Abstract
Telomerase is a specialized chromosome end-replicating enzyme required for genome duplication in many eukaryotes. An RNA and reverse transcriptase protein subunit comprise its enzymatic core. Telomerase is evolving rapidly, particularly its RNA component. Nevertheless, nearly all telomerase RNAs, including those of H. sapiens and S. cerevisiae, share four conserved structural elements: a core-enclosing helix (CEH), template-boundary element, template, and pseudoknot, in this order along the RNA. It is not clear how these elements coordinate telomerase activity. We find that although rearranging the order of the four conserved elements in the yeast telomerase RNA subunit, TLC1, disrupts activity, the RNA ends can be moved between the template and pseudoknot in vitro and in vivo. However, the ends disrupt activity when inserted between the other structured elements, defining an Area of Required Connectivity (ARC). Within the ARC, we find that only the junction nucleotides between the pseudoknot and CEH are essential. Integrating all of our findings provides a basic map of functional connections in the core of the yeast telomerase RNP and a framework to understand conserved element coordination in telomerase mechanism.
Keywords: RNA, RNP, telomerase, telomere, TLC1
Introduction
Telomeres are repetitive sequences at the ends of eukaryotic chromosomes that protect the DNA termini from degradation and deleterious recombination (Wellinger and Zakian, 2012). Maintaining telomeric DNA requires a specialized system, since ends cannot be completely replicated by DNA polymerases that copy the bulk of chromosomes. This end-replication problem is solved in most eukaryotes by the enzyme telomerase, which can repeatedly append DNA to the 3′ end of chromosomes (Greider and Blackburn, 1985; Blackburn and Collins, 2011).
Telomerase is minimally composed of an RNA (TER; TLC1 in S. cerevisiae) and a catalytic reverse transcriptase protein (TERT; Est2 in S. cerevisiae) subunit, which can reconstitute basal telomerase activity in vitro (Autexier and Greider, 1994; Weinrich et al, 1997; Zappulla et al, 2005). How TER and TERT coordinate telomere addition remains poorly characterized. In vivo, telomerase forms a holoenzyme complex containing additional protein subunits that recruit telomerase to the telomere, regulate its activity, and promote RNA stability and processing. Loss of core or essential holoenzyme components of telomerase leads to an ever-shortening telomeres (Est) phenotype that triggers a G2/M cell-cycle arrest known as senescence (Lundblad and Szostak, 1989; Weinert et al, 1994; Lendvay et al, 1996).
While telomerase accessory protein subunits vary among species, in S. cerevisiae they include the essential proteins Est1 and Est3 (Lundblad and Szostak, 1989; Lendvay et al, 1996). Additionally, the RNP includes the Ku and Sm protein complexes, which are not essential subunits for telomere maintenance (Lendvay et al, 1996; Seto et al, 1999; Peterson et al, 2001). Evidence supports roles of Est1 and Ku in telomerase recruitment and activation (Evans and Lundblad, 1999; Hughes et al, 2000b; Evans and Lundblad, 2002; Stellwagen et al, 2003; Pfingsten et al, 2012; for review, see Nandakumar and Cech, 2013), and Est3 promotes telomerase function through an unknown mechanism (Lendvay et al, 1996; Hughes et al, 2000a; Talley et al, 2011). Binding of the Sm7 protein complex near the 3′ end of TLC1 is critical for RNA stability as mutation of the Sm-binding site in TLC1 leads to RNA abundance being reduced 10-fold (Seto et al, 1999). Both polyadenylation and Nrd1/Nab3 transcriptional termination pathways have been implicated in 3′ end biogenesis (Chapon et al, 1997; Seto et al, 1999; Noel et al, 2012), and it has been reported recently that the Sm site plays a role in exosome-mediated trimming of the 3′ polyadenylated end (Coy et al, 2013). The binding sites for Est1 and Ku in TLC1 can be dramatically repositioned and still provide function, demonstrating that their roles are modular in the RNP and that yeast telomerase RNA is a functionally flexible scaffold for these protein subunits (Zappulla and Cech, 2004, 2006; Zappulla et al, 2011).
Telomerase RNAs vary dramatically in length (from 147 nucleotides in ciliates to over 2425 nts in yeasts), sequence, and overall secondary structure among eukaryotes (Romero and Blackburn, 1991; McCormick-Graham and Romero, 1995; Chen et al, 2000; Qi et al, 2012). Nevertheless, despite the vast changes in TERs during evolution, some structural elements are conserved within the catalytic core. The Blackburn lab proposed a phylogenetic model containing four conserved structural core elements (Lin et al, 2004). These four elements are also always found in the same 5′ to 3′ order (see Figure 1A). The first of these elements is a core-enclosing helix (CEH), which has not been extensively studied but which maps to two stretches of nucleotides shown to be required for binding Est2 in S. cerevisiae (Livengood et al, 2002). Second, there is a template-boundary element (TBE) that defines the 5′ end of the template. In Kluyveromyces, Saccharomyces, and Schizosaccharomyces yeasts, the TBE is a helix located 5′ of the template end that sterically limits reverse transcription (Tzfati et al, 2000; Seto et al, 2003; Box et al, 2008). Third is the single-stranded RNA template that directs the addition of DNA repeats (Greider and Blackburn, 1989). The fourth and final element is a pseudoknot with base triples (Chen et al, 2000; Shefer et al, 2007; Ulyanov et al, 2007; Gunisova et al, 2009; Qi et al, 2012). In S. cerevisiae, the pseudoknot region of the 1157-nucleotide (nt) TLC1 RNA has been reported to bind Est2 (Livengood et al, 2002; Lin et al, 2004) while the base triples within it contribute to telomerase catalysis through a binding-independent mechanism (Qiao and Cech, 2008). Miniaturized TLC1 alleles ranging from 384 to 500 nts, containing all of the core elements plus accessory protein binding sites, function in vivo and permit reconstitution of telomerase activity in vitro (Zappulla et al, 2005). Mini-T can be further reduced to a 170-nt form (Micro-T) that contains only the conserved core nucleotides and still functions equally well in vitro (Qiao and Cech, 2008).
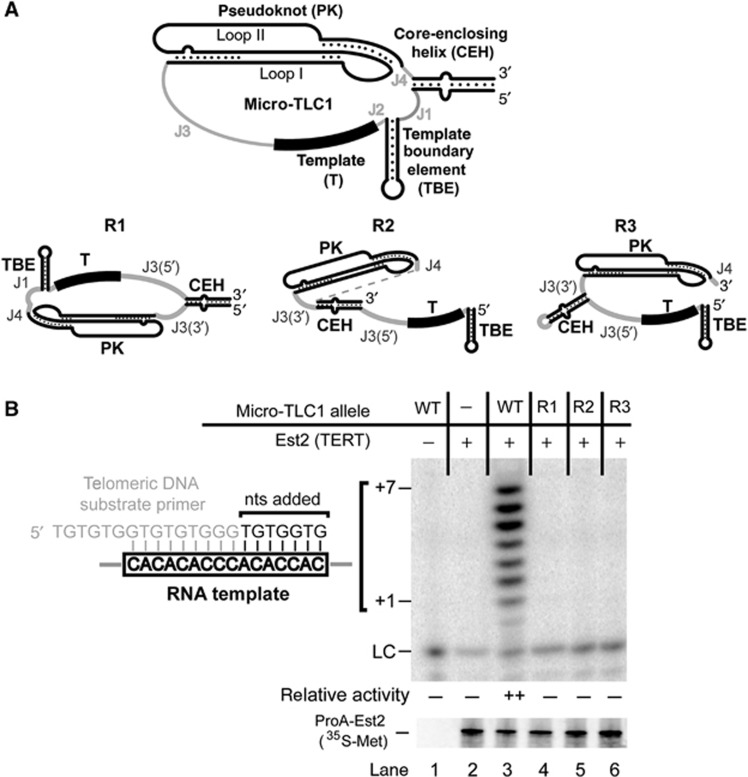
Figure 1.
Rearranging the relative order of conserved structural elements abolishes telomerase activity in vitro. (A) Schematic of the four conserved core elements retained in a miniaturized TLC1 allele, Micro-T(170) (top). Single-stranded RNA regions between the functional elements have been designated as junctions (J) and numbered 5′ to 3′. See Figure 2A for nucleotide sequence. Three rearranged Micro-T(170) derivatives (R1–R3; bottom) were tested in a reconstituted telomerase activity assay. Sequences of mutants are found in Supplementary Figure S1. (B) Rearranged mutants failed to reconstitute telomerase activity (top). Products of an in vitro telomerase activity assay are indicated (+1 to +7). LC, loading (and recovery) control: ∼1 nM [γ32P]-labelled primer added prior to telomerase reactions. The amount of immunopurified (IP’d) ProA-Est2p was monitored by 35S-methionine incorporation (bottom). The telomerase RNA template annealing to the telomeric DNA substrate primer, used throughout this work, and the sequence of products generated by telomerase activity are shown to the left. Telomerase activity was quantified by dividing the sum of the signal in the +0 to +7 band by the LC. The average activity (normalized to the loading control) from at least two independent telomerase preparations is summarized as follows: (–), no telomerase activity was detectable above background; (++), wild-type activity level.
Why there is a conserved ordering of the four core elements in telomerase RNAs is unknown. The individual core elements could be independent modules that would tolerate being repositioned, like the accessory Est1 and Ku binding sites (Zappulla and Cech, 2004; Zappulla et al, 2011), or they may be pieces of a larger functional unit that requires specific ordering of these elements (and potentially three-dimensional positioning) to support telomerase activity.
We hypothesize that the conserved ordering of core elements in telomerase RNA reflects functionally important linkages between the structural elements. In order to determine whether the conserved core structural elements contribute to—and potentially coordinate—telomerase activity, we performed physical connectivity mapping on S. cerevisiae telomerase RNA. We find that the core structural elements are not independent modules, but rather need to be linked together. Through the use of circular permutations (i.e., relocating the RNA ends), we show that physical connections are required from the pseudoknot to the template, defining an area of required connectivity (ARC). Furthermore, we find that the length of the junction between the pseudoknot and CEH of TLC1, as well as the CEH itself, are critically important features of the ARC. Our circular permutation mutagenesis also demonstrates that the Sm7 protein complex can function when dramatically repositioned on the Mini-TLC1 RNA, providing additional evidence that accessory proteins are independent functional modules in the telomerase RNP.
Results
Mapping relative positioning and connectivity requirements in the yeast telomerase core enzyme in vitro
The four conserved structural elements within the core of telomerase RNAs are always arranged in the same order. To test whether this layout is simply the result of a common evolutionary history or is required for telomerase mechanism, we designed and synthesized mutant yeast telomerase RNA alleles with altered ordering of the elements. We reconfigured the elements in the context of a miniaturized TLC1 allele, Micro-T(170) (Qiao and Cech, 2008), in three fundamentally different ways (Figure 1A, R1–R3). Since the relationship between the ssRNA template and the TBE helix is already established (Seto et al, 2003), we kept these two elements juxtaposed. In order to promote energetically favourable folding of each RNA element within the rearranged TLC1 core alleles, we used the RNA secondary structure prediction software Mfold (Zuker, 2003) as a major criterion in the design process (see Materials and methods). The Micro-T RNAs with rearranged elements were co-expressed in a rabbit reticulocyte transcription-translation expression system along with tagged Est2 (Friedman and Cech, 1999; Zappulla et al, 2005; Qiao and Cech, 2008). The telomerase core enzyme was then immunopurified and tested for activity. Although wild-type Micro-T(170) RNA showed the expected robust reconstituted activity (Qiao and Cech, 2008), none of the variant RNAs with the rearranged core elements were active (Figure 1B, lanes 4–6). We also tested the same rearrangements in the context of a 194-nt Micro-T predicted to have an 8-bp longer CEH and observed similar results. The fact that reordered core element TLC1 alleles do not function in vitro provides evidence that the wild-type order of the elements is required for telomerase activity. Furthermore, this result supports the hypothesis that the conserved elements act in a coordinated manner to provide enzyme activity.
We next examined the importance of the RNA junctions that connect the conserved elements in TLC1 (see Figure 1A). To disrupt these junctions, we repositioned the 5′ and 3′ ends to create ‘circularly permuted’ (cp) constructs, as has been done previously in ciliate telomerase (Miller and Collins, 2002; Mason et al, 2003), which introduce a break in the phosphate backbone. As shown in Figure 2A, we moved the ends to within each of the four single-stranded junctions as follows: J1, between the CEH and template boundary helix; J2, between the template boundary and template; J3, between the template and pseudoknot; and J4, between the pseudoknot and CEH. Those alleles with ends relocated to junctions J1, J2, or J4 completely abolished telomerase activity (Figure 2B, lanes 6–9). These results show that contiguous RNA from the pseudoknot through the CEH and template is essential for telomerase activity. These junctions that cannot be disrupted comprise an Area of Required Connectivity (ARC) in yeast telomerase RNA.
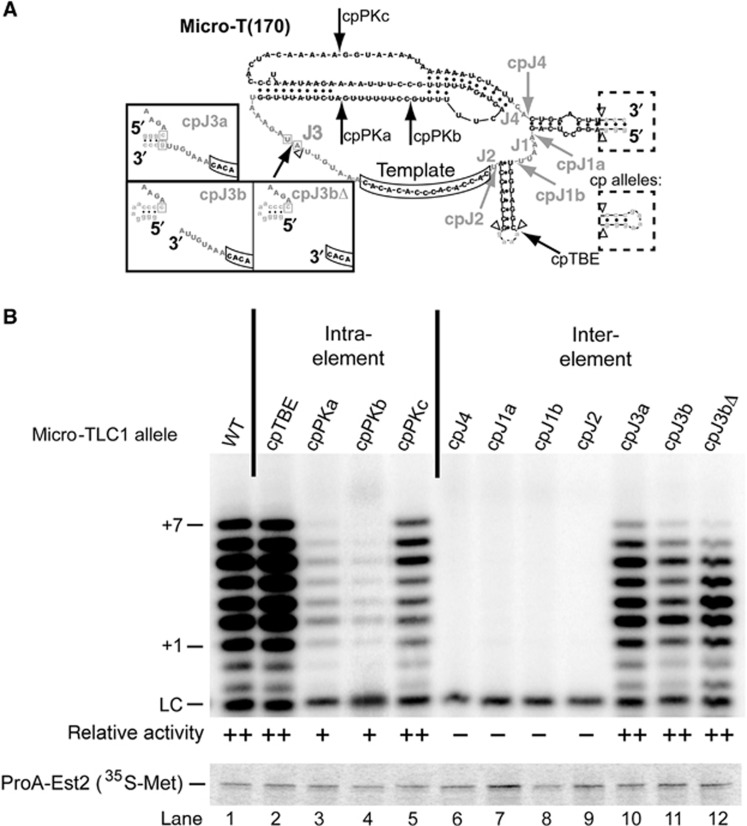
Figure 2.
In vitro activity of Micro-T circular permutants reveals an Area of Required Connectivity in yeast telomerase RNA. (A) Structure of Micro-T(170) with conserved core element nucleotides (black) and junction nucleotides (grey). The template is boxed. Open triangles denote the location of deletions compared to TLC1, while exogeneous sequences are lowercase. Arrows point to the location of the repositioned 5′ and 3′ ends in the circular permutation (cp) mutants, and the dotted boxes show the wild-type CEH and modified loop present in cp mutants. Solid boxes denote the predicted structures of the three cpJ3 mutants. Two nucleotides in J3 that were substituted in these cpJ3 mutants to prevent misfolding are boxed in grey. See Supplementary Figure S1 for complete sequences of all mutants. (B) In vitro telomerase activity assay, as in Figure 1, with Micro-T(170) circular permutants. Immunopurified Est2p is shown below. The average activity from at least two biological replicates is classified as: (−), no detectable telomerase activity; (+), detectable, but <50% of wild-type telomerase activity; (++), 50% or greater telomerase activity.
Unlike disrupting physical connections within the ARC, three alleles with circular permutations in J3, between the template and pseudoknot, reconstituted telomerase activity (Figure 2B, lanes 10–12), indicating that contiguous RNA is not required between all the conserved structural elements. Ciliates contain a template recognition element (TRE) and important physical connections 3′ of the template (Miller and Collins, 2002; Berman et al, 2011). To test whether a similar template-recognition function is provided by nucleotides in the homologous position in yeast telomerase RNA, we deleted eight nucleotides downstream of the template from cpJ3b to create cpJ3bΔ (Figure 2A). This mutant exhibited no significant change in telomerase activity (Figure 2B, lanes 11 and 12), showing that residues upstream of the template are not required in Micro-TLC1.
We also designed and tested TLC1 alleles with circular permutations within loops of structural elements. We repositioned the ends of the RNA to each of the three regions within elements predicted to be single stranded: the loop of the TBE and the loops of the pseudoknot. Moving the ends to these locations still permitted telomerase activity (Figure 2B, lanes 2–5). When the ends were positioned nearest to the conserved base-triple interactions (cpPKb), this caused the greatest reproducible reduction in telomerase activity (Figure 2B, lane 4). Together, these findings indicate that yeast telomerase RNA tolerates physical breaks within structural elements or J3.
In vivo experiments support the existence of the ARC in yeast telomerase RNA
To test for the importance of the ARC within the complete enzyme complex, we next examined the circular permutations tested in Micro-T(170) in the context of the 460-nt Mini-T RNA (Zappulla et al, 2005; Figure 3A; Supplementary Figure S2). Mini-T RNAs, unlike the further-truncated Micro-T RNA (Qiao and Cech, 2008), contain the binding sites for known holoenzyme components Est1, Ku and Sm7 and are capable of functioning in vivo (Zappulla et al, 2005; Figure 3A). A Mini-T allele therefore allows assessment of both in vitro function and genetic complementation in yeast. In order to test circularly permuted telomerase Mini-T RNAs in vivo, we moved the Sm-binding site along with the repositioned 3′ end, which we expected would be required for Sm7 to promote telomerase RNA processing and abundance (Figure 3A).
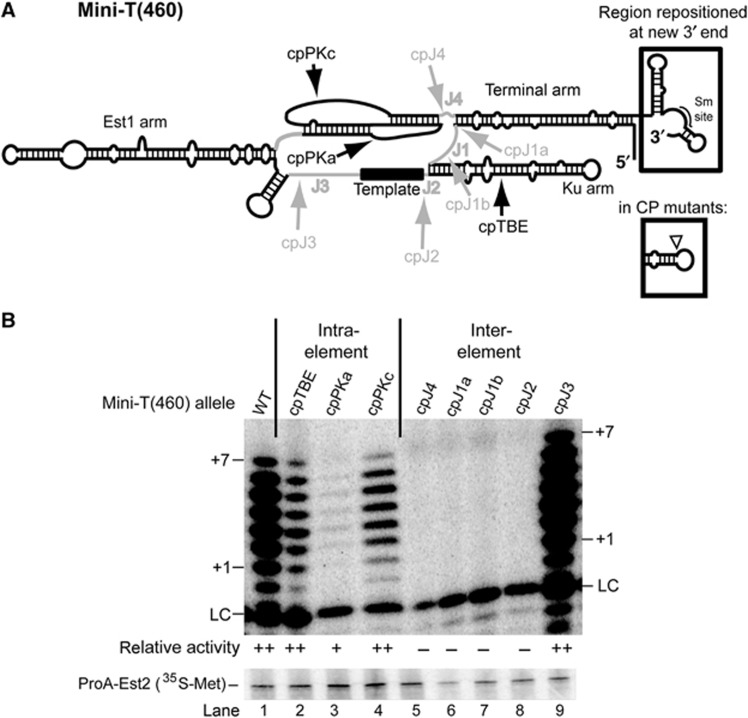
Figure 3.
Disrupting the ARC in Mini-T(460) abolishes telomerase activity in vitro. (A) Secondary structure of wild-type Mini-T(460). Structural elements are black and junctions are grey. Arrows indicate the position of the 5′ and 3′ ends in the CP mutants. The wild-type 5′ end of the Mini-T RNA is indicated with an open triangle, and the region of the 3′ end that was repositioned, including the associated Sm site, is boxed. Mutant sequences can be found in Supplementary Figure S1. A nucleotide resolution model of Mini-T(460) can be found in Supplementary Figure S2. (B) Circular permutations in the ARC abolish telomerase activity. In vitro telomerase activity assays, and average activity from at least two biological replicates, with cp Mini-T(460) alleles are shown, as in Figure 2.
Before testing functionality of Mini-T circular permutants in vivo, we tested whether they support core enzyme activity in vitro. Repositioning the ends to within the ARC of Mini-T(460) completely abolished activity (Figure 3B, lanes 5–8). By contrast, circular permutations outside of the ARC all exhibited telomerase activity (Figure 3B, lanes 2–4 and 9). Of these non-ARC circular permutants, cpPKa exhibited the most significantly reduced telomerase activity (Figure 3B, lane 3), similar to what we observed for the corresponding circular permutation in Micro-T(170) (Figure 2B, lane 3). This weak, yet detectable, telomerase activity was reproducible, whereas we never detected telomerase activity for any allele with a circular permutation within the ARC.
To determine whether the ARC defined by our in vitro tests with Micro-T and Mini-T is required for telomerase activity in vivo, we next aimed to test Mini-T circular permutants for complementation of tlc1Δ strain. The strain is also a rad52Δ mutant so telomeres cannot be maintained by alternate telomerase-independent means. To our knowledge, no circular permutant of a non-coding RNA has been tested for function in vivo, and we anticipated that circularly permuting an RNA in vivo could have a profound effect on RNA abundance. To try to retain appropriate TLC1 RNA bioaccumulation, we took the following steps. First, to prevent loss of Sm site function, which in wild-type TLC1 has been shown to cause an ∼10-fold decrease in TLC1 abundance (Seto et al, 1999), we repositioned the Sm site to the new 3′ end in the circular permutants. Second, we chose to express these circularly permuted Mini-T(460) RNAs from a multi-copy 2μ plasmid (Christianson et al, 1992; Mozdy and Cech, 2006), given that Mini-T(460) RNA in vivo is only ∼23% the level of wild-type TLC1 when it is expressed from a centromeric plasmid (Zappulla et al, 2005) and since we found that Mini-T circular permutants expressed from CEN plasmids were reproducibly undetectable by northern blotting (Supplementary Figure S3B) and did not complement a tlc1Δ mutant (Supplementary Figure S3A). With 2μ-based expression, we did detect by northern blotting that all Mini-T circular permutants migrate at the same relative position in the gel as wild-type Mini-T(460) (Figure 4B). The levels of these mature circularly permuted Mini-T RNAs were 22–65% the abundance of wild-type TLC1 expressed from a CEN plasmid (Supplementary Figure S4B). Notably, circular permutations in Mini-T caused an increased abundance of longer, presumably polyadenlylated (Chapon et al, 1997; Coy et al, 2013), forms of the RNA as compared to wild-type TLC1 or Mini-T, as well as sometimes also shorter forms, suggesting that repositioning the ends and Sm site may cause some alterations in RNA biogenesis. However, even the level of the mature form of the lowest abundance circular permutant, cpJ1b (Figure 4B, lane 11; 22%, Supplementary Figure S4B), is similar to the abundance we observed for wild-type Mini-T expressed from a CEN plasmid (Figure 4B, lane 4). This degree of telomerase RNA abundance in vivo has been shown to be sufficient for function in many instances (e.g., tlc1-Sm− at 1–10%; Seto et al, 1999; Zappulla et al, 2011; Lebo and Zappulla, 2012; Supplementary Figure S5B).
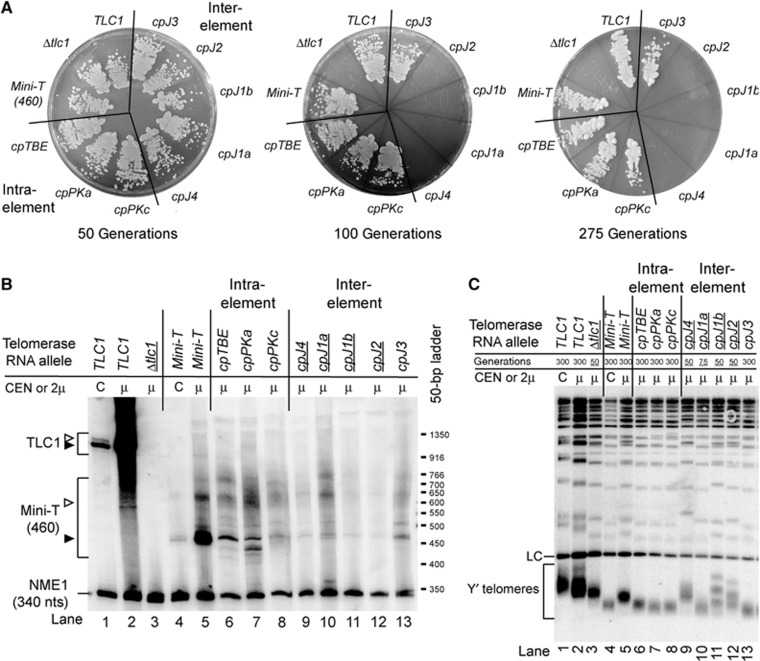
Figure 4.
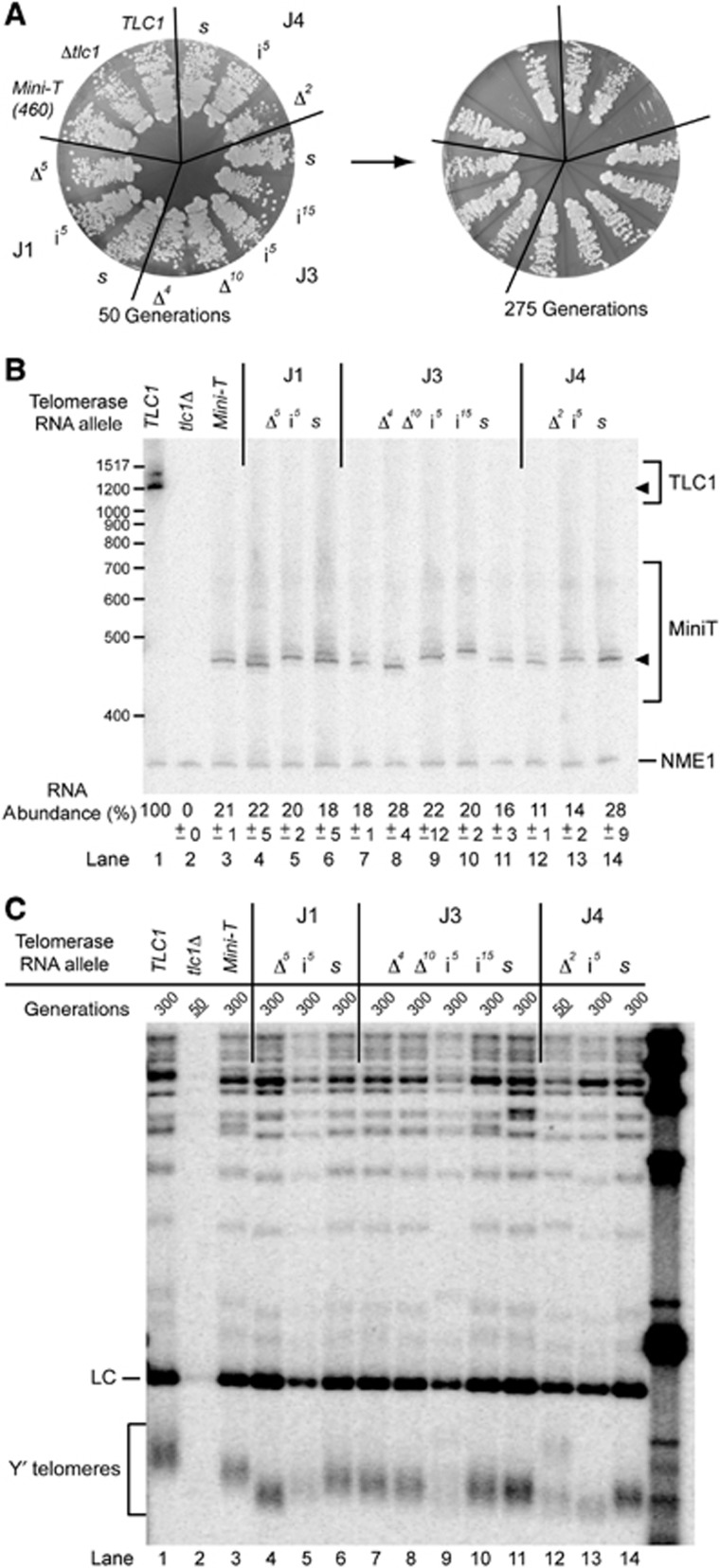
Circular permutations in the ARC of Mini-T cause senescence and repositioning the Sm site is tolerated but causes RNA abundance and processing defects. (A) Haploid yeast strains harbouring the indicated telomerase RNA allele, expressed from a 2μ plasmid, were passaged on plates up to 275 generations. (B) Mini-T circular permutant RNAs are detectable by northern blotting when expressed from 2μ plasmids. Control CEN samples were included to compare overexpression. Quantitation of the amounts of total and full-length RNA (black triangle) for two independent biological replicates can be found in Supplementary Figure S4. (C) Telomeres in ARC mutant strains begin to shorten at early time points. Southern blot on XhoI-digested genomic DNA probed for telomeric sequence and an internal chromosome IV fragment. Samples are shown at 300 generations, except for underlined samples where cells senesced (note generation listed above for each). For comparison, samples from CEN plasmids were also included.
Circular permutations within the ARC of Mini-T caused senescence by 100 generations (Figure 4A), consistent with a lack of telomerase activity in vitro (Figure 3B). To test whether the arrested cell proliferation caused by ARC-disrupting permutations was indeed senescence due to telomere shortening, we determined telomere length by Southern blotting. Compared to TLC1, wild-type Mini-T(460) exhibited 278-bp shorter telomeres at 300 generations when expressed from 2μ plasmids (Figure 4C, cf. lane 2 and lane 5). The circular permutations within the ARC of Mini-T caused shortening telomeres within 50 generations after a wild-type TLC1 cover plasmid was shuffled out of the strain prior to when these cells ceased to grow (Figure 4C, lanes 9–12). While the telomeres of some senescing ARC circular permutants appear to be the same length as wild type, it is important to note that these samples come from early-generation time points, where it can be seen that telomeres are already beginning to shorten (compare with tlc1Δ at 50 generations in lane 3). These data support the conclusion that the ARC is an essential feature of telomerase RNA in vivo.
In contrast, all of the Mini-T RNAs with circular permutations outside of the ARC that we tested functioned in vivo. Four circular permutants, with the ends within J3, the Ku arm, or the pseudoknot, supported growth through 275 generations (Figure 4A). The telomeres supported by these circular permutants are 148- to 177-bp shorter than wild-type Mini-T(460) (Figure 4C, lane 5 versus lanes 6–8 and 13). Although telomeres are shorter in these alleles, we consider it noteworthy that these circularly permuted Mini-T RNAs do maintain functional telomeres despite the following facts: (1) telomeres of cells expressing wild-type Mini-T from a 2μ plasmid are already shorter than wild-type TLC1 cells, making Mini-T-expressing cells more sensitive to further telomerase defects, (2) the Mini-T circular permutants are even less abundant than wild-type Mini-T, (3) the co-transcriptional RNA folding pathways differ with circular permutations, and (4) the Sm-binding site is repositioned.
The Sm-binding site in telomerase RNA functions when repositioned along with the RNA ends
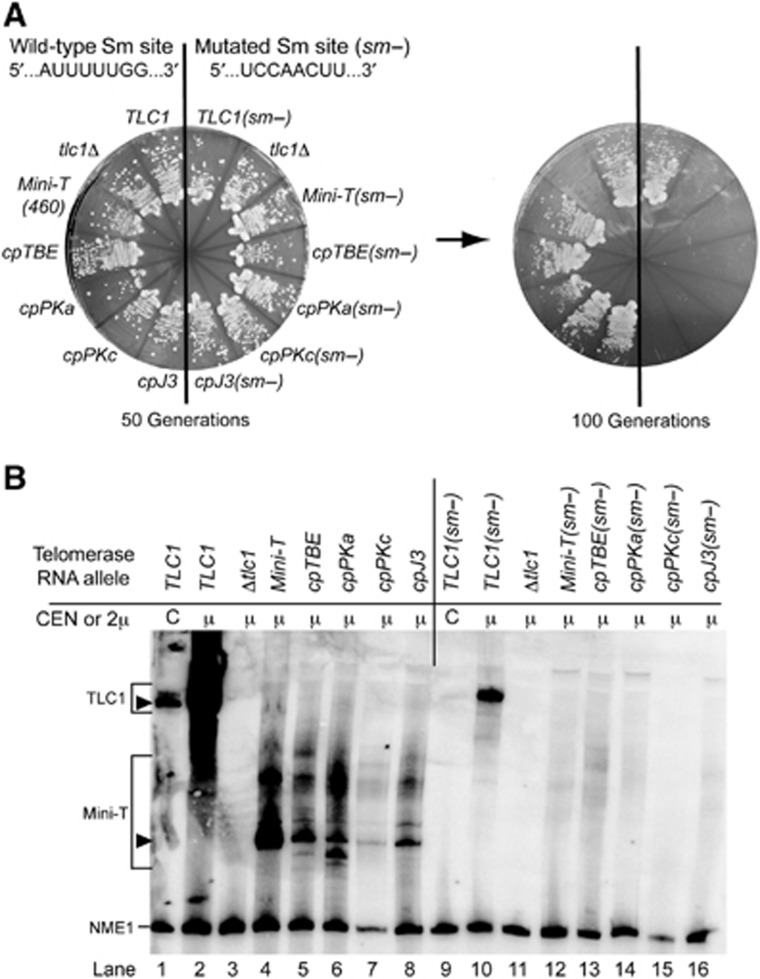
Repositioning the ends of yeast telomerase RNA along with the Sm site at the 3′ end supported growth in four of the circularly permuted Mini-TLC1 alleles we generated. This suggests that Sm7 is a functionally flexible subunit like Est1 and Ku (Zappulla and Cech, 2004; Zappulla et al, 2011). However, since the Sm site is not essential to TLC1 (despite mutations in this site reducing TLC1 abundance to under 10% of wild type; Seto et al, 1999), the fact that TLC1 circular permutations are viable does not necessarily demonstrate that Sm7 is functioning when its binding site on TLC1 is relocated. To test whether repositioned Sm-binding sites in viable circular permutants are functional, we mutated the site (5′-AUUUUUGG-3′) to a non-consensus sequence, 5′-UCCAACUU-3′ (Figure 5A) to inhibit Sm7 function (Guthrie and Patterson, 1988; Seto et al, 1999). When we introduced this mutant site into Mini-T(460), the mature RNA was no longer detectable, even when expressed from a 2μ plasmid (Figure 5B, lane 12; Supplementary Figure S5), and cells senesced by 100 generations (Figure 5A). Thus, while the Sm site is not essential in TLC1, it is required in Mini-T. When we mutated the Sm site in each of the four functional circular permutants, it also reduced mature telomerase RNA to levels undetectable by northern blotting (Figure 5B, lanes 13–16; Supplementary Figure S5), and cells senesced by 100 generations (Figure 5A). Thus, the abundance of circularly permuted RNAs depends on their repositioned Sm-binding sites. This shows that the repositioned Sm consensus sites in these circularly permuted telomerase RNAs are functional. Therefore, the Sm7 subunit complex binding position can be moved to very different locations within the yeast telomerase RNA while maintaining function, as previously observed for the binding sites for the Est1 (Zappulla and Cech, 2004) and Ku (Zappulla et al, 2011) subunits.
Figure 5.
Repositioned Sm sites in Mini-T(460) are required to prevent senescence and maintain RNA abundance. (A) Yeast strains harbouring the indicated telomerase RNA alleles on a 2μ plasmid were tested for senescence. Viable circular permutants with the wild-type TLC1 Sm consensus are on the left side of plates, while the same mutants with a previously characterized consensus mutation (sm−) (Seto et al, 1999) are on the right side of each plate. (B) Mature RNA is undetectable from all Mini-T alleles lacking the Sm-binding site consensus sequence (right side of the gel). Northern blotting was performed as in Figure 4. Closed triangles indicate full-length RNAs; quantitation of total and mature levels of TLC1 for duplicate samples are in Supplementary Figure S5. Note, TLC1(sm−) in lane 8 shows only the larger polyadenylated form, consistent with Seto et al (1999). NME1 was used as an internal control.
Junction length within the ARC is important for telomerase function
Having defined an ARC in TLC1 by circular permutations, we next tested whether specific characteristics of the single-stranded RNA junctions in the ARC are also important. In an alignment of 36 distinct sequences from 7 species of yeast in the sensu stricto clade (Supplementary Figure S6) the length and sequence of junctions within the ARC are highly conserved: J4 and J2 nucleotides are completely conserved (5′-AC-3′ and U, respectively) and 5 J1 nucleotides fit the pattern ADWUU (Figure 6A). Because J2 is the junction between the template and the TBE (Tzfati et al, 2000; Seto et al, 2003), we focused on the other junctions in the ARC. To begin to examine their role in telomerase mechanism, we tested Micro-T and Mini-T RNAs with deletions, insertions, and sequence changes within J1 and J4.
Figure 6.
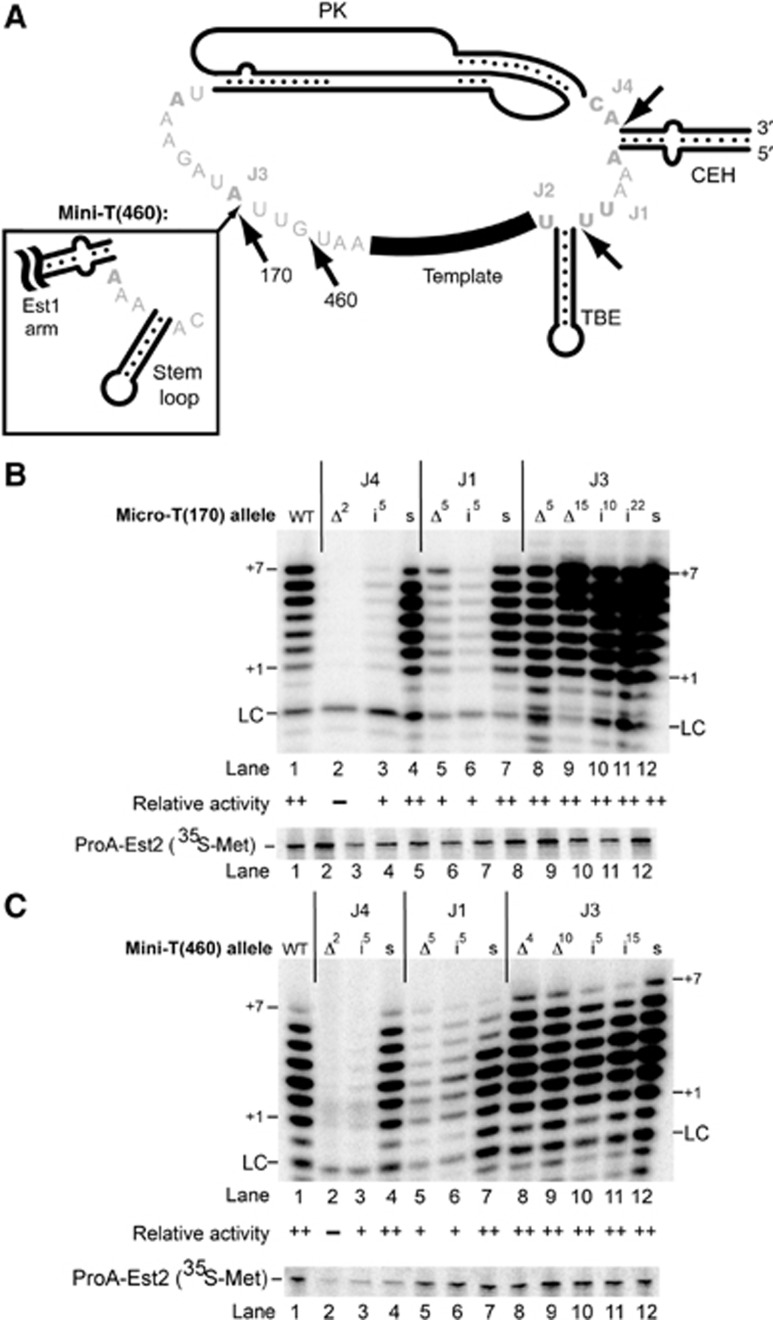
Changing the length of RNA within the ARC junctions J1 and J4 decreases telomerase activity in vitro. (A) Expanded view of the yeast telomerase RNA junction sequences. Nucleotides that are universally conserved among an alignment of 36 sequences from 7 Saccharomyces species are indicated in bold (see also Supplementary Figure S6). The inset in J3 shows sequences that are present in Mini-T(460) but not in Micro-T(170). Arrows indicate the location of insertions; sequences of insertions and substitutions are listed in Supplementary Figure S1. (B, C) Deletions and insertions in the ARC result in decreased telomerase activity. Standard in vitro telomerase activity assays with deletion (Δ), insertion (i), and sequence substitution (s) mutations in Micro-T(170) (B) and Mini-T(460) (C). Superscripts indicate the number of nucleotides that were deleted or inserted at a given location. Quantitation of two independent biological replicates is indicated, as in Figure 2.
Perturbations in J4, between the pseudoknot and CEH, had the most drastic effects. When the two J4 nucleotides were deleted (J4Δ2), in vitro telomerase activity was abolished in Micro-T(170) (Figure 6B, lane 2). Similarly, deleting these two J4 nucleotides from the larger Mini-T(460) also abolished in vitro telomerase activity (Figure 6C, lane 2). Insertion of five additional nucleotides at the 3′ end of J4 (J4i5) greatly decreased activity in Micro-T(170) and Mini-T (see lane 3 of Figures 6B and C). These data indicate that the length of the two-nucleotide junction between the pseudoknot and CEH is crucial for yeast telomerase activity in vitro. Altering the strictly conserved J4 sequence from CA to UG had a minor effect, causing no significant defects in Micro-T(170) or Mini-T(460) (lane 4 of Figures 6B and C), suggesting that sufficient length of J4, but not its sequence, is critical for telomerase function in vitro.
To determine whether the spacing in J4 was also important in vivo, we transformed Mini-T(460) mutants into the cell and tested for complementation. Deletion of J4 (J4Δ2) caused senescence irrespective of being expressed from a CEN or 2μ plasmid (Figure 7A; Supplementary Figure S7A). The J4 insertion mutant (J4i5) led to senescence when expressed from a CEN plasmid (Supplementary Figure S7A), but not when overexpressed from a 2μ plasmid (Figure 7A). Senescence of the insertion and deletion mutants expressed from CEN plasmids is likely due to defects in RNA accumulation, as essentially no RNA was detectable above background for these mutants (Supplementary Figure S7B). However, overexpression from the 2μ plasmid raised the RNA levels of these mutants to 11–14% of wild-type TLC1 (Figure 7B, lanes 12 and 13 versus lane 1). Thus, the senescence in cells expressing J4Δ2 from a 2μ plasmid is not simply due to low accumulation of RNA. The telomeres from J4Δ2 cells appear to be shortening at 50 generations (Figure 7C, lane 12), as expected in senescence. As opposed to altering the length of J4, changing the sequence of the J4 nucleotides (J4s) had no detectable effects on telomerase RNA levels (Figure 7B, lane 14; Supplementary Figure S7B) or growth (Figure 7A; Supplementary Figure S7A) in vivo.
Figure 7.
The two-nucleotide junction J4 is required for telomerase function in vivo. (A) Deletion of the two nucleotides in J4 causes senescence in vivo. Indicated junction mutations in Mini-T(460), expressed from 2μ plasmids, were serially passaged for 275 generations. (B) All Mini-T(460) junction mutants, when expressed from 2μ plasmids, are detectable by northern blotting. The abundance represents the average amount of mature RNA from two biological duplicates, with the error below indicating the range of expression from those duplicates. Sizes of full-length Mini-T RNAs (filled triangle) vary consistently with the predicted length of the RNA. (C) Telomere length of each junction mutant correlates with the in vitro activity (Figure 6C). A Southern blot probed for telomeric DNA and the chromosome IV loading control (LC) is shown. Note that J4Δ mutation causes senescence and so genomic DNA for that sample is from 50 generations, whereas others are from 300 generations, as listed above each lane (with senescence indicated by underlining).
Unlike J4, J1 is not essential for telomerase function. In vitro, changing the number of nucleotides in J1, either by deletion or by insertion (J1Δ5 and J1i5), decreased, but did not abolish, telomerase activity in both Micro-T(170) and Mini-T(460) (Figures 6B and C, lanes 5 and 6, respectively). In contrast, altering the nucleotide sequence of the five J1 nucleotides (J1s) had little effect on overall telomerase activity (lane 7 of Figures 6B and C). Altered J1 sequence did decrease nucleotide addition processivity in Mini-T(460), with extension reproducibly stalling after the fourth nucleotide added (Figure 6C, lane 7; see Figure 1B for primer-template alignment). This could result from J1 pairing with the template to form two isolated G-C base pairs at the base of the TBE (see Supplementary Figure S1 for sequence and potential extended pairing). Together, these data indicate that the length of J1 is more important than its sequence for telomerase activity in vitro.
In vivo, all J1 mutants supported cell proliferation through 275 generations when expressed from either a CEN (Supplementary Figure S7A) or 2μ (Figure 7A) plasmid, with no apparent defects in RNA abundance relative to wild-type Mini-T(460) (Figure 7B; Supplementary Figure S7B). The telomeres of J1 deletion and insertion mutants were reproducibly shorter than the J1 sequence mutant (Figure 7C, lanes 4–6), consistent with the in vitro phenotypes (Figure 6C). Thus, despite some decreases in telomerase activity in vitro upon changing the length and sequence of J1, telomerase maintains functional telomeres in vivo.
Although J3 is not a component of the ARC, its length is conserved among Saccharomyces species (Dandjinou et al, 2004; Zappulla and Cech, 2004; Supplementary Figure S6). However, J3 sequence is not conserved: only 4 of the 15 nucleotides found in J3 of Micro-T(170) are invariant (Figure 6A; Supplementary Figure S6). Despite the low conservation in J3, the corresponding region—just 3′ of the template—in ciliates contains an important TRE (Miller and Collins, 2002). Our results from studying circular permutants indicates that at least eight of the nucleotides in J3 are dispensable for function in vitro (Figure 2B, lane 12). To explore whether any other J3 nucleotides play a role in telomerase function, we designed and tested deletion, insertion, and substitution mutants in this junction. Both small and large deletions within J3 in Micro-T(170) and Mini-T(460) supported robust telomerase activity in vitro (Figures 6B and C, lanes 8 and 9). In fact, deletion of all 15 nucleotides between the template and pseudoknot in Micro-T(170) had wild-type telomerase activity in vitro (J3Δ15). Notably, this 155-nt RNA is the smallest yeast telomerase RNA reported so far that functions in vitro. Furthermore, adding 5 or up to 22 additional nucleotides to J3 supported robust telomerase activity (Figures 6B and C, lanes 10 and 11). Altering the sequence of nucleotides in J3 (J3s) also caused no defects in telomerase activity in vitro (Figures 6B and C, lane 12, respectively). Consistent with telomerase activity results in vitro, mutations in J3 supported telomerase function in vivo. When introduced in Mini-T(460), all five J3 mutations supported growth through 275 generations, irrespective of being expressed from either a CEN or the 2μ plasmid (Figure 7A; Supplementary Figure S7A). Compared to wild-type Mini-T(460), RNA abundance and telomere lengths were not further decreased by these J3 mutations (Figures 7B and C, lanes 7–11).
Connectivity is not the only essential feature of the ARC: the CEH is also required
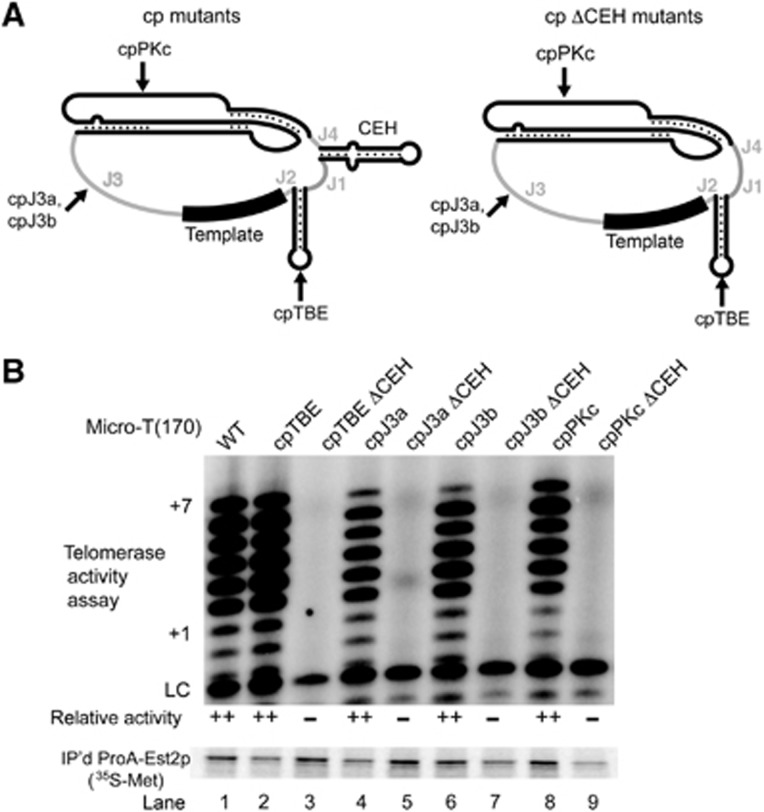
The CEH is the most poorly characterized conserved element in telomerase RNAs. To directly test whether the CEH provides some function beyond maintaining essential connectivity in the ARC, we deleted it while, importantly, still maintaining an intact ARC. To do this, we took advantage of four Micro-T(170) circular permutants that have wild-type levels of telomerase activity (cpTBE, cpJ3a, cpJ3b, cpPKc; see Figures 2B and 8). Deletion of the CEH nucleotides in these circular permutants maintains ARC connectivity (Figure 8A). Deleting the CEH from each of these four different circular permutants completely abolished function (Figure 8B, lanes 3, 5, 7, and 9). We conclude that the CEH has an essential function in enclosing the core to maintain the connectivity in the ARC, as well as a second essential ARC-independent role in telomerase activity.
Figure 8.
The core-enclosing helix (CEH) is required for telomerase RNA function in vitro. (A) Schematic of Micro-T(170) with indicated circular permutations (left, see also Figure 2A) and the corresponding allele with the core-enclosing helix deleted (right). Arrows indicate the location of the repositioned 5′ and 3′ ends in each mutant. Sequences of these RNAs are listed in Supplementary Figure S1. (B) Deletion of the core-enclosing helix abolished telomerase activity. Telomerase assays were conducted and quantified as described for Figures 1 and 2.
Discussion
A major outstanding question in the telomerase field is how the essential telomerase RNA and TERT protein work together to coordinate catalysis. Telomerase is unique among DNA-synthesizing enzymes in that it is an RNP that iteratively reverse transcribes a segment of its RNA subunit. To carry out nucleotide addition based on the repeated use of the RNA template, the interplay between TERT and the RNA (particularly non-templating nucleotides) is likely to be intricate. While we know that the Est1- and Ku-binding sites (Zappulla and Cech, 2004; Zappulla et al, 2011) can function independently of their position within the RNP, whether any functional flexibility exists in the TLC1 core had not been tested.
To address this question, we used a wide variety of mutagenic approaches to generate a comprehensive map of functional connections throughout the catalytic core of yeast telomerase RNA. Our in vitro and in vivo results identify regions of the core that tolerate substantial physical perturbations as well as those that must remain intact for telomerase to function. All mutations were tested in Micro-T(170) in vitro as well as in Mini-T(460), both in vitro and in vivo. Strikingly, for each particular mutation, the effect on telomerase function was indistinguishable in all assays in which it was tested (Figures 3 and 4; Figures 6 and 7). The perfect consistency of the results is despite the 170-nt Micro-T and 460-nt Mini-T RNAs representing quite different contexts for telomerase RNA folding and, in the case of Mini-T, also RNP assembly in vivo.
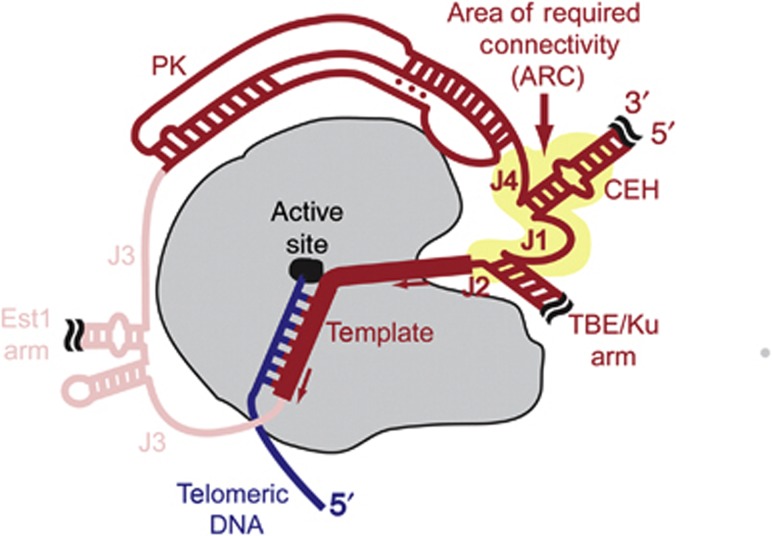
Most importantly, we discovered an essential region in the core of yeast telomerase RNA that must remain physically connected to function in vitro and in vivo (Figures 2, 3, 4). As shown in Figure 9, this ARC spans from the 3′ end of the pseudoknot through J4, the CEH, J1, the TBE, J2, and continuing to the template (Figure 9). Thus, the ARC connects the pseudoknot to the template through both direct RNA linkage and stable secondary structure (i.e., the CEH and TBE), and is likely to coordinate functionally important interactions among the core RNA elements as well as with TERT. The ARC is the first functionally important large-scale, overarching RNA region to be identified in telomerase RNA and its identification begins to provide the foundation for understanding how the conserved structural elements within it cooperate in telomerase mechanism.
Figure 9.
Schematic of yeast telomerase core enzyme highlighting the area of required connectivity (ARC) in the RNA. Repositioning the ends of telomerase RNA to J4, J1, or J2 abolishes telomerase activity, identifying the ARC in TLC1 RNA (yellow highlight). TERT, loosely based on the Triboleum castaneum structure (Mitchell et al, 2010), which lacks a TEN domain, is shown in grey with the active site as a black oval. Telomeric DNA is blue. Telomerase RNA is red, with the conserved functional elements encompassed by the ARC in dark red, and dispensable regions in light red. The telomerase RNA template is indicated with a thick line, and the direction of template movement relative to TERT during nucleotide addition is indicated with arrows. The ARC may include TERT-binding, template-positioning, and/or RNA-folding functions, and we hypothesize that it also fundamentally important for telomerase enzymatic mechanism.
Evidence for physical perturbability in the core of yeast telomerase
While the ARC defines a region of RNA in the catalytic core that cannot be interrupted, we find that non-ARC regions of the core accept significant structural perturbation. For instance, moving the ends to within structural elements is tolerated (Figures 2, 3, 4). Additionally, we find that disrupting connectivity in J3, 3′ of the template, is tolerated by telomerase in vitro and in vivo (Figures 2, 3, 4). This demonstrates that even within the catalytic core of telomerase RNA in yeast, there is a high capacity for enduring structural perturbations without loss of function. In fact, a Micro-TLC1 allele, lacking all 15 nucleotides between the template and pseudoknot exhibits full telomerase activity (Figure 6B), defining the smallest functional TLC1 core so far reported (155 nts). Similarly, Mini-Ts with deletion of four or ten nucleotides in J3, between the template and the small stem-loop 5′ of the Est1 arm, support telomerase activity in vivo (Figure 7). The lack of a requirement for any nucleotides 3′ of the template in vitro or in vivo suggests that no TRE exists in S. cerevisiae. Perhaps the TRE identified in ciliates (Miller and Collins, 2002) is related to repeat addition processivity (RAP), which is known to be lacking in the S. cerevisiae telomerase core enzyme in vitro using standard substrates (Cohn and Blackburn, 1995; Zappulla et al, 2005).
Not only can the ends of yeast telomerase RNA be repositioned to some locations within the core of TLC1, but the Sm-binding site can also be functionally repositioned to these regions. We have shown here that relocating the Sm site to four positions in the core of Mini-T supports telomerase function (Figures 4 and 5). Mutating the repositioned Sm site causes loss of detectable telomerase RNA and cellular senescence for each of these four alleles (Figure 5), demonstrating that the Sm7 subunit complex is functioning at its relocated position. These findings extend the scope of the flexible scaffold model as it applies to telomerase RNA in yeast by demonstrating that the Sm-binding site functions even when repositioned in TLC1, as was the case for binding sites for Est1 and Ku (Zappulla and Cech, 2004, 2006; Zappulla et al, 2011).
Even within the ARC junctions, where circular permutations are not tolerated, most mutations that alter length or sequence supported at least some telomerase activity (Figures 6 and 7). For instance, all deletions, insertions, and substitutions in J1 still permitted detectable telomerase activity in vitro (Figure 6) and maintained functional telomeres in vivo (Figure 7). The fact that J1 can be entirely deleted and, therefore, that the essential CEH and the important TBE can be immediately juxtaposed shows that the ARC can function despite significant physical perturbations, so long as the RNA backbone remains physically connected. Similarly, increasing the length or changing the sequence of J4 also supported telomerase activity (Figures 6 and 7). These findings underscore how the core of telomerase RNA, even within its ARC, can tolerate truncation, insertion, or substitution, without complete loss of function. This tolerance to mutations is further evidence that loss of telomerase activity in the ARC circular permutants is due to disrupting physical connectivity and not the extra nucleotides at the ends (e.g., for in vitro experiments, there are three additional G residues added to the 5′ end for efficient T7 RNA polymerase transcription initiation and up to two untemplated nucleotides could be added by T7 RNA polymerase to the 3′ end). Additionally, our findings suggest that there are not essential sequence-specific secondary or tertiary interactions formed by the junctions in the core of yeast telomerase RNA and, more generally, that telomerase RNA is notably physically flexible even in its catalytic core. Flexibility might be important for the mechanism of telomerase, which involves TERT reverse transcribing nucleotides of an RNA template that is intrinsic to the RNP. This necessitates the template threading through the active site of TERT (Mitchell et al, 2010) and, in cases where telomerase is processive, large RNA (and potentially also TERT) rearrangements associated with the translocation step.
J4 and the CEH are essential components of the ARC
Although we found the core of yeast telomerase tolerates some structural perturbations, we also find that deletion of one particular junction, J4, causes senescence (Figure 7). This result indicates that this junction, comprising two nucleotides between the pseudoknot and CEH, is an essential feature of the ARC. Although there are many possible explanations for why J4 is essential, perhaps deletion of J4 disrupts binding to TERT or prevents proper folding or orientation of the pseudoknot, which contains the catalytically important base triples (Qiao and Cech, 2008).
Evidence for another structured, critical element in the core of TLC1 comes from deletion of the CEH abolishing telomerase activity (Figure 8). If the CEH functions solely to enclose the core, then deleting it from four catalytically active enclosed-core circular permutants should not have disrupted function. Since large deletions on either side of the CEH have been shown to abolish binding to Est2 in vivo (Livengood et al, 2002), we hypothesize that the CEH is necessary for this interaction. However, the functional requirement for the CEH that we have discovered in yeast telomerase RNA does not appear to be universally conserved, since it is absent in some rodent species (Chen et al, 2000; Lin et al, 2004) and partially dispensable for function in humans and ciliates (Autexier et al, 1996; Beattie et al, 1998; Tesmer et al, 1999; Bachand and Autexier, 2001; Ly et al, 2003; Mason et al, 2003; Chen and Greider, 2005). The requirement for J4 and the CEH demonstrates that there are structural features within the ARC that are necessary for telomerase activity in addition to simple RNA strand connectivity.
The ARC is an evolutionarily conserved feature of telomerase RNAs
As mentioned above, we find that the ARC in S. cerevisiae is the area of the core that must be linked by way of covalent bonds and base-paired elements (Figure 9). We hypothesize that the conserved core elements proposed previously (Lin et al, 2004) are maintained in a particular order in telomerase to maintain the ARC and that the ARC represents an overarching conserved feature of nearly all telomerase RNAs. Examination of 107 available TER secondary structure models from ciliate, vertebrate, and fungal species (Lingner et al, 1994; McCormick-Graham and Romero, 1995; Chen et al, 2000; Dandjinou et al, 2004; Zappulla and Cech, 2004; Brown et al, 2007; Podlevsky et al, 2008; Webb and Zakian, 2008; Xie et al, 2008; Qi et al, 2012; Li et al, 2013) shows that 97% of telomerase RNAs have an intact ARC, as defined by having the pseudoknot connected to the template region via a CEH (Figure 9). As for the 3% (three species) without an intact ARC, they are a subset of the rodent lineage, which have their 5′ end just upstream of the template and thus lack a CEH. While all TERs contain extra helices, either within or outside the core, only four of the 103 ARC-containing species’ TERs have an additional helix within the ARC. These species are all Paramecia, with a helix between the predicted pseudoknot and CEH (McCormick-Graham and Romero, 1996). Thus, with just these few exceptions, RNA uninterrupted by additional secondary structural elements through the region corresponding to the ARC is a conserved physical feature in telomerase RNAs. Whether there is indeed functional conservation of the ARC remains currently under investigation.
In contrast to there being no extraneous base-paired elements found within the span of the ARC in telomerase RNAs, many fungal TERs have acquired one or more helices in J3 over the course of evolution. For example, the Est1-binding arm of S. cerevisiae, S. pombe, and K. lactis TERs is located in J3 (Dandjinou et al, 2004; Zappulla and Cech, 2004; Brown et al, 2007; Webb and Zakian, 2012). Additionally, N. crassa and K. lactis contain a helix in loop II of the pseudoknot (Brown et al, 2007; Podlevsky et al, 2008; Qi et al, 2012). Correspondingly, we have reported here that both J3 and loop II of the pseudoknot tolerate circular permutation (Figures 2, 3, 4). Thus, our experimental data support what is observed in nature.
The ARC in telomerase function
The ARC is a large-scale functional feature of the conserved core of telomerase RNA that encompasses all of the proposed conserved structural elements. Its existence strongly suggests that the elements that it encompasses function cooperatively during catalysis. We hypothesize that the ARC coordinates multiple key aspects of telomerase mechanism. As outlined below, the ARC may facilitate RNA folding and structure, bind TERT, position the template in the active site, and/or direct RNP dynamics in the catalytic cycle.
One possible role of the ARC could be to promote proper RNA structure and/or folding. For instance, the ARC could be a structurally important platform for high-order organization of the RNP catalytic core. Our mutant design process included careful inspection of RNA folding predictions to avoid off-target RNA secondary structure changes (see Materials and Methods); however, if higher order structure exists, then it could certainly be affected by the mutations we have tested. Also, it is possible that the altered order of RNA sequence in the ARC-disrupting circular permutants could affect co-transcriptional folding, which would suggest that the ARC helps direct TLC1 folding. However, the ARC circular permutants have less perturbed sequences than other functional alleles, such as those in the loops of the pseudoknot. Additionally, the nucleotide substitution mutants reported herein show that none of the junctions’ sequences are critical for function (Figures 6 and 7). This suggests that there are no sequence-specific interactions contributed by any of the junctions, including J1 and J4, which are within the ARC. Nevertheless, it remains possible that the ARC could contribute important secondary and tertiary structures in a sequence-nonspecific manner.
It seems very likely that the ARC contains binding sites for TERT. The pseudoknot and both sides of the CEH have previously been implicated in binding Est2 (Livengood et al, 2002; Lin et al, 2004), and we show that these regions need to be physically linked via the ARC (Figure 9). The entire ARC could comprise a single, large, high-affinity TERT-binding site, or multiple lower affinity binding sites. If there are multiple binding sites, perhaps a higher affinity site provides anchoring, while weaker, transiently used sites may optimally position the template and change during the progression of the catalytic cycle (see below). Thus, when connectivity within the ARC is disrupted by circular permutations, the overall affinity of TLC1 for Est2 is likely to decrease.
Another exciting possible function of the ARC is physically coordinating telomerase RNA so that the template can be properly threaded into the active site of TERT. If a region of the ARC anchors TLC1 to Est2, then the ARC may guide the RNA along additional contact points on the surface of the protein to correctly position the template at the catalytic centre, which is located deep within the palm of the protein (Figure 9; Mitchell et al, 2010). In this case, disrupting the physical interactions through the ARC may still allow binding to Est2, but could prevent proper template positioning. If there is a series of interactions between the ARC and TERT to align the template in the active site, then these contacts are likely transient throughout the catalytic cycle to permit template translocation through the active site during both addition of a single repeat and repositioning the template for additional rounds of repeat addition.
Finally, the ARC could be more directly involved in the dynamics of the RNP during the catalytic cycle. This involves the template translocating through the active site during a single round of telomere repeat addition, as well as a large-scale repositioning of the template for additional rounds of repeat addition. Further investigation will be required to understand whether the ARC plays a role in these aspects of telomerase mechanism.
In summary, our in vivo and in vitro results define specific structural perturbations that are not tolerated within certain portions of the core of yeast telomerase RNA, suggesting that structure underlies function in these regions. The major example is the ARC, unveiled by circular permutations. Identification of the ARC provides a large-scale functional feature in telomerase and a foundation for determining how telomerase RNA coordinates its conserved elements with TERT to synthesize telomeric DNA. Reciprocally, we have also identified regions within the core yeast of telomerase RNA that have a high capacity for structural perturbations, suggesting that these may normally possess noteworthy flexibility. Ultimately, knowing where flexibility exists will help inform our understanding of telomerase mechanism and dynamics.
Materials and methods
Design of mutant TLC1 alleles
To guide design of mutant alleles in miniaturized versions of TLC1, we utilized the Mfold RNA structure prediction software (Zuker and Jacobson, 1998; Zuker, 2003). When designing rearranged and circularly permuted alleles of Micro-T and Mini-T, we required that each individual secondary structural element adopt the same predicted conformation as in wild type; that is, the template being single stranded, and the CEH and TBE being properly paired. Since Mfold is unable to predict pseudoknots and no pseudoknot prediction programme correctly predicts the S. cerevisiae TER pseudoknot, we required that this region adopt the stem loops predicted by Mfold. These stem-loop structures have been reported to be in equilibrium with the pseudoknot (Liu et al, 2012). All mutant sequences can be found in Supplementary Figure S1.
Plasmids and yeast
A complete list of plasmids used in this work can be found in Supplementary Table S1. For in vitro expression, Micro- and Mini-T mutants were cloned into a pUC19 vector with a T7 promoter followed by three guanosine nucleotides for efficient transcription initiation, and a FokI site following the 3′ end for linearization for run-off transcription. For in vivo expression, Mini-T mutants were amplified by PCR and cloned into the BglII and NsiI sites of pSD107 (pTLC1-TRP-CEN; Chapon et al, 1997). To make Sm site mutants, the BglII to NsiI fragment (upstream of the Sm site) was cut from the pSD107 derivatives and ligated into pAS501 (pTLC1sm−-TRP-CEN; Seto et al, 1999). To create 2μ expression versions, the BamHI to PstI fragment containing Mini-T was excised from pSD107 or pAS501 derivatives and cloned into pRS424. All in vivo tests were performed in strain yVL1009 (MATα tlc1-Δ::LEU2 rad52-Δ::LYS ura3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ circ+ pSD120/pTLC1-URA- CEN; Chappell and Lundblad, 2004).
In vitro telomerase activity assays
Telomerase assays were performed as previously described (Zappulla et al, 2005). Briefly, template DNA encoding telomerase RNA and Pro-A-Est2 (Art2-11) were added to a rabbit reticulocyte lysate (RRL) transcription and translation system (Promega), along with [35S]-methionine. Telomerase was immunopurified with IgG Sepharose beads (GE Healthcare). To assess protein production and immunopurification, 2.5 μl of purified beads were separated on a 7.5% Mini-PROTEAN TGX precast gel (Bio-Rad) and [35S-Met]-ProA-Est2 was detected by phosphorimager analysis (GE Healthcare). To assess telomerase activity, 5 μl of purified bead-bound telomerase were incubated with 1 μM telomeric substrate DNA primer (DZ428, 5′-GGTGTGGTGTGGG-3′), ∼1 nM [γ-32P]-labelled DZ428 internal recovery and loading control (LC) (5000, c.p.m./reaction), 3.3 μM [α-32P]-TTP, 1 μM each dATP, dCTP, and dGTP, 38 mM Tris pH 8, 47 mM NaCl, 4.7% glycerol, 2.3 mM MgCl2, 0.5 mM spermidine, and 0.5 mM DTT. Reactions were incubated at 26°C for 10 min, then stopped by addition of ammonium acetate, and products were ethanol precipitated and resuspended in 1 × formamide loading buffer. The DNA products were then electrophoresed through a 10% polyacrylamide/1 × TBE/7 M urea denaturing gel at 90 W for 1.25 h, exposed on a phosphorimager screen, and imaged using a Typhoon 9410 Variable Mode Imager. To calculate the relative telomerase activity, the total amount of signal in the 0 to +7 bands was divided by the amount of LC after background subtraction (ImageQuant TL). We used the averaged values from two independent biological replicates to assign mutants to the following reproducible bins of activity: ‘–’ no detectable telomerase activity, ‘+’ <50% wild-type activity, ‘++’ >50% wild-type activity.
Senescence assays
To test Mini-T RNA function in vivo, TRP1-marked CEN or 2μ expression plasmids were transformed into yeast strain yVL1009 by standard lithium acetate yeast transformation methods (Schiestl and Gietz, 1989). Transformants were plated on synthetic medium lacking uracil and tryptophan, and subsequently streaked on medium lacking tryptophan and containing 5-FOA to counterselect for the URA3-marked wild-type cover plasmid. To test for senescence, two independent yeast strains containing either CEN or 2μ versions of Mini-T(460) alleles as the sole copy of TLC1 were serially restreaked 10 times (corresponding to ∼275 generations) on solid medium lacking tryptophan at 30°C.
Northern blots
Northern blots were performed as previously described (Zappulla et al, 2005). Briefly, total cellular RNA was isolated from log-phase yeast cultures using a hot phenol method (Kohrer and Domdey, 1991). In all, 30–50 μg of total RNA was boiled in 1 × formamide loading buffer and electrophoresed through a 4% polyacrylamide/1 × TBE/7 M urea denaturing gel at 35 W for ∼3 h. RNA was electrotransferred onto Hybond-N+ nylon membrane (GE Healthcare) and UV-crosslinked (SpectroLinker XL-1500). After pre-hybridization in Church buffer, blotted RNA was probed using 1 × 107 c.p.m. of the StuI-NsiI region of Mini-T, labelled by incorporation of [α-32P]-dCTP into random hexamer-primed Klenow DNA polymerase products. As an internal control, blots were also probed with 1 × 105 c.p.m. of NME1.
Southern blots
Southern blots were performed as previous described (Zappulla et al, 2011). Genomic DNA was prepared from yeast cultures at the indicated time points, using the Puregene system (Qiagen). Genomic DNA was quantified and digested with XhoI overnight. Digested DNA was electrophoresed through a 1.1% agarose gel at 70 V for 17 h. DNA was transferred onto Hybond-N+ nylon membrane by capillary action and UV crosslinked. The blot was probed for telomeric DNA and an internal control fragment from chromosome IV (Seto et al, 1999).
Supplementary Material
Acknowledgments
Thank you to members of the Zappulla laboratory for insightful comments on this manuscript. We thank Tom Cech, Feng Qiao, and Vicki Lundblad for providing plasmids and yeast strains. We also thank the JHU Integrated Imaging Center. This work was supported by US National Institutes of Health funding from R00 GM80400 to DCZ as well as startup funds from The Johns Hopkins University.
Author contributions: MAM and DCZ designed the experiments. QR performed the Southern blotting; MAM performed all other experiments. MAM and DCZ analysed the experimental data. MAM and DCZ prepared the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Autexier C, Greider CW (1994) Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes Dev 8: 563–575 [DOI] [PubMed] [Google Scholar]
- Autexier C, Pruzan R, Funk WD, Greider CW (1996) Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J 15: 5928–5935 [PMC free article] [PubMed] [Google Scholar]
- Bachand F, Autexier C (2001) Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol Cell Biol 21: 1888–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L (1998) Reconstitution of human telomerase activity in vitro. Curr Biol 8: 177–180 [DOI] [PubMed] [Google Scholar]
- Berman AJ, Akiyama BM, Stone MD, Cech TR (2011) The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat Struct Mol Biol 18: 1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Collins K (2011) Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3: a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Zappulla DC, Glynn EF, Baumann P (2008) A flexible template boundary element in the RNA subunit of fission yeast telomerase. J Biol Chem 283: 24224–24233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y (2007) A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res 35: 6280–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C, Cech TR, Zaug AJ (1997) Polyadenylation of telomerase RNA in budding yeast. RNA 3: 1337–1351 [PMC free article] [PubMed] [Google Scholar]
- Chappell AS, Lundblad V (2004) Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol Cell Biol 24: 7720–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW (2000) Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Chen JL, Greider CW (2005) Functional analysis of the pseudoknot structure in human telomerase RNA. Proc Natl Acad Sci USA 102: 8080–8085 discussion 8077-8089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122 [DOI] [PubMed] [Google Scholar]
- Cohn M, Blackburn EH (1995) Telomerase in yeast. Science 269: 396–400 [DOI] [PubMed] [Google Scholar]
- Coy S, Volanakis A, Shah S, Vasiljeva L (2013) The Sm complex is required for the processing of non-coding RNAs by the exosome. PLoS One 8: e65606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandjinou AT, Levesque N, Larose S, Lucier JF, Abou Elela S, Wellinger RJ (2004) A phylogenetically based secondary structure for the yeast telomerase RNA. Curr Biol 14: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (2002) The Est1 subunit of Saccharomyces cerevisiae telomerase makes multiple contributions to telomere length maintenance. Genetics 162: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Cech TR (1999) Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev 13: 2863–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L (2009) Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15: 546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Patterson B (1988) Spliceosomal snRNAs. Annu Rev Genet 22: 387–419 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Evans SK, Weilbaecher RG, Lundblad V (2000a) The Est3 protein is a subunit of yeast telomerase. Curr Biol 10: 809–812 [DOI] [PubMed] [Google Scholar]
- Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V (2000b) Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc Natl Acad Sci USA 97: 6457–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer K, Domdey H (1991) Preparation of high molecular weight RNA. Methods Enzymol 194: 398–405 [DOI] [PubMed] [Google Scholar]
- Lebo KJ, Zappulla DC (2012) Stiffened yeast telomerase RNA supports RNP function in vitro and in vivo. RNA 18: 1666–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V (1996) Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Podlevsky JD, Marz M, Qi X, Hoffmann S, Stadler PF, Chen JJ (2013) Identification of purple sea urchin telomerase RNA using a next-generation sequencing based approach. RNA 19: 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Ly H, Hussain A, Abraham M, Pearl S, Tzfati Y, Parslow TG, Blackburn EH (2004) A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. Proc Natl Acad Sci USA 101: 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR (1994) Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev 8: 1984–1998 [DOI] [PubMed] [Google Scholar]
- Liu F, Kim Y, Cruickshank C, Theimer CA (2012) Thermodynamic characterization of the Saccharomyces cerevisiae telomerase RNA pseudoknot domain in vitro. RNA 18: 973–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR (2002) Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol Cell Biol 22: 2366–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V, Szostak JW (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633–643 [DOI] [PubMed] [Google Scholar]
- Ly H, Blackburn EH, Parslow TG (2003) Comprehensive structure-function analysis of the core domain of human telomerase RNA. Mol Cell Biol 23: 6849–6856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DX, Goneska E, Greider CW (2003) Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol 23: 5606–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP (1995) Ciliate telomerase RNA structural features. Nucleic Acids Res 23: 1091–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP (1996) A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol Cell Biol 16: 1871–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MC, Collins K (2002) Telomerase recognizes its template by using an adjacent RNA motif. Proc Natl Acad Sci USA 99: 6585–6590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E (2010) Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol 17: 513–518 [DOI] [PubMed] [Google Scholar]
- Mozdy AD, Cech TR (2006) Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR (2013) Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 2: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JF, Larose S, Abou Elela S, Wellinger RJ (2012) Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res 40: 5625–5636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Stellwagen AE, Diede SJ, Singer MS, Haimberger ZW, Johnson CO, Tzoneva M, Gottschling DE (2001) The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat Genet 27: 64–67 [DOI] [PubMed] [Google Scholar]
- Pfingsten JS, Goodrich KJ, Taabazuing C, Ouenzar F, Chartrand P, Cech TR (2012) Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell 148: 922–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ (2008) The telomerase database. Nucleic Acids Res 36: D339–D343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li Y, Honda S, Hoffmann S, Marz M, Mosig A, Podlevsky JD, Stadler PF, Selker EU, Chen JJ (2012) The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res 1: 450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F, Cech TR (2008) Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH (1991) A conserved secondary structure for telomerase RNA. Cell 67: 343–353 [DOI] [PubMed] [Google Scholar]
- Schiestl RH, Gietz RD (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet 16: 339–346 [DOI] [PubMed] [Google Scholar]
- Seto AG, Umansky K, Tzfati Y, Zaug AJ, Blackburn EH, Cech TR (2003) A template-proximal RNA paired element contributes to Saccharomyces cerevisiae telomerase activity. RNA 9: 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR (1999) Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Shefer K, Brown Y, Gorkovoy V, Nussbaum T, Ulyanov NB, Tzfati Y (2007) A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol 27: 2130–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE (2003) Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL (2011) Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem 286: 26431–26439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer VM, Ford LP, Holt SE, Frank BC, Yi X, Aisner DL, Ouellette M, Shay JW, Wright WE (1999) Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol 19: 6207–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfati Y, Fulton TB, Roy J, Blackburn EH (2000) Template boundary in a yeast telomerase specified by RNA structure. Science 288: 863–867 [DOI] [PubMed] [Google Scholar]
- Ulyanov NB, Shefer K, James TL, Tzfati Y (2007) Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates. Nucleic Acids Res 35: 6150–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA (2008) Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol 15: 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA (2012) Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev 26: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev 8: 652–665 [DOI] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 17: 498–502 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Zakian VA (2012) Everything you ever wanted to know about Saccharomyces cerevisiae telomeres: beginning to end. Genetics 191: 1073–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ (2008) Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem 283: 2049–2059 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR (2004) Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci USA 101: 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR (2006) RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb Symp Quant Biol 71: 217–224 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR (2005) A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol 12: 1072–1077 [DOI] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich KJ, Arthur JR, Gurski LA, Denham EM, Stellwagen AE, Cech TR (2011) Ku can contribute to telomere lengthening in yeast at multiple positions in the telomerase RNP. RNA 17: 298–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Jacobson AB (1998) Using reliability information to annotate RNA secondary structures. RNA 4: 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.