Abstract
NCB (2013) 15: 1141–1152 ; DOI: 10.1038/ncb2839; published online September 08 2013
mRNA-splicing presents an important regulatory step that contributes to the functional repertoire of all cell types and tissues. Profound changes in alternative splicing have also been revealed during embryonic stem cell (ESC) differentiation. Based on a genome-wide screen for pluripotency regulators, Ng and colleagues now identify SON as a new splicing regulator in the maintenance of human ESC pluripotency and self-renewal (Lu et al, 2013).
RNA splicing is the process of removal of introns from nascent pre-mRNAs that results in the formation of mature mRNAs, with all exons joined together.
The human genome contains ∼40 000 genes that give rise to ∼200 000 transcripts, suggesting that each gene locus can generate several different transcripts. This phenomenon can be in part explained by alternative splicing. Alternative splicing occurs when an exon can be either included or excluded from the mature mRNA, giving rise to different transcripts called splice variants. Transcriptome analysis suggests that the vast majority of human multiexonic genes are alternatively spliced (Pan et al, 2008), and this occurs in a cell-type-specific manner. For these reasons splicing should not be considered a housekeeping cellular function, but rather a dynamically regulated process. Interestingly, little is known about the molecular mechanisms controlling it. The recent study on SON therefore provides one of the first examples of a protein that regulates alternative splicing in the context of ESC pluripotency.
The Ng lab at the Genome Institute of Singapore previously performed a genome-wide RNAi screen to identify genes required for self-renewal of human embryonic stem (hES) cells (Chia et al, 2010), and found among the top candidates the spliceosome-associated factor SON. SON is a nuclear protein that has been shown to interact with the spliceosome and to regulate splicing in HeLa cells (Sharma et al, 2010; Ahn et al, 2011).
First, Lu et al (2013) characterized the role of SON as a regulator of Pluripotency in hES cells and found that knockdown of SON leads to impaired self-renewal and downregulation of pluripotency markers such as Oct4, Nanog and Prdm14.
The reader might find those results not surprising, because a cell with a faulty splicing machinery should not be able to perform any cellular function—let alone maintaining pluripotency—but this would only be a rather superficial perspective:
Through a series of elegant experiments the authors showed that loss of SON impairs both self-renewal and cell survival, but the two processes are uncoupled. For instance, knockdown of SON in the presence of the ROCK inhibitor, a chemical commonly used to reduce cell death in hES cells, results in differentiation with negligible effects on survival.
The authors then showed that SON is expressed specifically in pluripotent cells and becomes downregulated upon differentiation. Moreover, when SON is reduced only ∼10% of hES cell-specific transcripts show aberrant splicing and, quite remarkably, among the same gene locus only a few introns are affected. In other words, SON is a very specific regulator of splicing in hES cells. But how is such exquisite specificity achieved?
To address this question, the authors deployed their powerful genomic technologies and found that the introns regulated by SON tend to be shorter, with a weaker splicing strength and more GC-rich than those not affected by SON depletion. These results indicate that the activity of SON is regulated by specific sequences, but the precise molecular mechanism call for further mechanistic investigation.
Loss of SON results mainly in intron inclusion but the authors investigated also the involvement of SON in alternative splicing. Notably, they observed that the alternative splicing of ∼700 genes was affected by SON knockdown, mainly by exon-skipping resulting in shorter splice variants.
Obvious questions resulting from these findings are ‘Is alternative splicing relevant for the biological activity of the regulated genes?’. In other words ‘Are different isoforms of the same gene functionally distinct?’.
The authors answered these crucial questions using PRDM14 as an example of a SON-regulated gene. SON knockdown causes the downregulation of the full-length PDRM14 mRNA, with a concomitant upregulation of a shorter PRDM14 isoform, which lacks exon 2. Strikingly, the short PRDM14 isoform is devoid of any activity in both a promoter assay and a reprogramming experiment, suggesting that indeed alternative splicing can have a dramatic effect on the biological activity of target genes.
In sum, this study describes a role of SON, a spliceosome-associated protein, in the regulation of constitutive and alternative splicing in hES cells and sheds new light on the regulation of alternative splicing in a cell-type-dependent manner (see Figure 1). With the eyes of a stem cell biologist, this paper adds an intriguing new layer of complexity to the regulation of one of the most fascinating biological phenomena: cellular pluripotency.
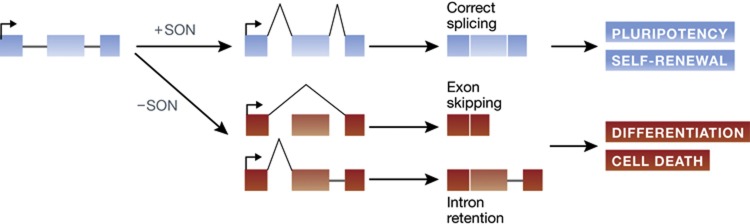
Figure 1.
The scheme summarizes the proposed role of SON in regulating splicing events in embryonic stem cells (ESCs). In the presence of SON, recognition of a unique set of splice targets facilitates express of transcripts that are essential for the maintenance of ESC self-renewal and pluripotency. In its absence, exon skipping and/or retention can trigger cellular differentiation or cell death. (Figure redrawn from Lu et al (2013), with permission from authors and NPG.)
Footnotes
The author declares that he has no conflict of interest.
References
- Ahn E-Y, DeKelver RC, Lo M-C, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu X-D, Zhang D-E (2011) SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell 42: 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, Huss M, Soh BS, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng H-H (2010) A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468: 316–320 [DOI] [PubMed] [Google Scholar]
- Lu X, Göke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH (2013) SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol 15: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara K-I, Bubulya A, Bubulya PA (2010) Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell 21: 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]