Abstract
Small RNAs use a diversity of well-characterized mechanisms to repress mRNAs, but how they activate gene expression at the mRNA level remains not well understood. The predominant activation mechanism of Hfq-associated small RNAs has been translational control whereby base pairing with the target prevents the formation of an intrinsic inhibitory structure in the mRNA and promotes translation initiation. Here, we report a translation-independent mechanism whereby the small RNA RydC selectively activates the longer of two isoforms of cfa mRNA (encoding cyclopropane fatty acid synthase) in Salmonella enterica. Target activation is achieved through seed pairing of the pseudoknot-exposed, conserved 5′ end of RydC to an upstream region of the cfa mRNA. The seed pairing stabilizes the messenger, likely by interfering directly with RNase E-mediated decay in the 5′ untranslated region. Intriguingly, this mechanism is generic such that the activation is equally achieved by seed pairing of unrelated small RNAs, suggesting that this mechanism may be utilized in the design of RNA-controlled synthetic circuits. Physiologically, RydC is the first small RNA known to regulate membrane stability.
Keywords: fatty acid synthesis, Hfq, mRNA activation, noncoding RNA, small RNA
Introduction
The intense study of eukaryotic microRNAs and bacterial small regulatory RNAs (sRNAs) over the last decade has provided tremendous insight into how small RNAs use base-pairing mechanisms to control large post-transcriptional regulons. These studies focussed predominantly on the repression of mRNAs targets, which is the primary mode of action of both the eukaryotic microRNAs and the bacterial sRNAs (Lioliou et al, 2010; Storz et al, 2011; Pasquinelli, 2012). The bacterial base-pairing sRNAs include the prominent class of Hfq-associated sRNAs, which are regarded as the largest group of post-transcriptional regulators in Gram-negative bacteria (Vogel and Luisi, 2011; De Lay et al, 2013). Model bacteria such as Escherichia coli and Salmonella enterica (hereafter referred to as Salmonella) may express >100 sRNAs (Chao et al, 2012; Zhang et al, 2013), most of which act on multiple targets in a large set of cellular pathways. Hfq, the RNA-binding protein from which this class of sRNAs derives its name, has multiple functions in this regulation: it stabilizes many sRNAs prior to target recognition, generally facilitates the short seed interactions between sRNAs and mRNAs, and recruits auxiliary factors such as the major mRNA decay enzyme RNase E (Morita and Aiba, 2011; Vogel and Luisi, 2011).
Regarding the mechanisms of target repression by Hfq and sRNAs, current evidence supports two general scenarios, namely the inhibition of translation and direct mRNA destabilization. In the former case, an sRNA binds near its target’s ribosome binding site (RBS) to sterically interfere with 30S ribosome association. This primary event of translational repression may subsequently be rendered irreversible through mRNA decay either by concomitant recruitment of RNase E as part of a tripartite sRNA:Hfq:RNase E complex (Morita et al, 2005; Prevost et al, 2011), or simply by the increased vulnerability of the untranslated mRNA to the RNase E-containing degradosome (Belasco, 2010). As a variation on the theme, translation may be repressed by sRNA-guided loading of Hfq near the RBS rather than by the sRNA-mRNA duplex itself (Desnoyers and Masse, 2012). In the other general scenario, translational control is bypassed and mRNA destruction is the primary event in target repression. In this case, RNase E is recruited to either the sRNA-mRNA duplex itself, or a nearby site that becomes accessible due to a structural rearrangement in the target mRNA (Afonyushkin et al, 2005; Desnoyers et al, 2009; Pfeiffer et al, 2009; Bandyra et al, 2012; Mackie, 2013a).
In contrast to eukaryotic microRNAs that almost exclusively cause mRNA repression, bacterial sRNAs are also known to activate mRNAs. A recurring theme among Hfq-associated sRNAs has been an anti-antisense mechanism whereby the sRNA acts as a competitive binder to prevent the formation of an intrinsic inhibitory structure around the RBS of the target mRNA. As a result, protein synthesis is upregulated, and the increased translation usually indirectly stabilizes the target mRNA (Fröhlich and Vogel, 2009; Soper et al, 2010). Other positive modes of regulation by Hfq and sRNAs have been reported too. For example, the cis-encoded antisense GadY RNA of E. coli promotes RNase III-mediated cleavage of the dicistronic gadXW mRNA, resulting in a more stable monocistronic gadX transcript and higher synthesis of GadX protein (Opdyke et al, 2004, 2011). In E. coli and Salmonella, there is also positive regulation in trans by the chitobiose operon mRNA that traps the ChiX/MicM sRNA and prevents the latter from repressing an unrelated chitoporin mRNA (Figueroa-Bossi et al, 2009; Overgaard et al, 2009). A different decoy mechanism for indirect activation was recently shown to control biofilm formation in E. coli. Here, the sRNA McaS sequesters the RNA-binding protein CsrA to alleviate CrsA-mediated translational repression of the pgaA mRNA (Jorgensen et al, 2013).
Notwithstanding these diverse modes of positive regulation, it is surprising that a pathway whereby sRNAs activate targets by direct interference with the RNase E-mediated decay has not been reported until recently. RNase E degrades many cellular mRNAs by recognizing the 5′ end, that is, the region of the mRNA where sRNAs commonly bind, followed by endonucleolytic cleavage and subsequent 3′ to 5′ exonucleolytic degradation by other enzymes (Belasco, 2010; Mackie, 2013b). Moreover, mRNA duplexes—both intrinsic and resulting from cis-antisense transcription in the 5′ UTR—can protect mRNAs from RNase E (Bouvet and Belasco, 1992; Stazic et al, 2011). Similarly, there are several reports of positive regulation in which 30S ribosomes or CsrA occlude RNase E recognition sites in the 5′ UTR of an mRNA (Braun et al, 1998; Lodato et al, 2012; Yakhnin et al, 2013). Collectively, this led us to hypothesize the existence of a translation-independent activation mechanism whereby Hfq and sRNAs act to activate targets directly by site-specific interference with RNase E-mediated mRNA decay. In support of this hypothesis, we have recently reported that the Hfq-associated SgrS sRNA selectively activates the synthesis of the phosphatase YigL by the selective capture and stabilization of an RNase E decay intermediate of the pldB-yigL operon mRNA (Papenfort et al, 2013). Here, we present evidence for a direct activation mechanism that operates in the 5′ region of a full-length monocistronic mRNA.
We report that the conserved sRNA RydC activates the synthesis of cyclopropane fatty acid (CFA) synthase in Salmonella. RydC was originally discovered by its co-immunoprecipitation (coIP) with Hfq in E. coli (Zhang et al, 2003). In vitro structure probing predicted an unusual pseudoknot structure of this sRNA, and an initial functional analysis suggested that RydC may be involved in the regulation of the yejABEF mRNA which encodes a putative ABC transport system (Antal et al, 2005). However, direct targets of RydC have been unknown, as has a conserved function of this sRNA. Using a combination of biochemical and genetic approaches, we demonstrate that RydC pairs with the 5′ UTR of a longer isoform of cfa mRNA. Formation of this RNA interaction, which occurs far upstream of the cfa start codon, is independent of the actual seed sequence of RydC, alters RNase E-dependent decay and stabilizes the target, even in the absence of mRNA translation. There is also evidence to suggest that independent of the base pairing, the sRNA-guided recruitment of the Hfq protein as an effector contributes to stabilization of the cfa mRNA. Physiologically, RydC may be the first regulatory sRNA known to influence bacterial membrane stability.
Results
Expression and molecular architecture of RydC
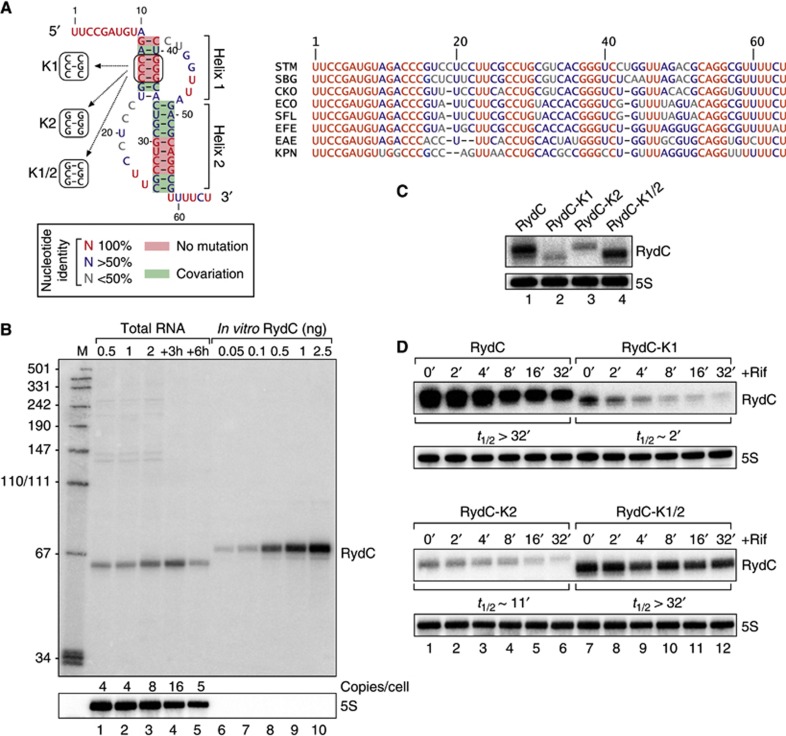
RydC is an ∼65-nt sRNA originally discovered in E. coli (Zhang et al, 2003; Antal et al, 2005) whose sequence and 3′ flanking gene (cybB) are conserved in many other enterobacterial species (Figure 1A; Supplementary Figure S1). Northern blot analysis of RydC in Salmonella revealed its expression throughout growth in L-broth, and as single RNA species with the length predicted for the primary RydC transcript (Figure 1B). The expression profile of RydC matches its previously reported association pattern with Hfq (Chao et al, 2012). Using in vitro synthesized RNA as a concentration standard, we estimated that under the conditions tested, there were ∼4–16 copies of RydC present per cell (Figure 1B).
Figure 1.
Conservation, structure and expression of RydC. (A) Alignment and pseudoknot structure of RydC RNA. STM: Salmonella Typhimurium; SBG: Salmonella bongori; CKO: Citrobacter koseri; ECO: Escherichia coli; SFL: Shigella flexneri; EFE: Escherichia fergusoni; EAE: Enterobacter aerogenes; KPN: Klebsiella pneumoniae. Mutations K1, K2 and K1/2 introduced to alter pseudoknot formation are indicated. (B) RydC levels in total RNA samples (corresponding to 1 OD600) of wild-type Salmonella at indicated time points of growth were compared on northern blots to signals of in vitro transcribed RydC to estimate the in vivo copy number. 5S RNA served as a loading control. (C) Expression levels of RydC, RydC-K1, RydC-K2 and RydC-K1/2 (as described in (A); expressed from the constitutive PL promoter) were determined in ΔrydC Salmonella by northern blot analysis of total RNA samples (OD600 of 1). (D) Stabilities of RydC, RydC-K1, RydC-K2 and RydC-K1/2 were determined in ΔrydC Salmonella by northern blot analysis of total RNA samples withdrawn prior to and at indicated time points after inhibition of transcription by rifampicin at an OD600 of 1. See Supplementary Figure S4 for quantification.
Source data for this figure is available on the online supplementary information page.
RydC is a representative example of a Hfq-associated sRNA with a potential pseudoknot fold (Antal et al, 2005), which is a structural motif more commonly found in larger transcripts or tRNAs (Brierley et al, 2007). While this pseudoknot was inferred from in vitro structure probing and sequence comparison (Antal et al, 2005), its impact on RydC expression or function in vivo remained unknown. Our alignment of currently available rydC sequences suggests an intriguing architecture whereby several compensatory mutations have structurally maintained a central core based on two pseudoknot helices, which are preceded by 10 ultra-conserved single-stranded nucleotides at the 5′ end of the sRNA (Figure 1A; Supplementary Figure S2).
To address the relevance of the pseudoknot fold in vivo, we disrupted helix 1 by changing the two conserved guanosines at positions 37 and 39 to cytosines (mutant RydC-K1; Figure 1A; Supplementary Figure S3). We observed an ∼5-fold decrease in RNA steady-state levels compared to wild-type RydC (Figure 1C), which was accompanied by a reduction in the half-life of the RNA from >32 min to <2 min (Figure 1D; Supplementary Figure S4). The reciprocal mutation of two cytosines at positions 13 and 15 to guanosines (RydC-K2; Figure 1A; Supplementary Figure S3), which similarly breaks helix 1, also reduced both the steady-state levels and stability of the sRNA. However, rescuing helix formation in the mutant RydC-K1/2 fully restored steady-state RNA levels and stability to that of the wild type (Figure 1C and D; Supplementary Figure S4). These results indicate that the RydC pseudoknot fold is required for sRNA stability in vivo.
RydC upregulates CFA synthase
To identify RydC targets in Salmonella, we first compared the protein expression patterns of strains in which the rydC gene was deleted (ΔrydC), complemented with a multi-copy plasmid (pPLRydC) that causes RydC overproduction, and of wild-type bacteria. We did not detect major changes in protein levels on a Coomassie-stained SDS–PAGE gel, suggesting that RydC was not a pleiotropic regulator, or regulated highly abundant proteins such as porins (Supplementary Figure S5).
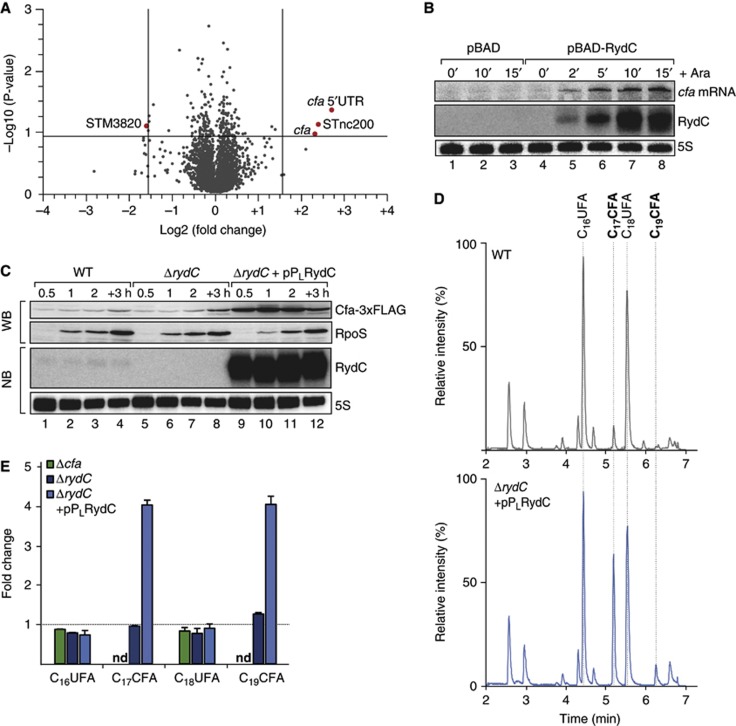
However, using sRNA pulse expression (Masse et al, 2005; Papenfort et al, 2006) from the inducible pBAD promoter of plasmid pBAD-RydC followed by microarray analysis of global gene expression changes, we identified two mRNAs whose levels were significantly (≥3-fold) altered in a RydC-dependent manner (Figure 2A; Supplementary Table S1). The strongest regulation was observed for the cfa mRNA encoding CFA synthase, which was upregulated ∼5-fold or ∼6.5-fold as detected with two probes corresponding to the mRNA’s coding sequence (CDS) or the 5′ UTR, respectively (Figure 2A). In addition, RydC upregulated the candidate sRNA STnc200 (Pfeiffer et al, 2007), and downregulated the STM3820 mRNA encoding a putative cytochrome C peroxidase.
Figure 2.
RydC induces cfa expression and activity. (A) Microarray analysis of Salmonella genes affected by pulse overexpression of RydC. RydC expression was induced by addition of arabinose (final concentration: 0.2%) to rydC mutant cells carrying pBAD-RydC or control plasmid pBAD. Changes in transcript abundances were scored on Salmonella-specific microarrays; genes displaying >3-fold change (P-value<0.15) are marked in red. (B) RydC and cfa mRNA levels were determined on northern blots of total RNA extracted from rydC mutant cells carrying plasmids pBAD or pBAD-RydC at indicated time points prior to and after addition of arabinose (Ara). The oligo directed against the 5′ UTR of cfa specifically recognizes cfa1 mRNA. (C) Expression of Cfa-3xFLAG in wild-type and ΔrydC mutant Salmonella either carrying a control construct or a plasmid for the constitutive overexpression of RydC from the PL promoter was monitored over growth on western blots. (D) Total ion chromatograms of Salmonella wild-type cells carrying a control plasmid or a ΔrydC mutant transformed with the RydC overexpression plasmid pPLRydC. Cells were grown in M9 minimal medium to exponential phase (OD600 of 0.5), and after alkaline hydrolysis, total fatty acids were analysed by LC/MS. Peaks assigned to C16UFA, C17CFA, C18UFA and C19CFA are indicated. (E) Relative quantification of C16UFA, C17CFA, C18UFA and C19CFA in Salmonella Δcfa or ΔrydC carrying either a control plasmid or pPLRydC. All measurements were normalized to wild type; error bars represent the standard deviation calculated from three independent biological replicates; nd: not detected.
Source data for this figure is available on the online supplementary information page.
We focussed on cfa as the target showing strongest regulation. A time-course experiment in which changes in cfa mRNA levels were monitored by northern blot analysis revealed the activation to be rapid, leading to ∼2.2-fold and ∼7.3-fold higher cfa mRNA levels at 2 and 15 min post pBAD-RydC induction, respectively (Figure 2B, lanes 4–8). This activation was specific to RydC, that is, not seen with the empty pBAD control vector (Figure 2B, lanes 1–3). To monitor Cfa protein levels by quantitative western blot, we tagged the cfa gene in the Salmonella chromosome with a C-terminal 3 × FLAG epitope. While the levels of Cfa protein did not significantly differ between the wild-type and ΔrydC strains at various stages of growth examined (likely due to low basal expression of RydC; Figure 2C, lanes 1–4 versus 5–8), overexpression of RydC increased the abundance of this protein up to ∼11-fold (lanes 1–4 versus 9–12).
This observed increase in Cfa protein was corroborated by measuring the effect of RydC expression on cellular lipid composition. The CFA synthase converts the cis double bond of pre-existing unsaturated fatty acids (UFAs) of membrane phospholipids into a more stable methylene bridge (Grogan, 1997). This modification increases the stability of the bacterial membrane and can be detected by liquid chromatography/mass spectrometry (LC/MS) analysis (Nagy et al, 2004). In comparing the total fatty acid content by LC/MS, using a Salmonella cfa mutant as a negative control, we detected an ∼4-fold increase in CFA (C17CFA; C19CFA) levels in the RydC overproducing strain, as compared to the wild-type (Figure 2D and E). Thus, RydC can affect bacterial membrane composition through activation of Cfa protein.
RydC selectively activates the longer of two cfa mRNA isoforms
Previous studies have identified two conserved transcription start sites (TSSs) for cfa in E. coli, Salmonella and other related γ-proteobacteria (Wang and Cronan, 1994; Kim et al, 2005), and both TSSs were confirmed in a recent dRNA-seq analysis of the Salmonella strain SL1344 used in this study (Kröger et al, 2012). Transcription from the distal TSS associated with a σ70-dependent promoter yields the cfa1 mRNA with an unusually long (210 nt) 5′ UTR. A shorter mRNA, cfa2, originates from the proximal TSS, 33 bp upstream of the start codon (Figure 3A; Supplementary Figure S6). Transcription of cfa2 requires the major stress σ-factor σS, encoded by rpoS, and is considered to account for the increased Cfa levels in stationary phase bacteria (Cronan, 2002).
Figure 3.
RydC acts on one of the two isoforms of cfa mRNA. (A) Sequence of the Salmonella cfa upstream region. Transcription initiates from two start sites indicated by arrows at −210 bp (σ70-dependent; cfa1 mRNA) or −33 bp (σS-dependent; cfa2 mRNA) relative to the translational start site, respectively. Promoter elements are highlighted in grey, and the start codon is boxed. (B) RydC specifically acts on the longer cfa mRNA isoform. Salmonella cfa::3xFLAG ΔrydC cells or an isogenic ΔrpoS mutant were transformed with the pBAD control plasmid (−) or the pBAD-RydC overexpression plasmid (+); Salmonella Δcfa ΔrydC served as a negative control. All strains were grown to an OD600 of 2, and total RNA samples withdrawn prior to and 15′ after arabinose addition were used as templates for primer extension. Transcripts originating from either TSS1 or TSS2 were identified using gene-specific sequencing ladders. (C) Schematic representation of the cfa gene including the upstream promoter region. Translational cfa::gfp fusions (under control of the constitutive PLTetO-1 promoter) were constructed comprising the 5′ upstream region from the distal (cfa1::gfp) or the proximal start site (cfa2::gfp) plus the first 45 nucleotides of the cfa CDS. (D) Regulation of reporter fusions was monitored by western blot analysis. At an OD600 of 1, total protein samples were prepared from Salmonella ΔrydC ΔrpoS mutants carrying plasmids to express cfa2::gfp or cfa1::gfp in combination with a control plasmid (−) or pPLRydC (+). GroEL served as a loading control. Expression of RydC was validated on a northern blot.
Source data for this figure is available on the online supplementary information page.
To understand which of the two isoforms of the cfa mRNA is controlled by RydC, we examined their abundance using primer extension experiments, prior to and 15 min after pBAD-RydC induction. Strikingly, RydC significantly activated the longer cfa1 mRNA but had no effect on the shorter cfa2 transcript (Figure 3B, lanes 1–4). As expected, activation of the σ70-dependent cfa1 mRNA was unaffected by a ΔrpoS mutation; by contrast, the σS-dependent cfa2 transcript was no longer detected in the ΔrpoS strain, with or without RydC (lanes 5–8). Importantly, the activation of cfa1 expression in wild-type cells reflects the effect of RydC on the total cfa mRNA pool, that is, the sum of products from both TSSs. Using quantitative real-time PCR (qRT–PCR) and primers binding in the codon sequence (which is common to both the cfa1 and cfa2 transcript) total cfa mRNA levels were observed to increase ∼3-fold upon RydC expression for 15 min (unpublished results).
The rapid activation of the cfa mRNA by RydC (Figure 2B) suggested control at the mRNA level. To address this, we cloned the 5′ UTRs of cfa1 or cfa2, along with the first 15 codons, into a GFP reporter plasmid (Urban and Vogel, 2007) and tested reporter activation by RydC. Note that these GFP reporters are transcribed from a constitutive (PLtetO) promoter, so changes in reporter activity indicate post-transcriptional regulation. Again, RydC overexpression did not affect the cfa2::gfp reporter but upregulated the cfa1::gfp reporter 7-fold (Figure 3D); similarly, the levels of only the cfa1::gfp and not the cfa2::gfp mRNA increased in the presence of RydC (Supplementary Figure S7). In further support of post-transcriptional regulation, an intact hfq gene was essential for the observed activation (Supplementary Figure S8). Interestingly, basal reporter activity of the cfa1::gfp construct was ∼10-fold lower than that of cfa2::gfp (Figure 3D, lane 1 versus 3). Thus, the shorter cfa2 mRNA yields higher levels of Cfa protein, whereas the long 5′ UTR of the cfa1 mRNA limits Cfa synthesis. However, using a cfa1-specific mechanism of post-transcriptional activation, RydC can achieve the same levels of Cfa synthesis as stress-induced transcription from the cfa2-associated σS promoter.
RydC activates Cfa expression by seed pairing
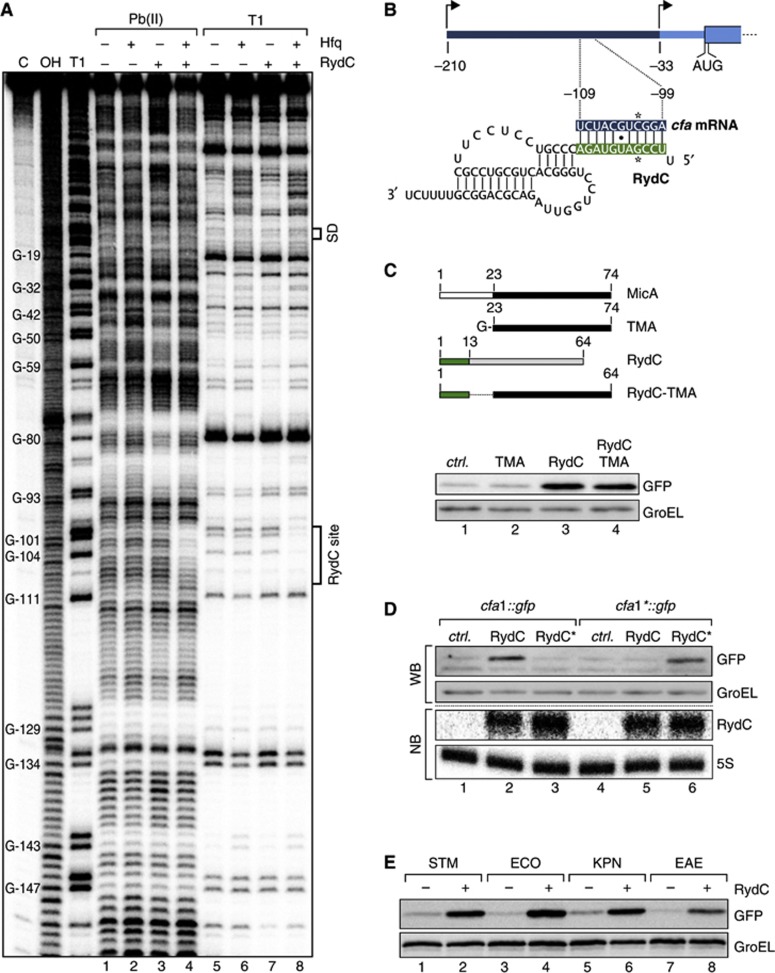
Given that mRNA regulation by Hfq-associated sRNAs typically involves base-pairing interactions (Vogel and Luisi, 2011), we experimentally and biocomputationally searched for a potential RydC binding site in the cfa1 mRNA. First, we subjected a radiolabelled, ∼300 nt RNA fragment (from the distal cfa TSS to residue 70 of the CDS) to structure probing with lead(II) acetate or RNase T1. Addition of RydC did not induce significant changes in the cleavage patterns in the cfa mRNA fragment (Figure 4A, lanes 4 versus 6 and 8 versus 10), likely because it requires Hfq for annealing to the mRNA. Indeed, concomitant addition of RydC and Hfq—but not Hfq alone—protected region −99 to −109 nt relative to the cfa start codon (lanes 7 and 11). This putative RydC site lies ∼60 nt upstream of where the cfa2 mRNA begins and is consistent with the observed selective regulation of cfa1 mRNA.
Figure 4.
RydC employs its conserved, single-stranded 5′ end to base pair with cfa1 mRNA. (A) In vitro structure probing using 5′ end-labelled cfa mRNA (TSS1 to nt 70 of the CDS; 20 nM) with lead(II) acetate (lanes 1–4) and RNase T1 (lanes 5–8) in the presence and absence of Hfq (20 nM) and RydC (200 nM). RNase T1 and alkaline ladders of cfa mRNA were used to map cleaved fragments. Positions of G-residues are indicated relative to the translational start site. The RydC binding site and the Shine-Dalgarno (SD) region are marked. (B) Predicted duplex forming between RydC (nts 2–11) and cfa mRNA (nts −109 to −99 relative to the translational start site). Positions of single-nucleotide exchanges generating the compensatory mutants RydC* and cfa* mRNA are indicated. (C) Schematic representation of wild-type MicA and RydC as well as the derivative constructs. The first 13 nts of RydC were fused to the 3′ part of MicA (nts 23–74; TMA) to construct RydC-TMA. The 5′ end of RydC is required to interact with cfa mRNA. GFP levels were determined on western blots of total protein samples isolated from Salmonella ΔrydC ΔrpoS mutants carrying plasmids for cfa1::gfp and either a control or plasmids for PL-driven overexpression of TMA, RydC or RydC-TMA. (D) Validation of the RydC-cfa mRNA interaction. Salmonella ΔrydC ΔrpoS mutants carrying plasmids for cfa1::gfp and cfa1*::gfp in combination with a control plasmid or RydC overexpression plasmids pPLRydC and pPLRydC*. Expression of GFP-fusion proteins was monitored on western blots of total protein samples prepared from cells in exponential growth (OD600 of 1). Equal expression of RydC and RydC* was controlled by northern blot analysis. (E) The regulation of cfa by RydC is conserved. GFP expression was monitored by western blot analysis in Salmonella ΔrydC mutant cells carrying translational Salmonella (STM), E. coli (ECO), K. pneumoniae (KPN) or E. aerogenes (EAE) cfa1::gfp reporter fusions in combination with either a control (−) or a plasmid to overexpress RydC versions of the indicated species (+).
Source data for this figure is available on the online supplementary information page.
Second, computational analysis using the RNAhybrid algorithm (Rehmsmeier et al, 2004) predicted a potential 11-bp RNA duplex to form by the pairing of nucleotides 2–12 of RydC with the region of cfa that is protected in the above structure probing experiments (Figure 4B). With a predicted change in free energy of −22.8 kcal/mol, this RNA duplex was well within the range of previously observed seed pairings (Papenfort et al, 2010). This interaction involved the highly conserved nucleotides of the single-stranded 5′ end of RydC (compare to Figure 1A), reminiscent of the 5′ terminal seed pairing reported for numerous other Hfq-associated sRNAs (Guillier and Gottesman, 2008; Pfeiffer et al, 2009; Papenfort et al, 2010; Corcoran et al, 2012; Holmqvist et al, 2012; Shao et al, 2013). Moreover, a chimaeric RydC-TMA sRNA in which the first 13 nucleotides of RydC are fused to an unrelated sRNA (TMA; truncated MicA; Bouvier et al, 2008) also activated the cfa1::gfp reporter indistinguishably from wild-type RydC, indicating that the 5′ end of RydC is sufficient for recognition of the cfa target (Figure 4C). For further proof that the predicted seed pairing underlies cfa regulation, we changed cytosine −102 to guanosine in the cfa1::gfp reporter. As expected, this mutation to which we refer as cfa1* abolished reporter activation by RydC (Figure 4D, lanes 1–2 versus 5–6). Likewise, mutation of RydC at position 5 from guanosine to cytosine (RydC*) abolished regulation of the wild-type cfa::gfp reporter (Figure 4D, lanes 1–3). However, the combination of these two compensatory mutations restored reporter activation to wild-type levels (Figure 4D, lane 6).
The described RydC-cfa seed pairing seemed conserved in many other γ-proteobacterial species, as seen in sequence alignments of RydC sRNA (Figure 1A) or cfa mRNA (Supplementary Figure S6). To test whether the conservation of the interaction sites extends to conservation of regulation, we constructed additional cfa1::gfp reporters with the sequences of E. coli, Enterobacter aerogenes and Klebsiella pneumoniae, and co-expressed each fusion together with RydC of the cognate species. Each of these reporters was found to be activated by the cognate RydC sRNA (Figure 4E). Thus, RydC activates cfa synthesis by a conserved seed pairing interaction with the long isoform of the cfa mRNA, far upstream of the canonical translation control elements.
Translation-independent target activation
Activation by a bacterial sRNA typically occurs through a targeted disruption of inhibitory secondary structures in the 5′ UTR of the translationally inactive target mRNA. These structural changes render the RBS more accessible, thereby promoting translational initiation (Fröhlich and Vogel, 2009). However, sequence inspection of the 5′ region of cfa revealed no conserved regions expected to sequester the RBS through intramolecular base pairing (Supplementary Figure S6). Likewise, RydC pairing had no affect on RBS accessibility in our structure probing experiments (Figure 4A), and addition of RydC did not increase ternary complex formation in 30S ribosome toeprinting assays with the cfa1 mRNA (unpublished results).
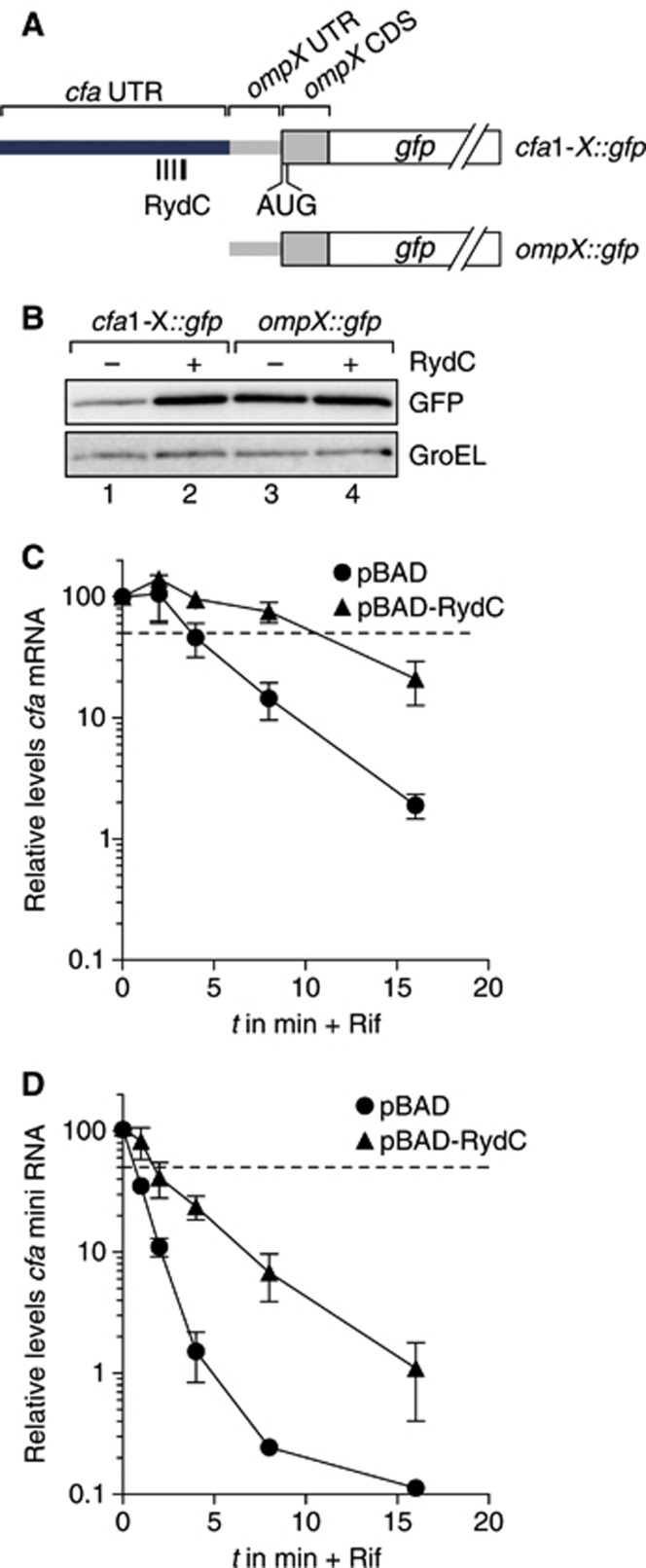
To further exclude the possibility of activation by the canonical anti-antisense principle, we fused the first 149 nt of the cfa1 mRNA (TSS1 to −62), that is, without the RBS of cfa, to the 5′ UTR of an unrelated ompX::gfp fusion (Figure 5A). While the parental ompX::gfp reporter was not regulated by RydC, the grafted 5′ region of cfa augmented the chimaeric cfa1-X::gfp reporter with strong (∼6-fold) activation by RydC (Figure 5B). Of note, as with the cfa1::gfp versus cfa2::gfp reporters above (Figure 3D), the inclusion of the 5′ region of cfa1 reduced the basal reporter activity of cfa1-X::gfp as compared to ompX::gfp (Figure 5B). Altogether, these results disfavoured a model of translational activation whereby RydC would positively affect the accessibility of the cfa RBS by resolving an inhibitory RNA structure.
Figure 5.
RydC can stabilize cfa mRNA. (A) Schematic representation of gfp fusion plasmids encompassing indicated fragments of the proximal cfa 5′ UTR (TSS1 to −62) inserted upstream of the 33-nt long 5′ UTR and the first 10 amino acids of ompX. (B) Regulation of reporter fusions as described in (A) was monitored by western blot analysis of total protein samples prepared from Salmonella ΔrydC ΔrpoS mutants in the absence (−) and presence (+) of RydC. (C) Stability of cfa mRNA in the presence of RydC. Salmonella ΔrydC ΔrpoS cells carrying either plasmids pBAD or pBAD-RydC were grown to OD600 of 1.0 when L-arabinose was added to induce RydC expression. After 15 min of induction, cultures were treated with rifampicin, and RNA samples were prepared from cells prior to and at indicated time points post treatment. Abundance of cfa mRNA was determined by qRT–PCR analysis. The signal obtained at 0 min was set to 100%, and the percentage of mRNA remaining at each time point was plotted on the y axis versus time on the x axis. The time point at which 50% of cfa mRNA had been decayed (dashed line) was calculated to determine the half-life (t1/2). Error bars represent the standard deviation calculated from three independent biological replicates. (D) Stability of cfa mini RNA in the presence of RydC. The cfa mini RNA was constitutively expressed in Salmonella ΔrydC Δcfa cells carrying either plasmids pBAD or pBAD-RydC, and its decay was analysed as described in (B).
Source data for this figure is available on the online supplementary information page.
Consequently, we tested whether RydC interfered with cfa mRNA decay by examining RydC-dependent changes in mRNA stability. The sRNA was induced from plasmid pBAD-RydC in a ΔrydC/ΔrpoS strain (to eliminate expression from the cfa2 promoter) for 15 min, before transcription was stopped with rifampicin treatment, and cfa mRNA decay was monitored by qRT–PCR. RydC increased the half-life of cfa1 mRNA by ∼3-fold, as compared to cells with induced control vector (t1/2 ∼4 min versus ∼12 min, Figure 5C). Next, to exclude that a higher translation rate contributed to the increase in mRNA stability, we constructed a non-coding cfa mini gene where the first 149 nt of the cfa1 mRNA (TSS1 to −62) are fused to a Rho-independent transcription terminator; this construct harbours the RydC site but lacks the RBS. A RydC effect on the stability of this cfa mini transcript was determined as above but in a Salmonella ΔrydC/Δcfa strain (Figure 5D). The cfa mini transcript was less stable than the translated full-length cfa1 mRNA (t1/2 of ∼0.6 min versus ∼4 min; Figure 5C and D), which may be explained by the absence of ribosomes that are known to protect from degradation by ribonucleases such as RNase E (e.g., Arnold et al, 1998; Braun et al, 1998). Nonetheless, RydC caused an equivalent 3-fold increase in the RNA half-life (to ∼1.9 min; Figure 5D). Altogether, while the 3-fold change in mRNA stability may not fully explain the observed 7-fold increase in cfa mRNA levels (see Figure 2B), we have shown that RydC can stabilize a cfa-derived transcript in the absence of translational control.
RydC and Hfq protect cfa mRNA from endonucleolytic cleavage by RNase E
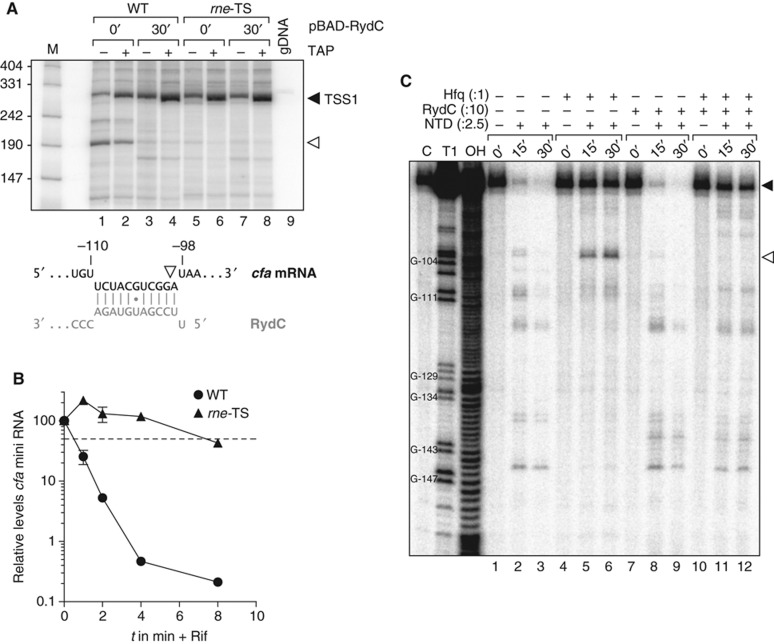
Understanding the basis for the RydC-mediated stabilization of the cfa mRNA requires knowledge of how this mRNA is degraded. In Gram-negative enterobacteria such as E. coli and Salmonella, the major enzyme to degrade mRNAs is the endoribonuclease RNase E (Carpousis et al, 2009; Belasco, 2010; Mackie, 2013b). Since RNase E is encoded by the essential rne gene, we made use of an available rne-ts allele (Figueroa-Bossi et al, 2009), which enables selective inactivation of the protein during growth at 44°C to examine RNase E effects on cfa mRNA decay.
Salmonella strains carrying the rne (wild type) or rne-ts alleles in a ΔrydC/ΔrpoS genetic background were grown to early stationary phase (OD600 of 1), and cells were shifted to 44°C for 30 min, before RydC expression was induced from pBAD-RydC. We analysed RNA samples taken prior to and 30 min after sRNA induction by a differential 5′ rapid amplification of cDNA ends (RACE) technique that distinguishes between primary 5′ ends and mRNA cleavage products (Bensing et al, 1996). Using a gene-specific primer in the cfa CDS, we observed two major 5′ ends (Figure 6A). The first represented the primary transcript of the distal TSS of cfa1 and, as expected, showed TAP-dependent enrichment (filled arrowhead). The second was an mRNA cleavage product, as inferred from its insensitivity to TAP treatment, which mapped to the RydC-cfa interaction site (open arrowhead); this signal disappeared when either RydC was expressed or RNase E was inactivated, suggesting it to be an RNase E cleavage site that was masked upon formation of the RydC-cfa RNA duplex.
Figure 6.
Binding of RydC interferes with RNase E cleavage of cfa mRNA. (A) 5′ RACE experiments were performed to determine the 5′ ends of cfa mRNA fragments in the absence and presence of RydC. Salmonella ΔrydC ΔrpoS Ptet-cfa::3xFLAG rne-ctrl (WT) and its isogenic rne-ts strain (rne-TS) carrying plasmid pBAD-RydC were grown at 44°C for 30 min. RNA isolated from culture samples withdrawn prior to and 30 min after induction of RydC was tobacco acid pyrophosphatase (TAP)-treated (+) or mock-treated (−) and used to prepare cDNA. Cleavage of the 5′ triphosphate group that is characteristic of primary transcripts by TAP renders the RNA a preferred substrate in the adjacent step of RNA linker ligation. A TAP-dependent elongation product (black arrowhead) and a second, TAP-independent PCR fragment (open arrowhead) were extracted, cloned and sequenced. Salmonella genomic DNA (gDNA) served as a negative control. The 5′ end of the cfa fragment (white arrowhead) is located within the region base-pairing RydC. (B) Inactivation of RNase E stabilizes the cfa mini RNA. The cfa mini RNA was constitutively expressed in Salmonella ΔrydC Ptet-cfa::3xFLAG rne-ctrl (WT) and its isogenic rne-ts strain (rne-TS) at the non-permissive temperature of 44°C. Decay of cfa mini RNA upon rifampicin addition was analysed as described in Figure 5B. (C) Mapping of cfa mRNA cleavage sites in vitro. Time-course experiment monitoring RNase E-mediated decay of 5′ labelled in vitro transcribed cfa mRNA fragment (TSS1 to −72). In all, 20 nM of mRNA was incubated with 50 nM purified RNase E N-terminal catalytic domain (NTD) in the absence or presence of Hfq (20 nM), and RydC or RydC* sRNAs (200 nM) as indicated. Reactions were stopped prior to and 15 or 30 min post addition of NTD. RNase T1 and alkaline ladders of cfa mRNA were employed to map cleavage products. The cleavage intermediate of cfa mRNA is indicated by an open arrowhead and mapped to position −99 relative to the cfa translational start site.
Source data for this figure is available on the online supplementary information page.
As expected if RNase E was the main nucleolytic factor involved in cfa mRNA decay, the basal levels of the full-length cfa1 mRNA increased ∼3-fold upon inactivation of RNase E, that is, independent of RydC expression (compare lanes 1 with 5, or 2 with 6 in Figure 6A). The inactivation of RNase E is epistatic to stabilization of the cfa1 transcript by RydC, as evidenced by the failure of RydC overproduction to significantly increase the concentration of cfa1 mRNA in rne-ts cells at 44°C (compare lanes 8 and 6 versus lanes 4 and 2). Furthermore, this increase in steady-state levels was also accompanied by increased transcript stability independent of translation; upon inactivation of RNase E, the half-life of the cfa mini RNA increased from ∼0.5 to 7.5 min (Figure 6B).
To assess the activities of RNase E and its potential antagonist RydC in more detail, we subjected the 5′ region of the cfa1 mRNA (TSS1 to −72) to RNase E cleavage in vitro (Figure 6C). Under the conditions used, the target mRNA fragment was fully digested by RNase E within 30 min (lanes 1–3). The presence of Hfq in equimolar concentration to the mRNA protected the latter from degradation (lanes 4–6) but also caused a pronounced cleavage (open arrowhead) at precisely the position that had been observed in vivo, that is, at −100 relative to the AUG (cf. Figure 6A). Intriguingly, the additional presence of RydC (lanes 10–12) suppressed the cleavage at this site in favour of other sites seen in the RNase E only reaction (compare to lanes 1–3). Since RydC alone had no effect (lanes 7–9), we interpret these results to mean that the combined action of RydC and Hfq inhibits RNase E cleavage in a site-specific manner. This may lead to the stabilization of the cfa1 mRNA and ultimately an increase in the levels of Cfa protein.
mRNA stabilization with interchangeable seed sequence
The suppressed RNase E cleavage site is located within the RydC-cfa RNA duplex, which at first glance suggests a simple model in which Hfq helps anneal RydC to the target to prevent this site from being recognized by RNase E (Figure 6). The chimaeric RydC-TMA sRNA would similarly sequester this RNase E site, which would explain its comparable potency as a cfa1 activator (Figure 4C).
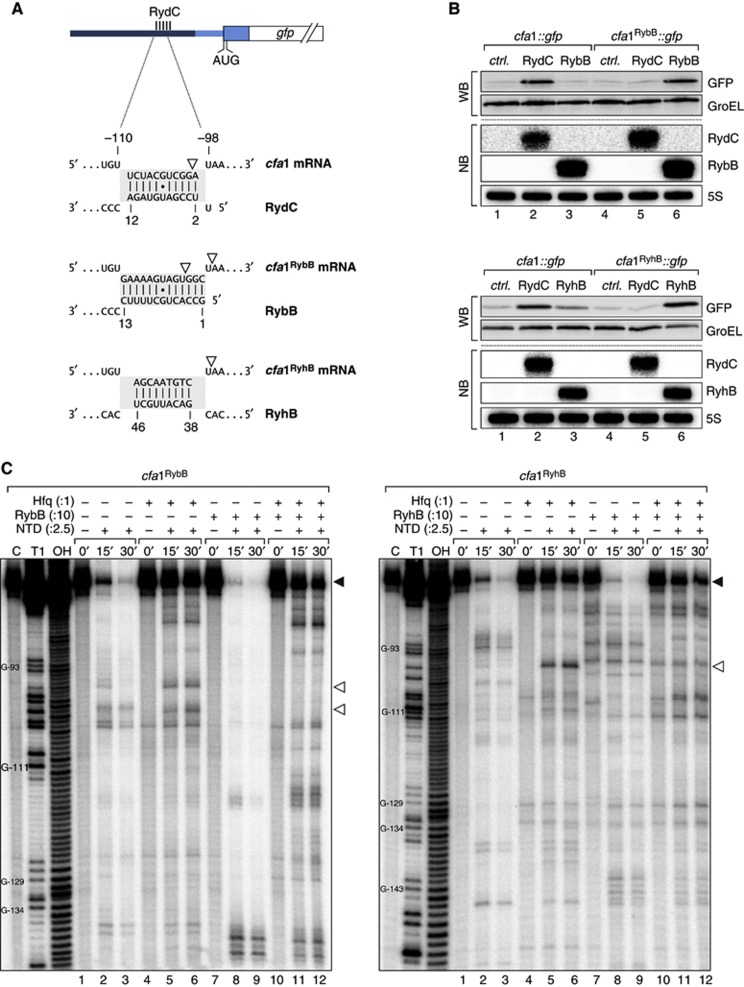
To test this RNase E site sequestration model, we replaced the 11-nucleotide RydC site (positions −99 to −109, Figure 4B) with the binding sites of the unrelated RybB or RyhB sRNAs (reporters cfa1RybB::gfp and cfa1RyhB::gfp, respectively; Figure 7A). As expected, these changes eliminated regulation by RydC (Figure 7B; Supplementary Figure S8). However, co-expression of RybB or RyhB selectively and fully restored mRNA activation (Figure 7B), arguing that the seed sequence is fully interchangeable.
Figure 7.
(A) Schematic representation of the base-pairing regions in cfa1, cfa1RybB or cfa1RyhB reporter fusions and the expected base-pairing interactions to RydC, RybB or RyhB, respectively. RNase E cleavage sites identified in in vitro assays are indicated by open arrowheads. (B) The regulation of cfa expression is independent of the actual seed sequence. Salmonella ΔrydC ΔrybB or ΔryhB mutants were transformed with cfa1::gfp and either cfa1RybB::gfp or cfa1RyhB::gfp reporter fusions, respectively. GFP levels were determined by western blot analysis in the presence of either a control plasmid or constructs to constitutively overexpress RydC, RybB or RyhB. See Supplementary Figure S9 for quantification. Expression of RydC, RybB and RyhB sRNAs was monitored on northern blots. (C) Determination of cfa1RybB (left panel) and cfa1RyhB (right panel) cleavage sites in vitro. Time-course experiment of RNase E-mediated decay of cfa1 variants as described in Figure 6C but using cfa1RybB or cfa1RyhB RNA and RybB or RyhB sRNAs, respectively. Mapping of the cleavage intermediates indicated by an open arrowhead is marked in (A).
Source data for this figure is available on the online supplementary information page.
Nonetheless, the successful seed sequence-independent activation seemed to be incongruent with the above cleavage sequestration model since it involved a dramatic change to the relevant RNase E site in the cfa1 mRNA (Figure 7A). To address this, we also examined the cfa1 mRNA variants with altered seed sequences in RNase E in vitro cleavage assays (Figure 7C). Both the cfa1RybB and cfa1RyhB mRNA fragments exhibited a prominent RNase E cleavage site but in either case it differed by a few nucleotides from the one in wild-type cfa1 (positions marked in Figure 7A). However, either of these new sites could be repressed by the cognate sRNA, that is, RybB or RyhB, respectively (Figure 7C). It therefore appears that this region in the 5′ UTR of cfa1 is generally sensitive to RNase E cleavage, and that sRNA pairing interferes with such cleavage in a generic manner.
Discussion
The regulatory process of gene activation has traditionally been associated with the activity of proteins such as transcription or σ-factors that guide the RNA polymerase to target promoter sequences. A protein-guided activation mechanism involving σS and transcription from the cfa2 promoter (Figure 3A) increases the synthesis of CFA synthase under numerous stress conditions (Grogan and Cronan, 1997). Judging from the strong sequence conservation of the cfa2 promoter (Supplementary Figure S6) this protein-dependent activation likely occurs in many different enterobacterial species. In the present work, we have discovered that the cfa gene can also be activated by a post-transcriptional mechanism whereby a regulatory RNA stabilizes the second isoform of the cfa mRNA that results from transcription of the constitutive cfa1 promoter (Figure 8). This mechanism has also been maintained during evolution, as suggested by both sequence alignments and our reporter gene assays (Figure 4E; Supplementary Figure S6). Thus, the cfa gene with its two mRNA variants that are activated by two distinct conserved mechanisms offers an experimental model to understand dual regulation at the transcriptional or post-transcriptional levels. Moreover, as discussed below, our investigation of cfa regulation suggests a new mechanism of post-transcriptional activation and the first conserved target of RydC, a highly conserved enterobacterial sRNA.
Figure 8.
RydC interacts with one of the two isoforms of cfa mRNA to increase Cfa. Two independent promoter sites controlled by either σ70 or the alternative σ-factor σS control the transcription of cfa. By base pairing with its conserved 5′ end to the longer of the two mRNA isoforms, RydC increases cfa1 mRNA stability and levels. The increased protein levels lead to an alteration of membrane stability, as Cfa converts the double bond of unsaturated fatty acid side chains into cyclopropane rings.
A generic mechanism to activate gene expression
Although repressed target genes still outnumber activated ones, there has been a steady increase in reported cases of positive regulation among Hfq-associated sRNAs. Typically, sRNA-mediated activation of targets occurs through base pairing with and alteration of a secondary structure in the 5′ UTR to make the target’s RBS more accessible (Fröhlich and Vogel, 2009). By contrast, the RydC-mediated activation of cfa differs from this previously reported mode of activation; we present multiple lines of genetic and biochemical evidence supporting a mechanism that involves minor if any control at the level of translation. Specifically, the cfa RBS is not sequestered by an intrinsic structure (Figure 4A), and is not required for RydC-mediated activation, as evidenced from our cfa1-X::gfp fusion (Figure 5B). Moreover, the cfa mRNA (Figure 5C), as well as the non-coding cfa mini gene (Figure 5D), are readily stabilized by RydC. This mode of activation is reminiscent of the recently described activation of yigL mRNA by the phosphosugar stress induced SgrS sRNA (Papenfort et al, 2013). However, while SgrS acts by protecting an internal RNase E decay intermediate of the pldB-yigL operon mRNA, RydC acts far upstream in the 5′ UTR of its monocistronic cfa target.
Given the overarching role of RNase E in general mRNA turnover and its known interaction with Hfq (Ikeda et al, 2011) it may not be surprising that sRNAs have ‘hijacked’ this pathway to control gene expression at the post-transcriptional level. In fact, negative regulation by sRNAs has long been known to require both Hfq and RNase E (Moll et al, 2003; Morita et al, 2005) for degradation of the target alone or along with an sRNA (Masse et al, 2003; Figueroa-Bossi et al, 2009) but this was nonetheless considered as a secondary event to translational repression (Maki et al, 2008). Intriguingly, recent studies have produced plenty of evidence for direct mRNA destabilization by sRNAs and RNase E, especially in the CDS or intergenic region of mRNAs (Desnoyers et al, 2009; Pfeiffer et al, 2009; Bandyra et al, 2012; Rice et al, 2012). This second aspect of the mechanistic duality in target repression has now been mirrored by SgrS and RydC that activate gene expression independent of translational control in the coding or non-coding sequence of targets, respectively.
We nonetheless caution that mRNA stabilization by inhibition of RNase E-mediated decay may not constitute the sole mechanism of cfa activation by RydC. RydC pulse expression increased the cfa mRNA levels by ∼7-fold (Figure 2B), while cfa stability experiments indicated only a 3-fold increase in mRNA half-life (Figure 5C). A similar change in RNA stability was observed for the cfa mini gene (Figure 5D). Since we can exclude transcriptional control by σS as an auxiliary factor influencing cfa levels (the stability experiments were conducted in ΔrpoS bacteria), counteraction of transcription attenuation may be an additional mechanism of cfa activation by RydC. It is yet unclear whether RydC affects transcription progression and given that we did not observe global changes in cfa structure in our footprinting experiments (Figure 4A), active transcription might be required to recapitulate such mechanism in vitro.
Of note, interaction of Hfq with the transcriptional terminator protein Rho has recently been reported (Rabhi et al, 2011), and a possible Hfq interaction with Rho may in part explain the strict requirement of Hfq for RydC-mediated activation of cfa (Supplementary Figure S8). In addition, a recent report showed that repression of the sdhC mRNA by Spot42 solely depends on the delivery of Hfq to the mRNA (Desnoyers and Masse, 2012). In principle, one may envisage a reciprocal mechanism whereby RydC stabilizes the cfa1 mRNA by depositing Hfq for interference with RNase E. However, Hfq alone does not confer full resistance to RNase E-mediated cleavage at position −100 of the cfa 5′ UTR (Figure 6C). Likewise, activation can also be achieved with a cfa reporter in which the sRNA site is changed to be recognized by the Hfq-independent sRNA IstR (Supplementary Figure S10). However, the ∼2-fold upregulation achieved by IstR is not as strong as the activations observed with the Hfq-binding sRNAs (Figure 7). This leads us to predict that for full regulation, several factors are at play in the RydC-mediated activation of Cfa synthesis. Concerning other mechanistic scenarios observed with Hfq-independent sRNAs, target activation has been reported for the VR-RNA and FasX sRNAs of the Gram-positive species Clostridium perfringens and Streptococcus pyogenes, respectively. Binding of VR-RNA to the 5′ UTR of the colA transcript induces a conformational change, which opens the RBS and facilitates cleavage of the mRNA into a truncated and more stable isoform (Obana et al, 2010). FasX sRNA binds to the extreme 5′ end of the ska mRNA where it prevents transcript decay leading to higher protein expression (Ramirez-Pena et al, 2010). This would be similar to our model whereby RydC blocks cleavage of the cfa mRNA by RNase E to increase the mRNA half-life and upregulate Cfa production. However, the S. pyogenes RNase whose activity is counteracted by FasX for mRNA stabilization is unknown.
Our demonstration that the RydC site within the cfa 5′ UTR can be replaced by the fully unrelated sequences of RyhB and RybB binding sites (Figure 7) suggests a generic mechanism of target activation. Approximately 60% of all transcripts matching annotated genes display increased stability in the absence of RNase E which includes the cfa mRNA (Stead et al, 2011). Given that RNase E cleavage sites can now be globally determined using RNA-seq analysis, our proposed mechanism of post-transcriptional activation through interference with RNase E cleavage might lend itself for the design of synthetic regulatory RNAs. These artificial regulators would be designed to recognize genuine RNase E target sites within 5′ UTRs or upstream genes within polycistronic mRNAs increasing mRNA stability by inhibiting RNase E cleavage and progression at these specific sites.
Physiological consequences of Cfa regulation
Bacteria actively modify their membranes to adapt to complex stresses such as changes in osmolarity, temperature or pH. It is the lipid matrix, which is mainly built of glycerolipids containing two fatty acyl side chains, that determines the properties of the membrane. Although membrane composition can be adjusted through the regulated de novo synthesis of fatty acids, certain environmental changes require the post-synthetic modification of existing phospholipids (Zhang and Rock, 2008). The Cfa protein plays a crucial role in these modifications; using S-adenosylmethionine (SAM) as a methyl donor it converts the UFA moiety of a phospholipid into the corresponding CFA (Grogan and Cronan, 1997). This cyclopropanation only mildly affects the biophysical properties of the membrane, but protects the acyl chain from oxidation or chemical destruction (Grogan and Cronan, 1997), rendering the membrane less reactive and cells more resistant to heat or acidic stress (Chang and Cronan, 1999).
Although CFA overproduction is tolerated (Grogan and Cronan, 1984), it is energetically costly and irreversible (Grogan and Cronan, 1997), and unsurprisingly bacteria tightly restrict the expression of Cfa synthase according to their needs. In E. coli, Salmonella and several related enterobacteria, transcription of the cfa mRNA is driven from two independent promoters (Wang and Cronan, 1994; Kim et al, 2005). The proximal promoter is highly dependent on σS, ensuring cfa expression during stationary phase and under various stress conditions including heat shock, osmotic stress, acidic pH and the stringent response upon amino-acid starvation (Grogan and Cronan, 1997). By contrast, the distal promoter element, located >200 bp upstream of the start codon, is transcribed constitutively by the housekeeping σ70 version of RNA polymerase (Cronan, 2002). Nonetheless, although active transcription occurs at all phases of growth, Cfa protein levels remain low (Kim et al, 2005), and even in stationary phase there is no linear correlation between the increase in σS and Cfa protein levels (Figure 2C). RydC, at least when overexpressed, is the first factor that achieves higher levels of Cfa production than σS.
What may be the physiological meaning of the additional control by RydC? σS is a very general transcription factor that (in E. coli) controls >10% of all genes and that only attains full activity in stationary phase or other stress conditions (Weber et al, 2005; Maciag et al, 2011). Thus, we speculate that the post-transcriptional activation by RydC provides a more selective route for Cfa synthesis, enabling remodelling of phospholipids without the global effects on gene expression by σS. We conceded that a full understanding of the physiological importance of the post-transcriptional activation of cfa will only be achieved through the identification of the stimulus that triggers RydC expression. Both the low copy number of RydC itself and its little variation during growth in standard media argue that we are yet to identify a physiological condition that induces RydC for full function. Thus far, a screening of 22 different growth conditions has not yet yielded a single one in which RydC would be strongly and/or selectively induced (JCD Hinton, personal communication).
Similarly elusive has been a transcription factor for the rydC gene. We interpret the strong conservation of nucleotides outside the classic −10 and −35 elements of the rydC promoter (Supplementary Figure S11) as evidence for a conserved interaction site of a DNA-binding protein, for example, a transcriptional activator. However, several genetic screens in Salmonella and E. coli using genome-wide transposon or multicopy plasmids libraries have thus far failed to identify such a regulator (Fröhlich, 2013). These open questions notwithstanding, the duality of transcriptional (through σS) and post-transcriptional (RydC) activation of cfa expression is likely to be conserved in many bacteria.
The bacterial envelope acts as both protective barrier and interaction site with the environment. While it has long been known that sRNAs modulate general membrane permeability by targeting the mRNAs of outer membrane porins, more recent studies have also discovered sRNA-mediated regulations of enzymes that modify the lipopolysaccharides decorating the outside of the envelope (Moon and Gottesman, 2009; Corcoran et al, 2012; Moon et al, 2013). The positive regulation by RydC identified here offers a unique opportunity to understand how RNA-based regulation is utilized to regulate another important trait of the bacterial envelope, that is, the stability of its membranes.
Materials and methods
DNA/RNA oligonucleotides and plasmids
Sequences of all oligonucleotides employed in this study are listed in Supplementary Table S2. All plasmids used in this study are summarized in Supplementary Table S3.
Plasmid pBAD-RydC (pKF41-2) was constructed as described previously (Papenfort et al, 2006), but using primers JVO-4532/JVO-0376 to amplify rydC from its transcriptional start site to 111 bp downstream of the terminator T-stretch from gDNA. The same insert was used to construct pPLRydC (pKF42-1) following the cloning strategy described in Urban and Vogel (2007). Plasmids expressing E. coli, K. pneumoniae and E. aerogenes RydC under the control of the PL promoter were cloned accordingly using inserts amplified from gDNA with JVO-10125/JVO-10126 (pKF203), JVO-10130/JVO-10131 (pKF204) and JVO-10134/JVO-10135 (pKF205), respectively. For plasmids expressing variants of Salmonella RydC from the PL promoter, pKF42-1 served as a template for PCR amplification with primer pairs JVO-4536/4537 (pPLRydC-K1; pKF60-1), JVO-5081/JVO-5082 (pPLRydC-K2; pKF61-1), and JVO-7035/PLLacOC (pPLRydC*; pKF86-1), and self-ligation was carried out as in Sharma et al (2011). Similarly, pPLRydC-K1/2 (pKF62-1) and pPLRydC-TMA (pKF38) were constructed by self-ligation of PCR products of JVO-4536/4537 on pKF61-1 and JVO-4420/PLLacOC on pFS135, respectively. Translational GFP fusions of cfa mRNA variants were constructed as described in Urban and Vogel (2007) using PCR products amplified from gDNA with primer pairs JVO-4055/JVO-4057 (cfa2::gfp; pKF30-1), JVO-4056/JVO-4057 (pKF31-1), JVO-10123/JVO-10124 (pKF206), JVO-10127/JVO-10129 (pKF208) and JVO-10132/10133 (pKF210). For the construction of cfa1::gfp variants, plasmid pKF31-1 was used as a template in PCR amplification with primer sets JVO-7033/JVO-7034 (cfa1*::gfp; pKF83-1), JVO-9401/JVO-9402 (cfa1RybB::gfp; pKF153-1), JVO-9553/JVO-9402 (cfa1RyhB::gfp; pKF165-1) or JVO-10174/JVO-9402 (cfa1IstR::gfp; pKF201), and obtained fragments were self-ligated. To obtain pKF133-1 (cfa1-X::gfp), a PCR fragment amplified from pKF31 using oligos pZE-Cat/JVO-8689 was digested with NsiI, and ligated into pKP60-1 via the same restriction site. The cfa mini fragment was amplified from pKF31 (JVO-4055/JVO-7032), digested with NsiI/XbaI, and ligated into correspondingly digested pXG10 to obtain plasmid pKF151 or pKF200-1 to obtain pKF215, respectively. Plasmid pKF200-1 is a variant of the pXG10-SF plasmid (Corcoran et al, 2012) in which the chloramphenicol resistance cassette is replaced by a kanamycin resistance cassette amplified from pJV-960 via JVO-10136/JVO-10137, digested with AatII/KpnI and ligated into a backbone obtained by PCR (pZECat/JVO-10138 on pXG10-SF) that was restricted with the same enzymes. Competent E. coli TOP10 was used for all cloning purposes.
Bacterial strains
A complete list of bacterial strains employed in this study is provided in Supplementary Table S4. Salmonella enterica serovar Typhimurium strain SL1344 (JVS-0007) is referred to as wild-type strain and was used for mutant construction. Single mutant derivatives were constructed by the λRed recombinase one-step inactivation method using pKD4 as the template. To eliminate the KanR cassette of λRed-derived mutants, cells were transformed with the FLP recombinase expression plasmid pCP20 (Datsenko and Wanner, 2000). Phage P22 transduction (using standard protocols) was employed to transfer chromosomal modifications to a fresh Salmonella wild-type background as well as to obtain strains carrying multiple mutations.
Briefly, to obtain JVS-8435 (SL1344 Δcfa), Salmonella cells carrying the pKD46 helper plasmid were transformed with a DNA fragment amplified from pKD4 using JVO-5773/JVO-4828. Similarly, the Δhfq Δrnc double mutant (JVS-10500) was constructed by transformation of JVS-0584 (+pKD46) with a PCR product amplified from pKD4 using JVO-0737/JVO-0738. A modified λRed approach based on Uzzau et al (2001) using primers JVO-4186/JVO-4187 and pSUB11 as a template was employed to construct a strain expressing Cfa with a C-terminal 3xFLAG-tag (JVS-4690). Chromosomal integration of the constitutive PLtet-O1 promoter marked with a DHFR resistance cassette upstream of the Salmonella cfa gene (JVS-9908/JVS-9909/JVS-10606/JVS-10607) was obtained by λRed-mediated recombination using a DNA fragment generated by PCR amplification (JVO-9009/JVO-9010) using strain JVS-9711 as a template. For JVS-9711, a fragment comprising the chloramphenicol resistance cassette, the Ptet promoter and the cfa1::gfp fusion was amplified from pKF31-1 using JVO-8739/JVO-8740, and integrated into the Salmonella put locus. To replace the cat gene, the strain was transformed in the presence of pKD46 with a linear fragment comprising a DHFR cassette plus flanking sites amplified from an EZ-Tn5 transposon (Epicentre) using JVO-5796/JVO-5797. The resulting mutants were selected in the presence of trimethoprim, screened for chloramphenicol sensitivity, and the modification was transferred to JVS-9675 by P22 transduction (to obtain JVS-9711).
Bacterial growth conditions
Cells were grown aerobically in Luria Broth (LB) medium at 37°C unless stated otherwise. A final concentration of 0.2% L-arabinose was added to cultures to induce expression from pBAD-derived plasmids. Where appropriate, liquid and solid media were supplemented with antibiotics at the following concentrations: 100 μg/ml ampicillin, 50 μg/ml kanamycin, 20 μg/ml chloramphenicol and 5 μg/ml trimethoprim.
Protein sample analysis
To prepare whole-cell protein samples, bacteria were collected by centrifugation (16 000 r.c.f.; 2 min; 4°C), and pellets were resuspended in 1 × protein loading buffer (Fermentas) to a final concentration of 0.01 OD/μl. To visualize total protein expression patterns by 12% SDS–PAGE, 0.1 OD was loaded per lane and gels were stained overnight with Coomassie Blue. To analyse protein levels by western blot, 0.01 OD (detection of OmpX-GFP fusion proteins) or 0.1 OD (all other samples) was loaded per lane and resolved by SDS–PAGE, after which proteins were transferred onto PVDF membranes as described in Sittka et al (2007). GFP fusion proteins and RpoS were detected using commercially available antibodies (GFP: 1:5000; mouse; Roche and RpoS: 1:1000; mouse; Neoclone), respectively. Anti-mouse or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (1:10 000; GE Healthcare) were used in all cases. Signals were visualized using the Western Lightning reagent (Perkin-Elmer) and an ImageQuant LAS 4000 CCD camera (GE Healthcare).
Analysis of cellular fatty acids
Fatty acids were analyzed by LC/MS; method reliability was verified a GC/MS-based protocol described earlier (Kim et al, 2005).
Cells were cultured (in triplicates) at 37°C to an OD600 of 0.5 in M9 minimal medium (1 × M9 salts, 2 mM MgSO4, 0.1 mM CaCl2, 0.4% (v/v) glycerol, supplemented with thiamine (0.5 μg/ml), L-histidine (40 μg/ml) and cas amino acids (0.2%)). Bacteria were harvested by centrifugation and snap frozen in liquid N2. Pellets were resuspended in 10% (w/v) potassium hydroxide in isopropanol, and lysed in the presence of glass beads (0.1 mm diameter) in a Retsch Mixer Mill MM 400 for 10′ at 30 Hz. Samples were hydrolysed for 1 h at 60°C, and adjacently acidified to pH 3–4 with formic acid.
Fatty acid profiling was performed on an ultra performance liquid chromatography system (UPLC) coupled to a Synapt G2 HDMS time-of-flight mass spectrometer (MS) equipped with an electrospray ion source (ESI; all Waters, Germany). Chromatographic separation was carried out on a Waters BEH C18 column (2.1 × 100 mm ID, 1.7 μm particle size) with gradient elution over 5 min from 70 to 100% acetonitrile (ACN) acidified with 0.1% formic acid, thereafter rinsed with 100% acidified ACN for 0.5 min and equilibrated with 75% acidified ACN for 2.5 min The mobile phase (flow rate: 0.3 ml/min) was directly introduced into the MS via the ESI source operated at a negative mode. The capillary voltage was set to 0.8 kV and nitrogen (at 350°C; flow rate: 800 l/h) was used as desolvation gas. The system was equipped with an integral LockSpray unit with its own reference sprayer for external calibration during the measurements; leucine encephalin (m/z of 554.2615) was used as the internal reference. The quadrupole was operated in a wide band RF mode, and data were acquired over the mass range of 50−1200 Da. Processing of chromatograms and peak detection were performed using the MassLynx software (version 4.1; Waters). The fatty acids were identified by their retention times (RT)±0.02 min in the extracted ion chromatogram (XIC) with a mass window of 0.03 Da. The RT of hexadecenoic acid (C16 UFA; m/z of 253.217), methylenehexadecanoate (C17 CFA; m/z of 267.233), octadecenoic acid (C18 UFA; m/z of 281.248) and methyleneoctadecanoate (C19 CFA; m/z of 295.264) were 4.45, 5.21, 5.56 and 6.28 min, respectively. Samples were spiked with internal standards (10 μg of linoleic acid prior to alkaline hydrolysis and 10 μg of linolenic acid prior to sample injection). The method was validated by determining the performance characteristic of repeatability (inter-day RSD<8%) and linearity of peak area in function of concentration (regression coefficient>0.991).
Microarray
A detailed protocol of the microarray experiment has been published previously (Sharma et al, 2011). Salmonella strain JVS-0291 carrying either control plasmid pKP8-35 or pKF42-1 was grown in LB to OD600 of 1.5 when expression from the pBAD promoter was induced by addition of arabinose. Samples collected (in duplicates) prior to and 15 min post arabinose treatment were used in the microarray experiment. Microarray data have been deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession code GSE48221).
In vitro RNA synthesis and RydC in vivo copy number
For RNA in vitro synthesis, DNA templates were amplified by PCR from Salmonella gDNA (RydC: JVO-4721/JVO-4722; RybB: JVO-1242/JVO-1243; RyhB: JVO-10439/JVO-10440; cfa(TSS1-CDS+70): JVO-4558/JVO-6516 cfa: JVO-4558/JVO-9044) or plasmids pKF153 (cfaRybB: JVO-4558/JVO-9044) and pKF165 (cfaRyhB: JVO-4558/JVO-9044), respectively. Template DNA (∼200 ng) was reverse transcribed employing the MEGAscript kit (Ambion) according to the manufacturer’s instructions. Bodylabelled cfa, cfaRybB and cfaRyhB transcripts were synthesized in the presence of [32P]-α-UTP. Size and integrity of the RNA were confirmed on a denaturing polyacrylamide gel. To estimate the in vivo abundance of RydC (copy number per cell), total RNA corresponding to 1 OD of wild-type Salmonella was compared to serial dilutions of the RydC in vitro transcript (0.05/0.1/0.5/1.0 and 2.5 ng) by northern blot analysis and hybridization with a RydC-specific riboprobe. Calculations of RNA levels per cell were based on the determination of viable cell counts per OD600 in Sittka et al (2007).
RNA half-life
To monitor RNA half-life of RydC mutant variants, cells were grown to an OD600 of 1, and treated with rifampicin (final concentration: 500 μg/ml) to abrogate transcription. RNA samples were withdrawn at indicated time points, and RNA decay was determined by northern blot analysis. To analyse the half-life of cfa mRNA and cfa mini RNA, cells were cultivated to an OD600 of 1 and RydC expression was induced by the addition of L-arabinose for 15′. Cells were treated with rifampicin, and RNA samples withdrawn at indicated time points were analysed by quantitative real-time PCR as described previously (Papenfort et al, 2006, 2012) using oligo sets JVO-1472/JVO-1473 (cfa mRNA), JVO-9484/JVO-9485 (cfa mini RNA), JVO-4636/JVO-4637 (gfp mRNA) and JVO-1977/JVO1978 (rrsA; standard).
In vitro structure probing
Structure probing and mapping of RNA/Hfq footprints was conducted on in vitro synthesized and 5′ end-labelled mRNA generated as described before (Papenfort et al, 2006). Upon denaturation at 70°C for 2 min, RNA was chilled on ice. Next, 0.2 pmol 5′ end-labelled mRNA was mixed with E. coli Hfq protein (0.2 pmol; kindly provided by K Bandyra and BF Luisi, University of Cambridge) or Hfq dilution buffer (1X structure buffer, 1% (v/v) glycerol, 0.1% (v/v) Triton X-100) in the presence of 1X structure buffer (0.01 M Tris pH 7, 0.1 M KCl, 0.01 M MgCl2) and 1 μg yeast RNA, and samples were incubated at 37°C for 10 min. Subsequently, unlabelled sRNA (2 pmol) or water was added, and reactions were kept at 37°C for an additional 10 min. For digestion, samples were treated with RNase T1 (0.1 U; Ambion, #AM2283) for 2.5 min or with lead(II) acetate (final concentration: 5 mM; Fluka #15319) for 1.5 min, respectively. Reactions were stopped by addition of 2 vol. equiv. of Precipitation/Inactivation buffer (Ambion), and precipitated at −20°C for 1 h. Pellets were washed with 70% ethanol, and resuspended in loading buffer GLII (95% formamide, 18 mM EDTA, 0.025% (w/v) SDS, 0.025% (w/v) xylene cyanol, 0.025% (w/v) bromophenol blue). To prepare RNase T1 sequencing ladders, 0.4 pmol 5′ end-labelled mRNA was denatured (95°C, 2 min) in the presence of 1X sequencing buffer (Ambion) and chilled on ice. RNase T1 was added (0.1 U), and RNA was digested for 5′ at 37°C. Alkaline (OH) sequencing ladders were prepared by incubating 0.4 pmol 5′ end-labelled mRNA at 95°C for 5 min in the presence of alkaline hydrolysis buffer (Ambion). Reactions were stopped by addition of 1 vol. equiv. of GLII. Samples were denatured prior to loading (95°C, 2 min) and separated by denaturing PAGE on 6% PAA/7M urea sequencing gels at constant power of 40 W. Gels were dried and signals were analysed on a Typhoon FLA 7000 phosphorimager and using the AIDA software (Raytest, Germany).
In vitro RNase E assays
Bodylabelled cfa, cfaRybB and cfaRyhB RNAs were denatured at 70°C for 2 min and adjacently incubated on ice for 3 min, and at room temperature for additional 3 min. In all, 2 pmol RNA was mixed with RNase E reaction buffer (25 mM Tris–HCl pH 7.5, 50 mM NaCl, 50 mM KCl, 10 mM MgCl2, 1 mM DDT), and either Hfq (2 pmol) or Hfq dilution buffer was added to the samples. Upon incubation at 37°C for 10 min, sRNA (20 pmol) or water was added, and reactions were kept at 37°C for additional 10 min. Next, 5 pmol purified RNase E NTD (N-terminal domain; kindly provided by K Bandyra and BF Luisi, University of Cambridge) was added. Samples were withdrawn prior to and at indicated time points after RNase E addition, and reactions were stopped with 2.5 vol. equiv. of Precipitation/Inactivation buffer and 0.5 vol. equiv. of 100 mM EDTA. Upon precipitation at −20°C for 1 h, samples were collected by centrifugation (30 min; 16 000 r.c.f.; 4°C), washed with 70% ethanol, and pellets were resuspended in GLII. Sample processing and analysis was done as described for RNA in vitro structure probing experiments.
RNA isolation and northern blot analysis
Total bacterial RNA was isolated from culture aliquots using the TRIzol reagent (Invitrogen) and analysed by northern blot as previously described (Urban and Vogel, 2007; Fröhlich et al, 2012). RydC, RybB, RyhB, the TMA chimaera, cfa1 mRNA and 5S rRNA were detected by the 5′ end-labelled oligonucleotides JVO-4363, JVO-1205, JVO-9609, JVO-0396, JVO-3707 and JVO-0322, respectively. The riboprobes to detect RydC and IstR-1 were synthesized by T7-mediated in vitro transcription of ∼200 ng of template DNA (amplified on Salmonella gDNA with JVO-4532/JVO-4378 [RydC] or on pJV3H-22 with JVO-10424/JVO-10425 [IstR-1]) in the presence of [32P]-α-UTP with the MAXIscript kit (Ambion).
Primer extension
For primer extension, 10 μg of RNA samples (in 8 μl H2O) prepared by the Hot Phenol method was denaturated in the presence of 1 pmol 5′ end-labelled primer (JVO-5769 for cfa1 mRNA; JVO-7022 for cfa2 mRNA) at 70°C for 2 min and adjacently chilled on ice for 5 min. Next, 5 μl of reaction mix (3 μl 5X first strand buffer, 5 mM DTT, 0.5 mM each dATP, dGTP, dCTP and dTTP) was mixed with the samples at 42°C, and 1 μl SuperScript III (100 U; diluted 1:1 in H2O) was added. cDNA synthesis was performed at 50°C for 60 min, followed by incubation at 70°C for 15 min to inactivate the enzyme. Samples were RNase H treated (1 μl; 2.5 U) for 15 min at 37°C and the reaction was stopped by the addition of 10 μl GLII loading buffer. In all, 12 μl of the samples was separated electrophoretically together with template-specific ladder (prepared using the SequiTherm EXCELII DNA Sequencing Kit; template amplification JVO-7030/JVO-7022 on Salmonella gDNA) on 6–8% sequencing gels.
5′ RACE
RACE to determine RNA 5′ ends was performed as in Argaman et al (2001) with modifications. In all, 5 μg of DNA-free RNA (in 5 μl H2O) was mixed with 5 μl of 10X TAP buffer and 0.25 μl of SUPERaseIn RNase inhibitor. Reactions were split into two, and either supplemented with 2.5 U tobacco acid pyrophosphatase (TAP) or H2O (negative control) before being incubated at 37°C for 30 min. In all, 150 pmol of RNA-linker A4 was added to both reactions prior to P:C:I extraction and precipitation with 3 vol. equiv. of 30:1 ethanol:sodium acetate (pH 6.5) mix (−20°C, 3 h). The RNA pellet was dissolved in 13.5 μl H2O, denatured for 5 min at 95°C and chilled on ice for 5 min. The RNA-linker ligation was performed o/n at 16°C in the presence of 10 U T4 RNA ligase, 1X RNA ligase buffer, 10% (v/v) DMSO and 10 U SUPERaseIn RNase Inhibitor. Following P:C:I extraction and precipitation with 3 vol. equiv. of 30:1 ethanol:sodium acetate (pH 6.5) mix (−20°C, 3 h), 2 μg linker-ligated RNA was denatured at 65°C for 5 min and adjacently converted to cDNA using 100 pmol random hexamer primers and 200 U Superscript III reverse transcriptase according to the manufacturer’s instructions. Prior to enzyme addition, samples were incubated at 25°C for 10 min. Reverse transcription was carried out in a series of incubation steps (41°C for 15 min; 50°C for 15 min; 55°C for 15 min; 60°C for 15 min). The enzyme was inactivated at 85°C for 5 min and RNA was digested in the presence of 1 U RNase H at 37°C for 20 min. 5′ fragments of RNAs were amplified with Taq polymerase by PCR from the cDNA templates (1 μl in 50 μl reactions) using a gene-specific primer (JVO-7022) in combination with JVO-0367 (antisense to the RNA linker) in the presence of [32P]-α-dCTP. Cycling conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 40 s, 56°C for 40 s, 72°C for 40 s; and 72°C for 8 min). The PCR products were separated on 6% native PAA gels in 0.5x TBE. Selected bands were purified and subcloned using the TOPO TA Cloning Kit as recommended by the manufacturer. Inserts of obtained clones were amplified by PCR (M13fwd/M13rev) and analysed by sequencing.
Sequence retrieval and bioinformatic predictions
Information for sequence alignments was collected using BlastN searches (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi) of the following genome sequences (accession numbers are given in parentheses): Citrobacter koseri ATCC BAA-895 (NC_009792), Escherichia coli K12 substr. MG1655 (NC_000913), Enterobacter aerogenes KCTC 2190 (NC_015663), Escherichia fergusonii ATCC 35469 (NC_011740), Klebsiella pneumoniae 342 (NC_011283), Salmonella bongori NCTC 12419 (NC_015761), Salmonella Typhimurium LT2 (NC_003197) and Shigella flexneri 2a str 301 (NC_004337). Alignments were calculated using MultAlin (Corpet, 1988); http://multalin.toulouse.inra.fr/multalin/multalin.html). Potential interactions between two RNA molecules were predicted using RNAhybrid (Rehmsmeier et al, 2004). Pseudoknot formation was predicted using pknotsRG (Reeder et al, 2007).
Supplementary Material
Acknowledgments
We thank members of the Vogel lab for comments on the manuscript; B Plaschke, T Achmedov and M Lesch for excellent technical assistance; BF Luisi for the gift of RNase E and Hfq preparations; SA Gorski for help with the manuscript; HJ Mollenkopf for help with microarray experiments. The lipid analysis was performed in cooperation with the Metabolomics Core Facility at Würzburg University. KP is supported by a postdoctoral fellowship of the Human Frontiers in Science Program. The Vogel laboratory received funds from DFG Priority Program SPP1258 (Vo875/3-2) and the Bavarian BioSysNet program.
Author contributions: KSF, KP and JV designed the study; KSF and AF performed the experiments; KSF, KP and AF analysed the data; KSF, KP and JV wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR (2005) Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res 33: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal M, Bordeau V, Douchin V, Felden B (2005) A small bacterial RNA regulates a putative ABC transporter. J Biol Chem 280: 7901–7908 [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Arnold TE, Yu J, Belasco JG (1998) mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA 4: 319–330 [PMC free article] [PubMed] [Google Scholar]
- Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, Luisi BF (2012) The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 47: 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco JG (2010) All things must pass: contrasts and commonalities in eukaryotic and bacterial mRNA decay. Nat Rev Mol Cell Biol 11: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing BA, Meyer BJ, Dunny GM (1996) Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci USA 93: 7794–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P, Belasco JG (1992) Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature 360: 488–491 [DOI] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J (2008) Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32: 827–837 [DOI] [PubMed] [Google Scholar]
- Braun F, Le Derout J, Regnier P (1998) Ribosomes inhibit an RNase E cleavage which induces the decay of the rpsO mRNA of Escherichia coli. EMBO J 17: 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Pennell S, Gilbert RJ (2007) Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 5: 598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpousis AJ, Luisi BF, McDowall KJ (2009) Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci 85: 91–135 [DOI] [PubMed] [Google Scholar]
- Chang YY Jr, Cronan JE (1999) Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol Microbiol 33: 249–259 [DOI] [PubMed] [Google Scholar]
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J (2012) An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31: 4005–4019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JC, Vogel J (2012) Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84: 428–445 [DOI] [PubMed] [Google Scholar]
- Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16: 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE Jr (2002) Phospholipid modifications in bacteria. Curr Opin Microbiol 5: 202–205 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S (2013) Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 288: 7996–8003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Masse E (2012) Noncanonical repression of translation initiation through small RNA recruitment of the RNA chaperone Hfq. Genes Dev 26: 726–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Morissette A, Prevost K, Masse E (2009) Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J 28: 1551–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L (2009) Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23: 2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS (2013) Assigning functions to Hfq-dependent small RNAs in the model pathogen Salmonella Typhimurium. Doctoral Thesis. Julius-Maximilians-University Würzburg, Würzburg
- Fröhlich KS, Papenfort K, Berger AA, Vogel J (2012) A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res 40: 3623–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich KS, Vogel J (2009) Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682 [DOI] [PubMed] [Google Scholar]
- Grogan DW, Cronan JE Jr (1984) Cloning and manipulation of the Escherichia coli cyclopropane fatty acid synthase gene: physiological aspects of enzyme overproduction. J Bacteriol 158: 286–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan DW, Cronan JE Jr (1997) Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev 61: 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S (2008) The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res 36: 6781–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Unoson C, Reimegard J, Wagner EG (2012) A mixed double negative feedback loop between the sRNA MicF and the global regulator Lrp. Mol Microbiol 84: 414–427 [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Yagi M, Morita T, Aiba H (2011) Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol Microbiol 79: 419–432 [DOI] [PubMed] [Google Scholar]
- Jorgensen MG, Thomason MK, Havelund J, Valentin-Hansen P, Storz G (2013) Dual function of the McaS small RNA in controlling biofilm formation. Genes Dev 27: 1132–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Kim S, Kim HG, Lee J, Lee IS, Park YK (2005) The formation of cyclopropane fatty acids in Salmonella enterica serovar Typhimurium. Microbiology 151: 209–218 [DOI] [PubMed] [Google Scholar]
- Kröger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J et al. (2012) The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA 109: E1277–E1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioliou E, Romilly C, Romby P, Fechter P (2010) RNA-mediated regulation in bacteria: from natural to artificial systems. Nat Biotechnol 27: 222–235 [DOI] [PubMed] [Google Scholar]
- Lodato PB, Hsieh PK, Belasco JG, Kaper JB (2012) The ribosome binding site of a mini-ORF protects a T3SS mRNA from degradation by RNase E. Mol Microbiol 86: 1167–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag A, Peano C, Pietrelli A, Egli T, De Bellis G, Landini P (2011) In vitro transcription profiling of the sigmaS subunit of bacterial RNA polymerase: re-definition of the sigmaS regulon and identification of sigmaS-specific promoter sequence elements. Nucleic Acids Res 39: 5338–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GA (2013a) Determinants in the rpsT mRNAs recognized by the 5′-sensor domain of RNase E. Mol Microbiol 89: 388–402 [DOI] [PubMed] [Google Scholar]
- Mackie GA (2013b) RNase E: at the interface of bacterial RNA processing and decay. Nat Rev Microbiol 11: 45–57 [DOI] [PubMed] [Google Scholar]
- Maki K, Uno K, Morita T, Aiba H (2008) RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc Natl Acad Sci USA 105: 10332–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE, Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Vanderpool CK, Gottesman S (2005) Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U (2003) Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 9: 1308–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S (2009) A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74: 1314–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Six DA, Lee HJ, Raetz CR, Gottesman S (2013) Complex transcriptional and post-transcriptional regulation of an enzyme for Lipopolysaccharide modification. Mol Microbiol 89: 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Aiba H (2011) RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev 25: 294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H (2005) RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy K, Jakab A, Fekete J, Vekey K (2004) An HPLC-MS approach for analysis of very long chain fatty acids and other apolar compounds on octadecyl-silica phase using partly miscible solvents. Anal Chem 76: 1935–1941 [DOI] [PubMed] [Google Scholar]
- Obana N, Shirahama Y, Abe K, Nakamura K (2010) Stabilization of Clostridium perfringens collagenase mRNA by VR-RNA-dependent cleavage in 5′ leader sequence. Mol Microbiol 77: 1416–1428 [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Fozo EM, Hemm MR, Storz G (2011) RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol 406: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G (2004) GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186: 6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P (2009) Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73: 790–800 [DOI] [PubMed] [Google Scholar]
- Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J (2010) Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci USA 107: 20435–20440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J (2006) SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62: 1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Podkaminski D, Hinton JC, Vogel J (2012) The ancestral SgrS RNA discriminates horizontally acquired Salmonella mRNAs through a single G-U wobble pair. Proc Natl Acad Sci USA 109: E757–E764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J (2013) Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153: 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282 [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J (2009) Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol 16: 840–846 [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J (2007) A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66: 1174–1191 [DOI] [PubMed] [Google Scholar]
- Prevost K, Desnoyers G, Jacques JF, Lavoie F, Masse E (2011) Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25: 385–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi M, Espeli O, Schwartz A, Cayrol B, Rahmouni AR, Arluison V, Boudvillain M (2011) The Sm-like RNA chaperone Hfq mediates transcription antitermination at Rho-dependent terminators. EMBO J 30: 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Pena E, Trevino J, Liu Z, Perez N, Sumby P (2010) The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 78: 1332–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Steffen P, Giegerich R (2007) pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res 35: W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB, Balasubramanian D, Vanderpool CK (2012) Small RNA binding-site multiplicity involved in translational regulation of a polycistronic mRNA. Proc Natl Acad Sci USA 109: E2691–E2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Feng L, Rutherford ST, Papenfort K, Bassler BL (2013) Functional determinants of the quorum-sensing non-coding RNAs and their roles in target regulation. Embo J 32: 2158–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J (2011) Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81: 1144–1165 [DOI] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J (2007) The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63: 193–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA (2010) Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA 107: 9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stazic D, Lindell D, Steglich C (2011) Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res 39: 4890–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead MB, Marshburn S, Mohanty BK, Mitra J, Pena Castillo L, Ray D, van Bakel H, Hughes TR, Kushner SR (2011) Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res 39: 3188–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43: 880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35: 1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L (2001) Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci USA 98: 15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AY, Cronan JE Jr (1994) The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol Microbiol 11: 1009–1017 [DOI] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R (2005) Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin AV, Baker CS, Vakulskas CA, Yakhnin H, Berezin I, Romeo T, Babitzke P (2013) CsrA activates flhDC expression by protecting flhDC mRNA from RNase E-mediated cleavage. Mol Microbiol 87: 851–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S (2013) Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425: 3678–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO (2008) Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6: 222–233 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.