than 15Å from FeMo-co), and is at a point where the pathway possibilities for NH3 are diverging (see next section). In the Kp protein the hydroxyl side chain of Ser51A is near the position of water 640A in Av, and is disordered, which is indicative of movements that could be part of NH3 passage here. These differences in the later section of the cyan pocket are consistent with the notion that as NH3 approaches the protein surface its mechanism for molecular translocation is less controlled.
Figure 5 also draws attention to the differentiation of main chain atoms (boxed) and side-chain groups in the hydrogen-bonding network, and the prevalent involvement of main chain carbonyl functions, as potential hydrogen bond acceptors. Residue Ile59A in Av is Val58A in Kp, but since it is the main chain carbonyl O that participates in the hydrogen bonding this difference of hydrophobic side chains is inconsequential. There are differences in the region of waters 674B (3U7Q) and 25 (1QH1), and their hydrogen bonding with Lys426/Lys424, but this is not significant because these functions are distant from the detailed pathway described in the next section.
Atomic level mechanism for NH3 movement
The preceding analysis has revealed a possible pathway for movement of NH3 away from FeMo-co towards the protein surface. The questions now are (a) whether there is space for this movement of NH3, (b) whether the environment is consistent with the hydrogen bonding expected of NH3, (c) what impediments might exist, and (d) what fluctuations in the tertiary structure of the protein are likely to be involved. Possible pathways for molecular NH3 to move from FeMo-co to and through the cyan cavity towards the surface of the protein were investigated by manual molecular modelling.
Since water and NH3 are similar in size and hydrogen bonding attributes, one question is whether NH3 might occupy the locations that are observed to contain water in the resting protein structures. [It is very unlikely that the crystallographically observed small molecules are NH3 (or NH4+) because the crystallisation medium for structure 3U7Q did not contain NH3/NH4+ buffers60.] Alternatively, is there space for NH3 to move around the water molecules in the resting protein? Would it be necessary for water molecules to move aside in order to allow passage of NH3?
The moving entity was modeled as NH3, because NH3 is generated at the surface of FeMo-co and is released into a hydrophobic anhydrous space. On diffusion into the water pool and beyond it is possible that NH4+ is formed. In the early stages of the path there is no evident source of a replenishable proton, and so NH4+ formation would be just a temporary relocation of one hydrogen atom across a hydrogen bond, and the following modeling of hydrogen bonds would be unchanged. Near the protein surface the more aqueous medium would generate NH4+.
Standard geometries were used for the three donor and one acceptor hydrogen bonds of NH3. The goal was to find a sequence of locations that engaged some or all of these possible hydrogen bonds, such that NH3 could swing from one location to the next, breaking and forming hydrogen bonds. The starting point had NH3 just dissociated from Fe6 (Fe6–NH3 = 2.7Å), and NH3 hydrogen bonded to both S2B and O1 of HCA. It soon became evident that NH3 can only move away underneath the side chain of Gln191A into the water pool. Through this first stage of NH3 movement the important surroundings are O1, O2 and O4 of HCA, the side-chain CH3 of Ala65A, and both the terminal amide group and the CH2 functions of the side chain of Gln191A. In the resting state the terminal NH2 of Gln191A is well positioned to donate hydrogen bonds to O1 of HCA and to the side chain carboxylate of Glu380A. During this stage it is probable that the side chain amide group of Gln191A relocates and reconfigures in order the improve the hydrogen bonding with passing NH3, at the expense of the hydrogen bond to O1 and OCO of Glu380A.
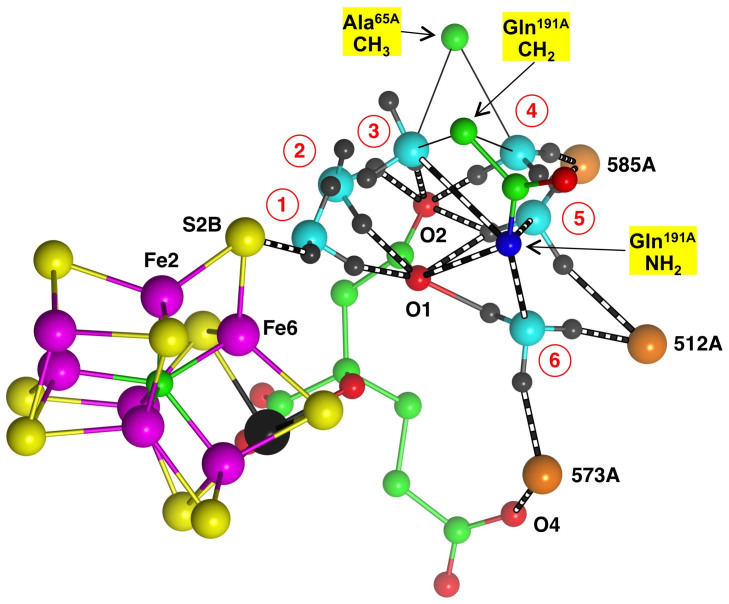
Figure 6 shows the first six possible positions for NH3. Position 1 has N-H hydrogen bonds to S2B and O1, then at position 2 the N-H hydrogen bonds are bent to O1 and O2 of HCA, then in position 3 breaking with O1 and in position 4 forming a new N-H hydrogen bond to water 585A. In position 5 the N-H hydrogen bond to water 585A is retained and the other N-H hydrogen bond bifurcates to O1 and O2, with a new hydrogen bond to water 512A. Finally, in position 6 the NH3 swings around its N-H hydrogen bond to O1, and forms a new hydrogen bond to water 573A. Several comments are warranted. First, the NH3 trajectory is a downwardly spiraling arc maintained by N-H hydrogen bonds to O1 and/or O2, and this carboxylate arm of HCA is regarded as a key controller of mechanism. Second, the side chain of Gln191A is closely involved, and with reconfiguration could form Gln191-NH2 → NH3 hydrogen bonds at positions 3, 5 and 6: these potential hydrogen bonds are marked on Fig. 6. Third, water molecules are used as hydrogen bond acceptors at positions 4, 5 and 6, with water molecules 512A and 573A moving by ca 0.7Å away from the approaching NH3 molecule. Sidechain β-CH3 of Ala65A and γ-CH2 of Gln191A provide steric boundaries.
Figure 6. The first six sequential positions (numbered 1–6) constructed for NH3.
This representation is simplified in order to maintain the clarity of the multiple positions for NH3 (N cyan, H gray) and its hydrogen bonds (black and white). The thin lines to β-CH3 of Ala65A and γ-CH2 of Gln191A signify steric boundaries for NH3 at positions 3 and 4.
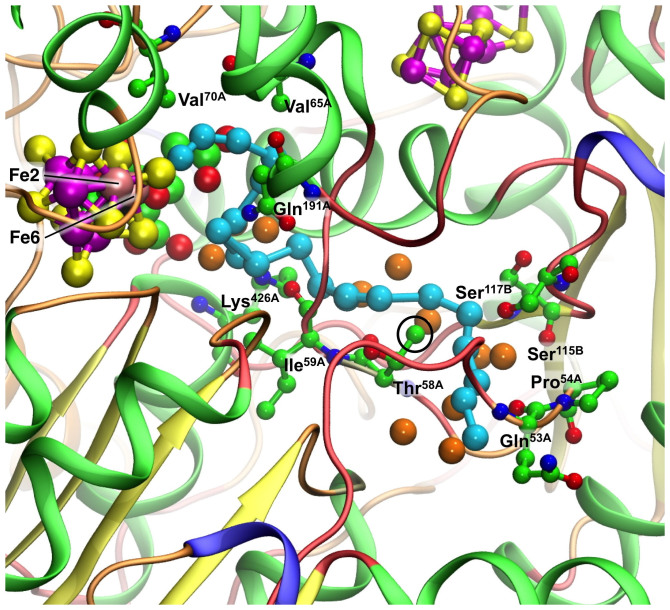
This modeling was continued, stepping the hydrogen bonds of NH3, to identify a total of 18 possible locations for NH3 towards the protein surface. These positions are shown in Fig. 7, together with the water molecules and significant amino-acid functions. Detail is provided in Supplementary Figs. S2 and S3. In the journey from the water pool to the cyan cavity NH3 passes between the side-chain of Lys426A and the main-chain oxygen atom of Ile59A. The terminal NH2 function of Lys426A is a hydrogen bond accepting pivot point, and because the distances from it are slightly short it is proposed that this side-chain function moves slightly away during NH3 passage. The main-chain oxygen atoms of neighbouring residues Thr58A, Ile59A and Arg60A function as hydrogen bond acceptors in positions 8 through 12. The side chain CH3 function of Thr58A is significant, because the lone pairs of NH3 in positions 12 through 15 are distant 2.6–2.9Å from it, with the potential for formation of weak C-H → NH3 hydrogen bonds. On the other side of this arced sequence of NH3 positions the side chains of Ser115 and Ser117 in chain B are involved. Beyond the last modeled position, near Gln53A, there is a ‘river-delta' of possibilities for NH3 to escape to the protein surface. Examination of the atom thermal parameters for 3U7Q and 1GQU reveals that above average values occur in the sections of chains A and B extending from the end of the NH3 pathway to the protein surface.
Figure 7. The sequence of modeled positions for NH3 from the reaction face of FeMo-co towards the protein surface.
Cyan spheres are the locations of the N atom: H atoms are not shown (but are included in Supplementary Figs. S2, S3). Orange spheres are water. Key interacting amino acids are marked and labeled. The side chain CH3 of Thr58A (circled) is significant, because the lone pairs of NH3 looping around it are 2.6–2.9Å distant, with the potential for formation of weak C-H → NH3 hydrogen bonds. All protein chains are included and are coloured by structure (α-helix green, extended-β yellow, turn orange, coil red), revealing the minimal secondary structure around most of the proposed NH3 pathway.
In positions through the cyan cavity domain, N-H → OH2 hydrogen bonds are part of the model. The ability of these water molecules to accept one such hydrogen bond at each position of NH3 is favourable in terms of hydrogen atom accounting, because each water can form two other donor O-H → O hydrogen bonds, one to the next water in the chain, and one to protein main chain carbonyl oxygen. Displacements of water molecules by 0.4–1.0Å are involved in the model, and it is clear that some local geometric reorganisation would be required as NH3 passed along this pathway.
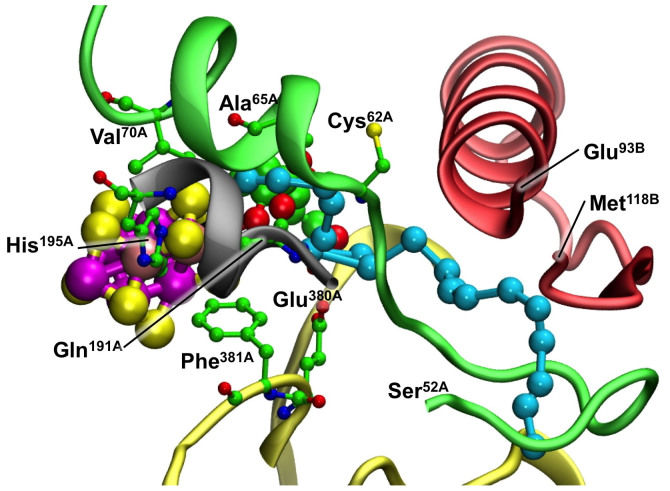
This leads to considerations of secondary and tertiary structure, and structural fluctuations during NH3 passage. Figure 7 portrays this structure of the protein, and reveals the lack of secondary structure on the viewer side of the proposed path. The path winds between the A and B chains (see also Fig. 5), with the B chain occurring as a structured backdrop and the A chain as a movable front curtain. Figure 8 is a different view of key structural elements in the surrounding protein. Three different sections of the A chain are involved. The part of chain A that lies over the reaction face and contains Val70A is α-helical only as far as residue 62, but then is largely unstructured along the ammonia path to residue 52 (green). The section containing influential residues His195A and Gln191A (grey) is α-helical only near FeMo-co. Glu380A and Lys426A, both near the ammonia path, are connected by a long loop (yellow). Chain B (red) is near the proposed ammonia path at residues 93 and 115–117. The chain from 93 to 108 is α-helical, directed well away from the ammonia pathway, then loops back to serines 115 and 117.
Figure 8. The modeled NH3 pathway, emphasising secondary structure in the surrounding protein.
Four different parts of chain A are involved. The A-chain helix over the reaction face (green) does not continue as chain A extends along the NH3 path from Cys62A to Ser52A. The α-helix of chain B (red) is near the NH3 pathway only at Glu93, then chain B loops back to flank the NH3 pathway near Met118.
The flexibility of these elements of the surroundings of the proposed ammonia pathway has been assessed via the crystallographic temperature factors. For crystal structure 1QGU (residue numbering as in Fig 5 (b)), the Cα atoms of the helical section of chain A over the reaction face (Val69 to Cys61 in 1QGU) have temperature factors well below average, but as the chain continues from residue 58 to lower numbered residues the Cα atom temperature factors increase to well above average, indicative of flexibility as the ammonia efflux extends towards the protein surface. In the B chain the below average temperature factors of the α-helix (Fig. 8) increase through the key serine residues (113, 115) that are involved in the proposed ammonia pathway.
Discussion
This exploration of the structures of nitrogenase proteins has uncovered a pathway for the movement of product NH3 from its site of formation on FeMo-co to the surface of the protein, and a molecular mechanism for the translocation of NH3 has been developed. This pathway is conserved in proteins from species Av, Kp and Cp. The integrity of the acid-base dimension of the physiological reaction of nitrogenase, consuming acid and generating base, is maintained, because the NH3 egress pathway is separate from and physically opposite to the proposed proton influx pathway. As in the glutamine amidotransferase enzymes, the moving entity in the early stages of the efflux is probably NH3, becoming NH4+ near the protein surface.
Water molecules occur along most of the NH3 pathway, at least in the resting protein as crystallised, and provide acceptor and donor hydrogen bonding sites for NH3. Polypeptide main chain atoms also provide hydrogen bonding sites, and the suggested mechanism is essentially a sequence of interchanges of hydrogen bonds over short distances. The mechanism does not involve a flow of water molecules accompanying NH3, but it does involve movements of some water molecules away from the main path by distances of up to 1Å, to create sufficient space for NH3 to skip from one hydrogen bond to the next.
At this point it is appropriate to make comparisons with the well-studied ammonia channels in glutamine amidotransferase (GAT) enzymes. In all of these, ammonia generated by hydrolysis of glutamine (or asparagine) in the amidase domain is translocated through tunnels to a distant synthase site: the ammonia is shielded from the medium and maintained as NH3 because NH3 is needed as nucleophile in the synthase domain. The GAT enzymes are diverse in product, function and structure, and include carbamoyl phosphate synthetase68, glutamine phosphoribosylpyrophosphate amidotransferase (GPAT)46,69, glutamate synthase (GltS)39, tRNA-dependent amidotransferases (or GatCAB). The ammonia tunnels are structured more by the residues of the synthase domain than those of the amidase domain, accounting for the variety of tunnel architectures39,40,70. In nitrogenase the formation of NH3 at FeMo-co is analogous to the formation of NH3 at the amidase site, but there is no requirement to prevent formation of NH4+ near the end of the pathway at the surface of the MoFe protein. Nevertheless, there are results and interpretations from the amidotransferase enzymes that can inform further investigation and understanding of NH3 efflux from nitrogenase.
In crystalline GatCAB amidotransferases a 30-35Å-long hydrophilic tunnel links the amidase and synthetase sites. The tunnel is ungated along its length, and contains 18 water molecules that are hydrogen bonded to each other or to surrounding polar residues. The published representations71,72 show a structure quite similar to that described here for nitrogenase. Molecular dynamics calculations have revealed gates and valves modulating the direction of NH3 transport47, and confirmed large barriers associated with NH4+ rather than NH3 as the moving entity. In glutamate synthase the tunnel is composed mainly of backbone atoms of hydrophilic residues, and water molecules fill the tunnel space39. There are some structural similarities between the various sections of the long intramolecular ammonia translocation tunnel in carbamoyl phosphate synthetase (CPS)68 and the path proposed here for nitrogenase. Molecular dynamics simulations of CPS reveal hydrogen bond stepping, movements of side-chains, and involvement of small water clusters. It is calculated that in the first stage of a long tunnel, NH3 is transported over a distance of ~13Å with a maximum barrier of 6 kcal mol−1 44,45. Further, it is suggested that water molecules have an important role in the transfer of NH3, because sites vacated by NH3 are refilled with water molecules such that the NH3 hydrogen bonds cannot reform, thereby forcing NH3 forward.
It is very probable that fluctuations of protein structure are part of the mechanism for nitrogenase, because the proposed route passes between different polypeptide chains, and those chain sections have minimal secondary structure and above average temperature factors. Gas transport channels in a variety of enzymes are characterised by structural flexibility73. Conformational variations are a primary feature of the ammonia-channeling glutamine amidotransferases41,42,43,46,47.
Amino acid Gln191A is a key component of the mechanism in nitrogenase, because it is normally hydrogen bonded through side-chain NH2 to O1 of HCA (another key component), and NH3 has to move around both of these atoms, and to avoid the side chain CH2 of Gln191A, before escaping from the water pool (Fig. 6). Therefore the consequences of modification of this residue provide an experimental approach to testing the validity of the mechanism. Mutation of Gln191A to lysine with a longer side chain shuts down NH3 production from N274, but NH3 is produced with azide as substrate by this mutant15, indicating that the NH3 efflux channel is not closed by Lys191A. The interpretation of the diminished activity of the Lys191A mutant, that is only high Km reduction of C2H2 and no reduction of N2, is probably that given previously25, which is that Lys blocks hydrogenation steps at Fe6 but not hydrogenation steps at Fe2. I am not aware of other experimental data that bears on the validity of the NH3 efflux mechanism proposed here.
In this paper I have presented a plausible mechanistic pathway for egress of NH3 produced by nitrogenase. How can this proposal be tested? A standard experimental approach is modification of key amino acids and assessment of reactivity. The early stages of the NH3 path involve the side-chain of Gln191A, in two ways: the terminal amide NH2 group engages some hydrogen bonds, while γ-CH2 provides a steric barrier. In view of the flexibility of the side-chain it is not surprising that lysine in this position does not fully shut down the escape of NH3, but large non-hydrogen bonding side-chains such as Phe or Trp at position 191A are predicted to impede the NH3 pathway. A large increase in size of the side-chain of residue 65A could also interfere. Another significant side-chain is that of Thr58A (Fig. 7), because the proposed pathway passes closely around its γ-CH3. A more voluminous side-chain, without hydrogen bonding capability, could interfere with NH3 passage. These are suggestions for mutations to clog the pathway, but the likely fluctuations in protein structure may open alternative paths. In this context, residue 191A in the tighter earlier part of the NH3 route would be the prime target.
Might it be possible to incorporate a reagent for NH3 into the protein near the putative pathway, and then crystallise the protein containing the captured escaping NH3?
Computationally, a normal modes analysis75,76 of the protein structure would be informative about its flexibility around the proposed pathway. Molecular dynamics simulations, enhanced with various techniques44,46,77, should provide further insight into ammonia transport in nitrogenase.
Methods
The initial search for cavities was undertaken with the program fpocket61, using a procedure which strips water and heteroatoms (but not FeMo-co, homocitrate, or the P-cluster) from the atom coordinates (pdb) file, and then searches for alpha spheres. An alpha sphere contacts four atoms on its boundary and contains no internal atom: the four atoms are equally distant from the sphere centre, defining the radius of the sphere. Pockets are clusters of intersecting spheres, and pocket searches are controlled by filtering sphere properties such as radius, separation, and chemical attributes of the defining atoms. The variables in fpocket were first evaluated: for the searches of nitrogenase FeMo proteins, minimum alpha sphere radii in the range 2.7–3.0Å were found to be most informative, with pockets required to contain at least 35 alpha spheres. After each search the water excluded from the fpocket calculation was added back into the output files, in order to examine the presence of water in or near the pockets. Calculations of pockets were made for all of the high-resolution crystal structures (PDB 3U7Q, 1M1N, 2MIN, 3MIN, 2AFH, 2AFI, 1MIO, 1QGU, 1QH1, 1QH8, 1H1L) that included the MoFe protein and a high proportion of resolved water molecules (the best structures contain ca 1300 water molecules per FeMo-co in the MoFe protein: structures 3K1A, 1M34 and 2AFK describe considerably less water).
A 3U7Q pdb file with the modeled NH3 positions included is available from the author.
Supplementary Material
Supplementary information
Acknowledgments
This research was supported by the University of New South Wales.
References
- Burgess B. K. & Lowe D. J. Mechanism of molybdenum nitrogenase. Chem. Rev. 96, 2983–3011 (1996). [DOI] [PubMed] [Google Scholar]
- Howard J. B. & Rees D. C. Structural basis of biological nitrogen fixation. Chem. Rev. 96, 2965–2982 (1996). [DOI] [PubMed] [Google Scholar]
- Smith B. E. Structure, function, and biosynthesis of the metallosulfur clusters in nitrogenases. Adv. Inorg. Chem. 47, 159–218 (1999). [Google Scholar]
- Rees D. C. & Howard J. B. Nitrogenase: standing at the crossroad. Curr. Opin. Chem. Biol. 4, 559–566 (2000). [DOI] [PubMed] [Google Scholar]
- Christiansen J., Dean D. R. & Seefeldt L. C. Mechanistic features of the Mo-containing nitrogenase. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 269–295 (2001). [DOI] [PubMed] [Google Scholar]
- Igarashi R. Y. & Seefeldt L. C. Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase. Crit. Rev. Biochem. Mol. Biol. 38, 351–384 (2003). [DOI] [PubMed] [Google Scholar]
- Rees D. C. et al. Structural basis of biological nitrogen fixation. Phil. Trans. Roy. Soc. A 363, 971–984 (2005). [DOI] [PubMed] [Google Scholar]
- Seefeldt L. C., Hoffman B. M. & Dean D. R. Mechanism of Mo-Dependent Nitrogenase. Annu. Rev. Biochem. 78, 701–722 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.-Y., Danyal K. & Seefeldt L. C. Mechanism of Mo-Dependent Nitrogenase. Methods Mol. Biol. 766, 9–29 (2011). [DOI] [PubMed] [Google Scholar]
- Seefeldt L. C., Dance I. G. & Dean D. R. Substrate Interactions with Nitrogenase: Fe versus Mo. Biochemistry 43, 1401–1409 (2004). [DOI] [PubMed] [Google Scholar]
- Dos Santos P. C. et al. Substrate Interactions with the Nitrogenase Active Site. Acc. Chem. Res. 38, 208–214 (2005). [DOI] [PubMed] [Google Scholar]
- Sarma R. et al. Insights into substrate binding at FeMo-cofactor in nitrogenase from the structure of an α-70Ile MoFe protein variant. J. Inorg. Biochem. 104, 385–389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth M. J., Fisher K., Kim C. H. & Newton W. E. Effects on substrate reduction of substitution of histidine-195 by glutamine in the alpha-subunit of the MoFe protein of Azotobacter vinelandii nitrogenase. Biochemistry 37, 17495–17505 (1998). [DOI] [PubMed] [Google Scholar]
- Fisher K., Dilworth M. J., Kim C.-H. & Newton W. E. Azotobacter Vinelandii Nitrogenases Containing Altered MoFe Proteins with Substitutions in the FeMo-Cofactor Environment: Effects on the Catalyzed Reduction of Acetylene and Ethylene. Biochemistry 39, 2970–2979 (2000). [DOI] [PubMed] [Google Scholar]
- Fisher K., Dilworth M. J. & Newton W. E. Differential Effects on N2 Binding and Reduction, HD Formation, and Azide Reduction with α-195His- and α-191Gln-Substituted MoFe Proteins of Azotobacter Vinelandii Nitrogenase. Biochemistry 39, 15570–15577 (2000). [DOI] [PubMed] [Google Scholar]
- Christiansen J., Cash V. L., Seefeldt L. C. & Dean D. R. Isolation and characterisation of an acetylene-resistant nitrogenase. J. Biol. Chem. 275, 11459–11464 (2000). [DOI] [PubMed] [Google Scholar]
- Christiansen J., Seefeldt L. C. & Dean D. R. Competitive Substrate and Inhibitor Interactions at the Physiologically Relevant Active Site of Nitrogenase. J. Biol. Chem. 275, 36104–36107 (2000). [DOI] [PubMed] [Google Scholar]
- Benton P. M. C., Christiansen J., Dean D. R. & Seefeldt L. C. Stereospecificity of Acetylene Reduction Catalyzed by Nitrogenase. J. Am. Chem. Soc. 123, 1822–1827 (2001). [DOI] [PubMed] [Google Scholar]
- Benton P. M. C. et al. Interaction of Acetylene and Cyanide with the Resting State of Nitrogenase α-96-Substituted MoFe Proteins. Biochemistry 40, 13816–13825 (2001). [DOI] [PubMed] [Google Scholar]
- Barney B. M. et al. Trapping a Hydrazine Reduction Intermediate on the Nitrogenase Active Site. Biochemistry 44, 8030–8037 (2005). [DOI] [PubMed] [Google Scholar]
- Barney B. M. et al. Breaking the N2 triple bond: insights into the nitrogenase mechanism. Dalton Trans. 2277–2284 (2006). [DOI] [PubMed] [Google Scholar]
- Fisher K. et al. Conformations generated during turnover of the Azotobacter vinelandii nitrogenase MoFe protein and their relationship to physiological function. J. Inorg. Biochem. 101, 1649–1656 (2007). [DOI] [PubMed] [Google Scholar]
- Dance I. The Hydrogen Chemistry of the FeMo-co Active Site of Nitrogenase. J. Am. Chem. Soc. 127, 10925–10942 (2005). [DOI] [PubMed] [Google Scholar]
- Dance I. The Correlation of Redox Potential, HOMO Energy, and Oxidation State in Metal Sulfide Clusters and Its Application to Determine the Redox Level of the FeMo-co Active-Site Cluster of Nitrogenase. Inorg. Chem. 45, 5084–5091 (2006). [DOI] [PubMed] [Google Scholar]
- Dance I. Mechanistic Significance of the Preparatory Migration of Hydrogen Atoms around the FeMo-co Active Site of Nitrogenase. Biochemistry 45, 6328–6340 (2006). [DOI] [PubMed] [Google Scholar]
- Dance I. The mechanistically significant coordination chemistry of dinitrogen at FeMo-co, the catalytic site of nitrogenase. J. Am. Chem. Soc. 129, 1076–1088 (2007). [DOI] [PubMed] [Google Scholar]
- Dance I. Elucidating the Coordination Chemistry and Mechanism of Biological Nitrogen Fixation. Chem. Asian J. 2, 936–946 (2007). [DOI] [PubMed] [Google Scholar]
- Dance I. The chemical mechanism of nitrogenase: calculated details of the intramolecular mechanism for hydrogenation of η2-N2 on FeMo-co to NH3. Dalton Trans., 5977–5991 (2008). [DOI] [PubMed] [Google Scholar]
- Dance I. The chemical mechanism of nitrogenase: hydrogen tunneling and further aspects of the intramolecular mechanism for hydrogenation of η2-N2 on FeMo-co to NH3. Dalton Trans., 5992–5998 (2008). [DOI] [PubMed] [Google Scholar]
- Dance I. Nitrogenase: a general hydrogenator of small molecules. Chem. Commun. 49, 10893–10907 10.1039/C3CC46864J (2013). [DOI] [PubMed] [Google Scholar]
- Dance I. The controlled relay of multiple protons required at the active site of nitrogenase. Dalton Trans. 41, 7647–7659 (2012). [DOI] [PubMed] [Google Scholar]
- Durrant M. C. Controlled protonation of iron–molybdenum cofactor by nitrogenase: a structural and theoretical analysis. Biochem. J. 355, 569–576 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khademi S. et al. Mechanism of Ammonia Transport by Amt/MEP/Rh: Structure of AmtB at 1.35 Å. Science 305, 1587–1594 (2004). [DOI] [PubMed] [Google Scholar]
- Andrade S. L. A., Dickmanns A., Ficner R. & Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. U.S.A. 102, 14994–14999 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade S. L. A. & Einsle O. The Amt/Mep/Rh family of ammonium transport proteins (Review). Mol. Membr. Biol. 24, 357–365 (2007). [DOI] [PubMed] [Google Scholar]
- Javelle A. et al. Structural and mechanistic aspects of Amt/Rh proteins. J. Struct. Biol. 158, 472–481 (2007). [DOI] [PubMed] [Google Scholar]
- Thoden J. B., Holden H. M., Wesenberg G., Raushel F. M. & Rayment I. Structure of Carbamoyl Phosphate Synthetase: A Journey of 96 Å from Substrate to Product. Biochemistry 36, 6305–6316 (1997). [DOI] [PubMed] [Google Scholar]
- Raushel F. M., Thoden J. B. & Holden H. M. Enzymes with Molecular Tunnels. Acc. Chem. Res. 36, 539–548 (2003). [DOI] [PubMed] [Google Scholar]
- van den Heuvel R. H., Curti B., Vanoni M. A. & Mattevi A. Glutamate synthase: a fascinating pathway from L-glutamine to L-glutamate. Cell. Mol. Life Sci. 61, 669–681 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks A., Lund L. & Raushel F. M. Tunneling of intermediates in enzyme-catalyzed reactions. Curr. Opin. Chem. Biol. 10, 465–472 (2006). [DOI] [PubMed] [Google Scholar]
- Mouilleron S. & Golinelli-Pimpaneau B. Conformational changes in ammonia-channeling glutamine amidotransferases. Curr. Opin. Struct. Biol. 17, 653–664 (2007). [DOI] [PubMed] [Google Scholar]
- Mouilleron S., Badet-Denisot M.-A., Badet B. & Golinelli-Pimpaneau B. Dynamics of glucosamine-6-phosphate synthase catalysis. Arch. Biochem. Biophys. 505, 1–12 (2011). [DOI] [PubMed] [Google Scholar]
- Mouilleron S., Badet-Denisot M.-A. & Golinelli-Pimpaneau B. Ordering of C-terminal Loop and Glutaminase Domains of Glucosamine-6-Phosphate Synthase Promotes Sugar Ring Opening and Formation of the Ammonia Channel. J. Mol. Biol. 377, 1174–1185 (2008). [DOI] [PubMed] [Google Scholar]
- Fan Y., Lund L., Yang L., Raushel F. M. & Gao Y.-Q. Mechanism for the Transport of Ammonia within Carbamoyl Phosphate Synthetase Determined by Molecular Dynamics Simulations. Biochemistry 47, 2935–2944 (2008). [DOI] [PubMed] [Google Scholar]
- Fan Y., Lund L., Shao Q., Gao Y.-Q. & Raushel F. M. A Combined Theoretical and Experimental Study of the Ammonia Tunnel in Carbamoyl Phosphate Synthetase. J. Am. Chem. Soc. 131, 10211–10219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. S., Roitberg A. E. & Richards N. G. J. Computational Studies of Ammonia Channel Function in Glutamine 5′-Phosphoribosylpyrophosphate Amidotransferase. Biochemistry 48, 12272–12282 (2009). [DOI] [PubMed] [Google Scholar]
- Kang J., Kuroyanagi S., Akisada T., Hagiwara Y. & Tateno M. Unidirectional Mechanistic Valved Mechanisms for Ammonia Transport in GatCAB. J. Chem. Theory Computation 8, 649–660 (2012). [DOI] [PubMed] [Google Scholar]
- Marini A.-M., Urrestarazu A., Beauwens R. & André B. The Rh (Rhesus) blood group polypeptides are related to NH4+ transporters. Trends Biochem. Sci. 22, 460–461 (1997). [DOI] [PubMed] [Google Scholar]
- Gruswitz F. et al. Function of human Rh based on structure of RhCG at 2.1 Å. Proc. Nat. Acad. Sci. USA 107, 9638–9643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Kostrewa D., Bernèche S., Winkler F. K. & Li X.-D. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 101, 17090–17095 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard T. P., Rovira C., Peters G. H. & Jensen M. Ø. Ammonium Recruitment and Ammonia Transport by E. coli Ammonia Channel AmtB. Biophys. J. 91, 4401–4412 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick D. L. & Brooks C. L. Deprotonation by Dehydration: The Origin of Ammonium Sensing in the AmtB Channel. PLoS Comput. Biol. 3, e22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruswitz F., O'Connell J. & Stroud R. M. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 Å. Proc. Nat. Acad. Sci. USA 104, 42–47 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikita H. & Knapp E.-W. Protonation States of Ammonia/Ammonium in the Hydrophobic Pore of Ammonia Transporter Protein AmtB. J. Am. Chem. Soc. 129, 1210–1215 (2007). [DOI] [PubMed] [Google Scholar]
- Javelle A. et al. Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB. Proc. Nat. Acad. Sci. USA 105, 5040–5045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. A. & Kustu S. The pivotal twin histidines and aromatic triad of the Escherichia coli ammonium channel AmtB can be replaced. Proc. Nat. Acad. Sci. USA 108, 13270–13274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Orabi E. A., Baday S., Bernèche S. & Lamoureux G. Ammonium Transporters Achieve Charge Transfer by Fragmenting Their Substrate. J. Am. Chem. Soc. 134, 10419–10427 (2012). [DOI] [PubMed] [Google Scholar]
- Ullmann R. T., Andrade S. L. A. & Ullmann G. M. Thermodynamics of Transport Through the Ammonium Transporter Amt-1 Investigated with Free Energy Calculations. J. Phys. Chem. B 116, 9690–9703 (2012). [DOI] [PubMed] [Google Scholar]
- Hall J. A. & Yan D. The Molecular Basis of K+ Exclusion by the Escherichia coli Ammonium Channel AmtB. J. Biol. Chem. 288, 14080–14086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatzal T. et al. Evidence for Interstitial Carbon in Nitrogenase FeMo Cofactor. Science 334, 940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilloux V. L., Schmidtke P. & Tuffery P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinformatics 10, 168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einsle O. et al. Nitrogenase MoFe-Protein at 1.16 Å Resolution: A Central Ligand in the FeMo-Cofactor. Science 297, 1696–1700 (2002). [DOI] [PubMed] [Google Scholar]
- Peters J. W. et al. Redox-Dependent Structural Changes in the Nitrogenase P-Cluster. Biochemistry 36, 1181–1187 (1997). [DOI] [PubMed] [Google Scholar]
- Tezcan F. A. et al. Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein. Science 309, 1377–1380 (2005). [DOI] [PubMed] [Google Scholar]
- Kim J., Woo D. & Rees D. C. X-ray Crystal Structure of the Nitrogenase Molybdenum-Iron Protein from Clostridium pasteurianum at 3.0Å Resolution. Biochemistry 32, 7104–7115 (1993). [DOI] [PubMed] [Google Scholar]
- Mayer S. M., Lawson D. M., Gormal C. A., Roe S. M. & Smith B. E. New insights into structure-function relationships in nitrogenase: a 1.6Å resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe protein. J. Mol. Biol. 292, 871–891 (1999). [DOI] [PubMed] [Google Scholar]
- Mayer S. M., Gormal C. A., Smith B. E. & Lawson D. M. Crystallographic Analysis of the MoFe Protein of Nitrogenase from a nifV Mutant of Klebsiella pneumoniae Identifies Citrate as a Ligand to the Molybdenum of Iron Molybdenum Cofactor (FeMoco). J. Biol. Chem. 277, 35263–35268 (2002). [DOI] [PubMed] [Google Scholar]
- Thoden J. B., Raushel F. M., Benning M. M., Rayment I. & Holden H. M. The structure of carbamoyl phosphate synthetase determined to 2.1Å resolution. Acta Crystallogr. D Biol. Crystallogr. 55, 8–24 (1999). [DOI] [PubMed] [Google Scholar]
- Smith J. L. Glutamine PRPP amidotransferase: snapshots of an enzyme in action. Curr. Opin. Struct. Biol. 8, 686–694 (1998). [DOI] [PubMed] [Google Scholar]
- Vanoni M. A. & Curti B. Structure–function studies of glutamate synthases: A class of self-regulated iron-sulfur flavoenzymes essential for nitrogen assimilation. IUBMB Life 60, 287–300 (2008). [DOI] [PubMed] [Google Scholar]
- Nakamura A., Yao M., Chimnaronk S., Sakai N. & Tanaka I. Ammonia Channel Couples Glutaminase with Transamidase Reactions in GatCAB. Science 312, 1954–1958 (2006). [DOI] [PubMed] [Google Scholar]
- Wu J. et al. Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme. J. Mol. Biol. 391, 703–716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecilla-Camps J. C., Amara P., Cavazza C., Nicolet Y. & Volbeda A. Structure–function relationships of anaerobic gas-processing metalloenzymes. Nature 460, 814–822 (2009). [DOI] [PubMed] [Google Scholar]
- Scott D. J., Dean D. R. & Newton W. E. Nitrogenase-catalyzed Ethane Production and CO-sensitive Hydrogen Evolution from MoFe Proteins Having Amino Acid Substitutions in an a-Subunit FeMo Cofactor-binding Domain. J. Biol. Chem. 267, 20002–20010 (1992). [PubMed] [Google Scholar]
- Berendsen H. J. C. & Hayward S. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 10, 165–169 (2000). [DOI] [PubMed] [Google Scholar]
- Floquet N. et al. Collective motions in Glucosamine-6-phosphate Synthase: Influence of Ligand Binding and role in Ammonia Channelling and Opening of the Fructose-6-Phosphate Binding Site. J. Mol. Biol. 385, 653–664 (2009). [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman K. & Kern D. Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information