Abstract
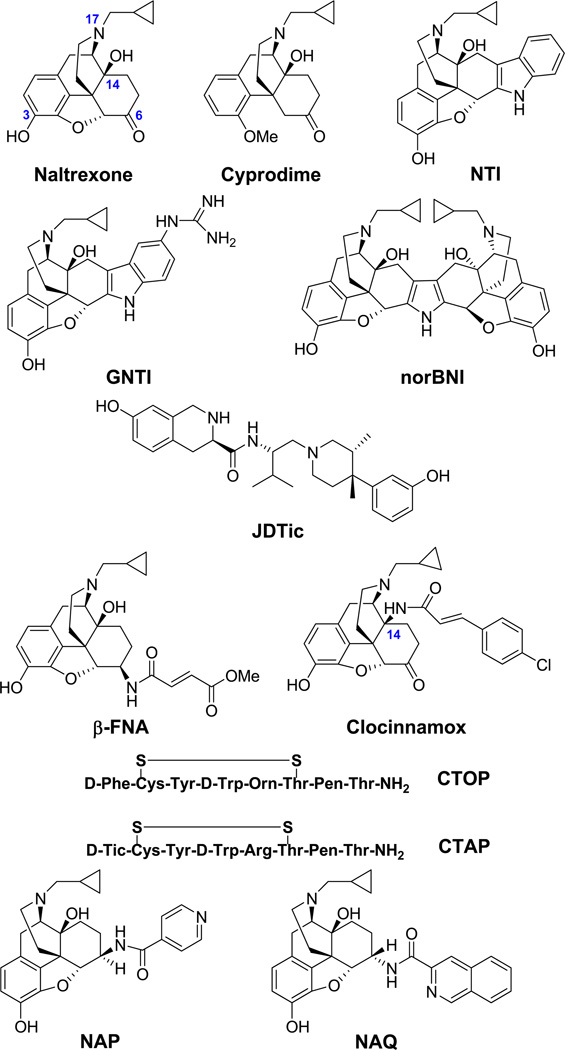
Highly selective opioid receptor antagonists are essential pharmacological probes in opioid receptor structural characterization and opioid agonist functional studies. Currently, there is no highly selective, non-peptidyl and reversible mu opioid receptor antagonist available. Among a series of naltrexamine derivatives that have been designed and synthesized, two compounds, NAP and NAQ, were previously identified as novel leads for this purpose based on their in vitro and in vivo pharmacological profiles. Both compounds displayed high binding affinity and selectivity to the mu opioid receptor. To further study the interaction of these two ligands with the three opioid receptors, the recently released opioid receptor crystal structures were employed in docking studies to further test our original hypothesis that the ligands recognize a unique “address” domain in the mu opioid receptor involving Trp318 that facilitates their selectivity. These modeling results were supported by site-directed mutagenesis studies on the mu opioid receptor, where the mutants Y210A and W318A confirmed the role of the latter in binding. Such work not only enriched the “message-address” concept, also facilitated our next generation ligand design and development.
Keywords: Mu opioid receptor, selective antagonists, crystal structures, automated docking, site-directed mutagenesis, GPCR
Introduction
Because receptor-selective opioid antagonists are vital tools for identifying the receptor types involved in interactions with selective opioid agonists, such antagonists have played very important roles in the study of opioid receptors. In general, an agonist interaction is characterized as opioid receptor-mediated only if its effect is competitively inhibited by an opioid antagonist.1–3 More specifically, characterization of the mu opioid receptor (MOR) structure–function relationship is essential because the analgesic effect, addictive properties, and notorious side effects (such as addiction/abuse liability, respiratory depression, and constipation) of the key drug morphine are abolished in MOR knock-out mice, indicating that these side effects are primarily due to its interaction with the MOR.4–6 Yet, the lack of a non-peptidyl, highly selective, reversible, and potent MOR antagonist limits our understanding of the structure–function relationship of the MOR, the interaction of nonpeptidyl MOR agonists with it, and more significantly, the activation mechanism of the receptor with respect to its role in drug abuse and addiction.
As one of the most serious chronic and relapsing medical disorders, heroin and prescription opioid abuse and dependence are very common, and still increasing.7 Another serious substance addiction disease is alcoholism. It is reported that disorders related to alcohol abuse affect 7–8% of Americans at any given time, or about 15 to 20 million adults, while these disorders account for $185 billion in U.S. health care costs, lost wages, bodily injury, and property damage annually.8–9 Some antagonists with relatively low selectivity for the MOR, e.g., naltrexone and naloxone, have been shown to be able to block relapse and curb drug craving in opiate addicts as well as to treat alcoholism.10–15 Although these antagonists have shown promise in these treatments, some severe side effects have been reported, likely due to their lack of selectivity. For example, patients receiving naltrexone for opioid dependence exhibited higher than expected rates of overdose and suicide, and some reports have linked this drug with depression and dysphoria.16–18 Evidence has accumulated that the delta opioid receptor (DOR) may be connected to mood-related behavior,19–21 while the kappa opioid receptor (KOR) may play an important role in the inhibition of glycinergic neurotransmission to cardiac vagal neurons.22 Therefore, a new antagonist with drug-like properties and high selectivity for the MOR may be a very promising medication for treatment of drug addiction and alcoholism.
Based on the “message–address concept”,23,24 highly selective non-peptide antagonists for the KOR (e.g., norBNI and GNTI)25,26 and for the DOR (e.g., NTI)27 were designed and synthesized more than two decades ago. These compounds are widely used as selective ligands in pharmacological studies. Thus far, however, no optimal antagonist has been developed for the MOR, though some moderately potent ligands, e.g., cyprodime,28 are in use. Compared with the high selectivity of GNTI26 for the KOR (Ki ratios: MOR/KOR≈120, DOR/KOR≈250) and NTI27 for the DOR (Ki ratios: MOR/DOR≈152, KOR/DOR≈276), cyprodime29 has only moderate selectivity for the MOR (Ki ratios: KOR/MOR≈45, DOR/MOR≈40). Another drawback of cyprodime is that it shows much lower affinity for the MOR than naloxone and naltrexone, which limits its application. Also, while selective, but irreversible, antagonists for the MOR such as β-FNA, clocinnamox and others have been reported,30–31 their potential covalent binding with the receptor limits their utility: reversible antagonists are usually preferred because they can temporarily “knock out” receptors for pharmacological studies, and then be washed out from the binding locus to “revive” the receptors.
Most of the highly selective and reversible MOR antagonists currently available are conformationally constrained peptides, e.g., d-Phe-Cys-Tyr-d-Trp-Orn-Thr-Pen-Thr-NH2 (CTOP) and d-Phe-Cys-Tyr-d-Trp-Arg-Thr-Pen-Thr-NH2 (CTAP). They are relatively stable metabolically and have been used to target the MOR with in vitro and in vivo studies.32 However, their limited bioavailability, i.e., relatively poor ability to cross the blood–brain barrier, render them not generally suitable for many types of in vivo studies and certainly not suitable for medical applications.33,34 The utility of an antagonist as a pharmacological tool is significantly enhanced if it has both in vitro and in vivo activity; thus, non-peptide ligands are preferred for their better ability to penetrate the CNS and for their lesser vulnerability to metabolic inactivation. Therefore, the development of a non-peptide, potent, selective and reversible antagonist for the MOR remains highly desirable.
We recently reported a series of novel ligands that were designed based on our homology modeling of the three opioid receptor subtypes.35 They were experimentally characterized as MOR selective antagonists in the in vitro and in vivo studies. In particular, two compounds (NAP and NAQ; Figure 1) showed predominant binding affinity to the MOR over both the DOR and the KOR, and possessed only marginal agonist efficacy at the MOR in the radioligand binding assays. In calcium flux functional assays, either ligand showed any significant agonist activity compared to the MOR full agonist DAMGO while in the DAMGO agonism inhibitory activity studies, NAP showed IC50 at 19.5 ± 5.5 nM and NAQ at 150 ± 9.4 nM. Such results were in line with their radioligand binding affinity though at a relatively lower level. Therefore, these two compounds are defined as our leads for further development of non-peptidyl MOR antagonists. For the next stage of molecular design, an understanding of the interaction of these two leads with the opioid receptors and the resulting MOR selectivity at an atomic level is critical. The many recent depositions of high-resolution GPCR crystal structures, including opsin, the human β2- and β1-adrenergic receptors, the human A2A adenosine receptor, chemokine receptor CXCR4, dopamine D3 receptor, sphingosine 1-phosphate receptor 1 and histamine receptor H1, among others,36 has transformed structure-based drug discovery for GPCR targets. The release of three opioid receptor subtype (MOR, KOR and DOR) crystal structures37–39 last year was one of the most exciting breakthroughs in opioid receptor research field in decades. Here, we report docking studies of NAP and NAQ into these three experimental structures, combined with primary site-directed mutagenesis studies that validate the modeling observations. These results facilitate our understanding of the receptor–ligand interactions involved in the observed MOR selectivity and will inform our future work.
Figure 1.
Representative opioid receptor-selective antagonists.
Results and Discussion
Sequence alignment analyses of three opioid receptors
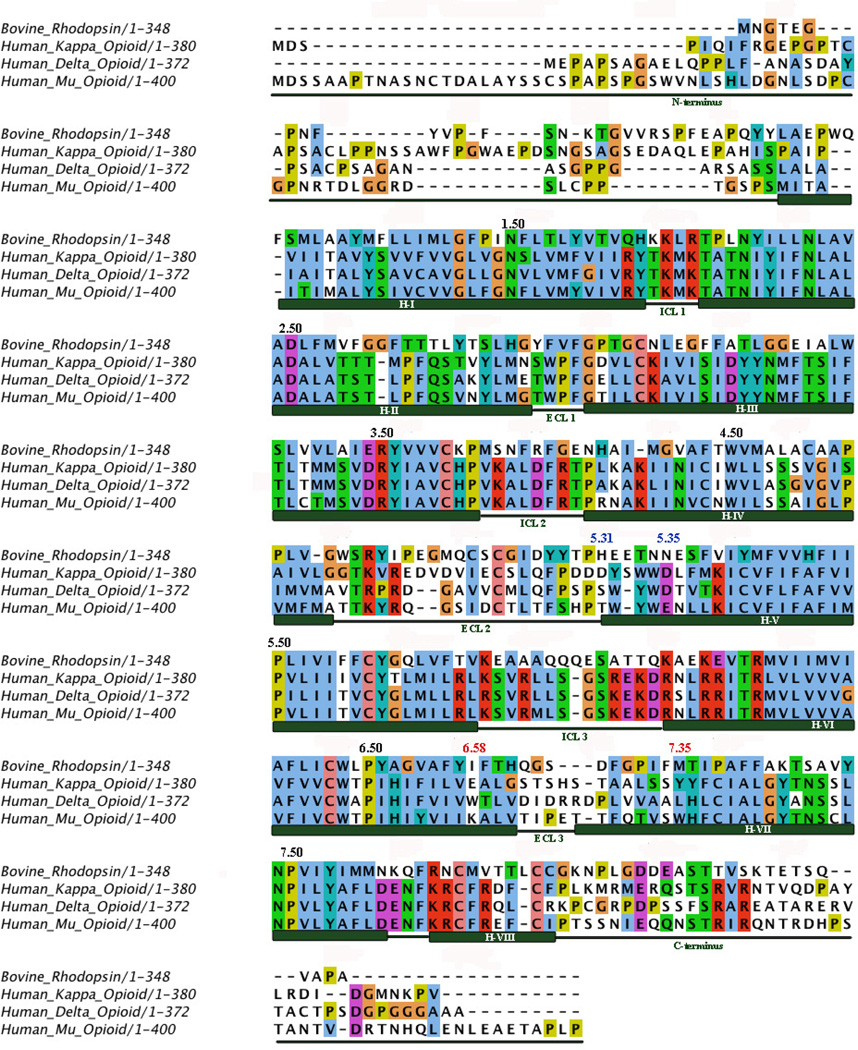
In our original efforts to conduct structure-based design of novel ligands as selective MOR antagonists, we adopted homology modeling methods simply because there were no x-ray crystal structures for any of the opioid receptors, and in fact, any G-protein coupled receptors (GPCR) other than bovine rhodopsin. Analysis of sequence alignments of all three opioid receptors along with bovine rhodopsin (Figure 2) not only provided us the three dimensional conformation motif, but also revealed that: 1) the three human opioid receptors share very high homology (over 60% sequence identity); 2) a generally higher sequence identity is observed for the ligand binding pockets believed to be formed primarily by transmembrane (TM) helices 2, 3, 6 and 7 (the so-called “message” domain of the receptor), and this is in line with the similar structural features of many opioid receptor ligands (Figure 1) representing the “message” moiety of these ligands; 3) an even higher identity (close to 90%) is seen for the intracellular loop (ICL) regions, which is because the three opioid receptors share the same family of G-proteins (Gi/o) for signal transduction, and the G-protein binding domain of the receptor is mainly on the ICL loci; and 4) much lower sequence identities were observed in the extracellular loop (ECL) regions, and for both N- and C termini. More strikingly, ECL3 of the three opioid receptors carried the lowest sequence identity of all domains (Table 1). The location of ECL3 directly above the “message” region of the binding site helped us to define a potential “address” domain in the MOR, which we used in designing our MOR-selective antagonists.35 Our ability, now, to compare the three opioid receptor crystal structures by domain gives us nearly identical results, which is further validation for our original molecular design strategy.
Figure 2.
Sequence alignment of opioid receptors and bovine rhodopsin. Ballesteros–Weinstein indices are indicated for the most conserved residues in each trans-membrane helix (black), important residues for Site 1 (red) and important residues for Site 2 (blue).
Table 1.
Comparison of the amino acid sequence identity among opioid receptors by domain.
| Percent sequence identity | |||
|---|---|---|---|
| Domain | Delta/mu | Delta/kappa | Mu/kappa |
| TM1 | 69 | 62 | 62 |
| TM2 | 95 | 86 | 82 |
| TM3 | 90 | 95 | 100 |
| TM4 | 43 | 57 | 33 |
| TM5 | 85 | 77 | 77 |
| TM6 | 73 | 64 | 73 |
| TM7 | 78 | 82 | 86 |
| EL1 | 73 | 67 | 67 |
| EL2 | 52 | 52 | 30 |
| EL3 | 21 | 13 | 21 |
| IL1 | 90 | 90 | 100 |
| IL2 | 91 | 95 | 91 |
| IL3 | 86 | 81 | 86 |
| N-terminus | 28 | 33 | 18 |
| C-terminus | 40 | 32 | 35 |
| Entire protein | 62 | 61 | 57 |
Small molecule construction and conformational analysis
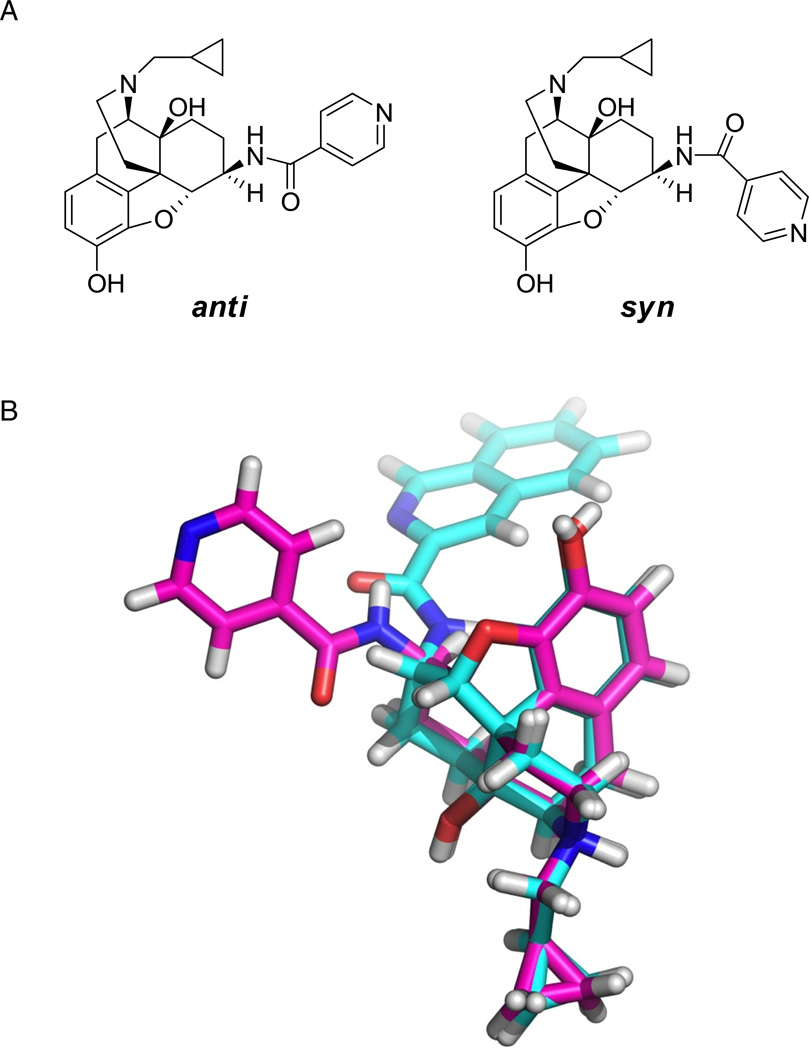
Models for the small molecules NAP, NAQ and naltrexone (NTX) were built prior to conformational analysis of the first two. Then, brief dynamics simulations were conducted for NAP and NAQ within a periodic water box. Due to the partial double bond in the amide linkage of the two compounds, they adopted either an “anti” or “syn” conformation, as shown for NAP (Figure 3). The averaged conformations measured from the last 10 ns of dynamics (of the total 100 ns) for both ligands were predominantly anti, which is expected to be more thermodynamically favorable of the two conformations.
Figure 3.
A) Representation of “anti” and “syn” conformations for NAP. B) Conformational analysis results for NAP (magenta) and NAQ (cyan). Superimposed average structure for last 10 ns of a 100 ns NVT dynamic simulation inside a water box with PBC. Both NAP and NAQ preferred “anti” conformation over “syn”.
Docking studies of the opioid universal antagonist NTX
Previously, we used NTX as a probe to propose the “address” binding domain within the MOR antagonist binding pocket with homology-derived models of the three opioid receptors.35 Similar regions have been implicated as the binding loci for various peptide and non-peptide ligands.40–42 In the present study, NTX was docked in a similar fashion within a pocket formed by helices 3, 6 and 7 in each receptor X-ray crystal structure using the GOLD43 docking program. The conformations of the ligands co-crystallized in the receptors were used as starting points for docking. In this way, the docking problem was transformed into the much simpler issue of energetically placing just the 6-position side chain NTX, as there is a common core for it and the ligands bound in the crystal structures; β-funaltrexamine in the MOR, naltrindole in the DOR and JDTic in the KOR (see Figure 1). This guided docking of NTX was followed by energy minimization to optimize the structural models for the ligand–receptor complexes. Two scoring systems were used to quantitatively characterize the binding poses. The CHEM-PLP score, the default scoring function of GOLD, which has been optimized for modeling steric complementarity between ligand and protein along with distance- and angle-dependent hydrogen bonding, was used to obtain plausible docking poses. The obtained poses were then rescored with HINT (Hydropathic INTeractions), an empirical free energy scoring tool based on the experimental measurements of logP for 1-octanol and water,44 to estimate atomic level free energies associated with non-covalent interactions.45,46 Optimal docking poses for each NTX–receptor complex were chosen by the highest CHEM-PLP and HINT scores, which were, in this case, generally in agreement (Table 3).
Table 3.
The GOLD (CHEM-PLP) and HINT scores for the best binding poses in the three opioid receptors.
| Compounds | Docking posea |
MOR | KOR | DOR | |||
|---|---|---|---|---|---|---|---|
| CHEM-PLP | HINTb | CHEM-PLP | HINTb | CHEM-PLP | HINTb | ||
| NTX | 79 | 1140 | 69 | 1009 | 85 | 1035 | |
| NAP | Site 1 | 85 | 1535 | 46 | 769 | 89 | 1263 |
| Site 2 | 88 | 1022 | 78 | 489 | 93 | 1034 | |
| NAQ | Site 1 | 80 | 1345 | 50 | −344 | 75 | 465 |
| Site 2 | 82 | 1125 | 56 | 616 | 73 | −10 | |
Clustering of docking poses revealed two unique binding modes. See text for more information.
HINT score differences have generally been shown to relate to ΔΔG such that 515 score units ≈ −1 kcal/mol.
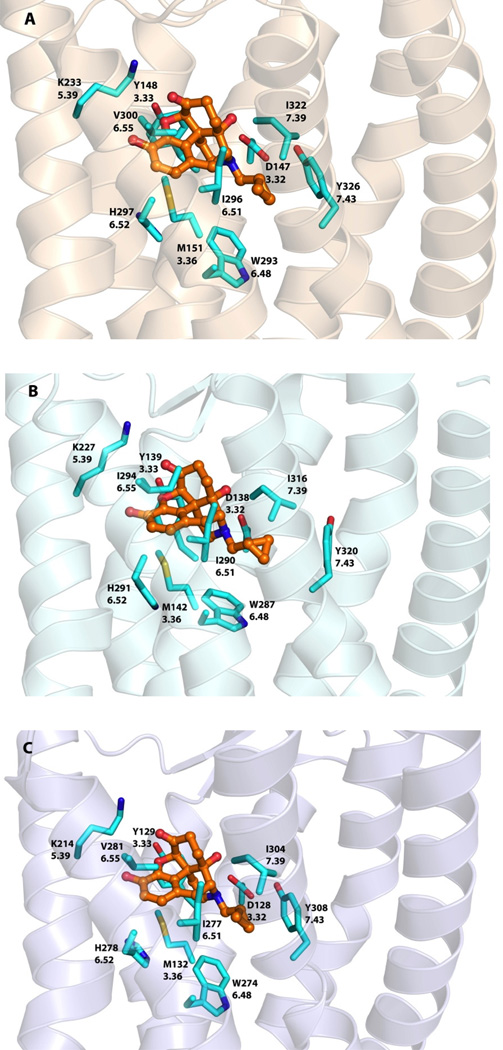
Amino acid residues shown to interact with the ligand were highly conserved among the three opioid receptors (Figure 4). Asp3.32 was involved in an ionic interaction with the 17-position tertiary amino group of the ligand while Tyr3.33 formed a hydrogen bond with the dihydrofuranyl oxygen (superscripts “x.yy” refer to Ballesteros–Weinstein numbering48). The orientation of the 3-phenolic hydroxyl group also indicated the likelihood of a hydrogen bonding interaction with His6.52 through two water molecules, as seen in MOR and KOR crystal structures. Lys5.39 may be involved in hydrogen bonding interactions with the 6-position carbonyl oxygen atom and Met3.36, Trp6.48, Ile6.51, Val/Ile6.55, Ile7.39 and Tyr7.43 formed a hydrophobic pocket to accommodate the morphinan skeleton of the molecule.
Figure 4.
Docked poses of NTX (orange carbon atoms) inside three opioid receptor crystal structures: A) MOR B) KOR and C) DOR. Amino acid residue atoms: carbon (cyan), oxygen (red), nitrogen (blue), sulfur (yellow).
Docking studies of NAP and NAQ
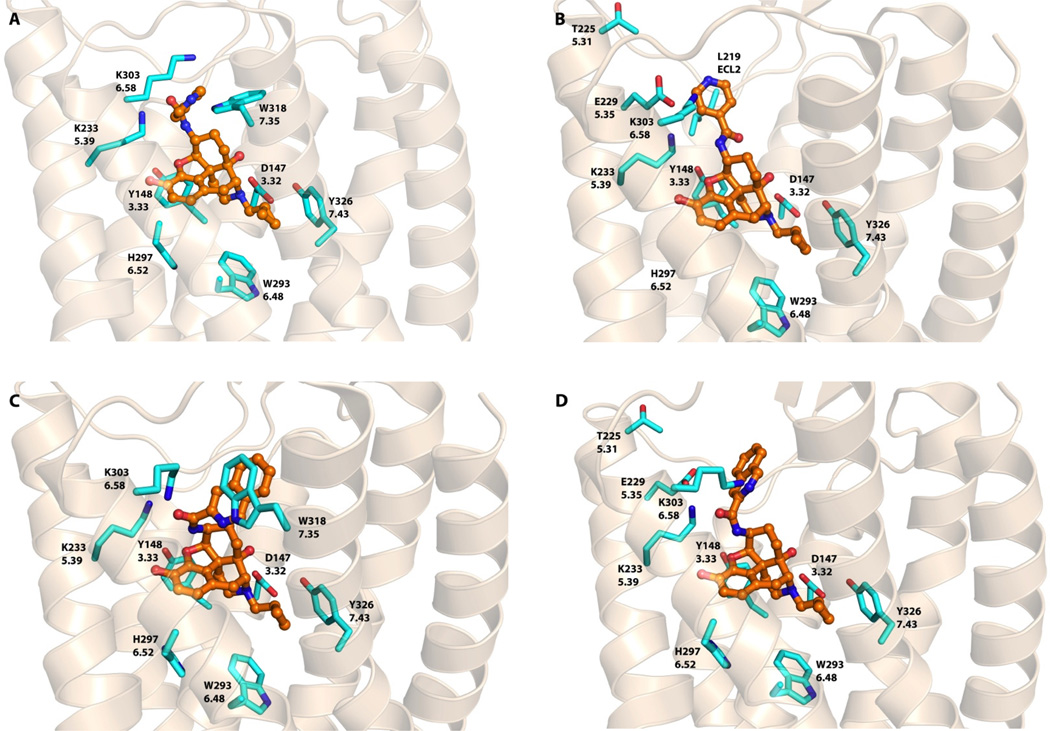
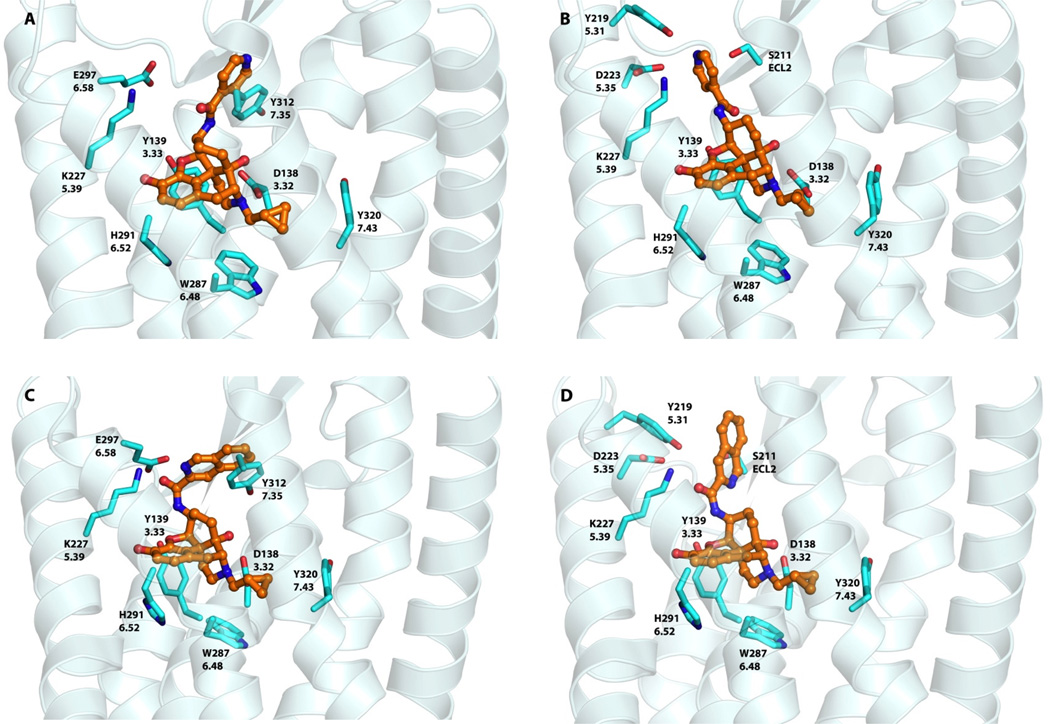
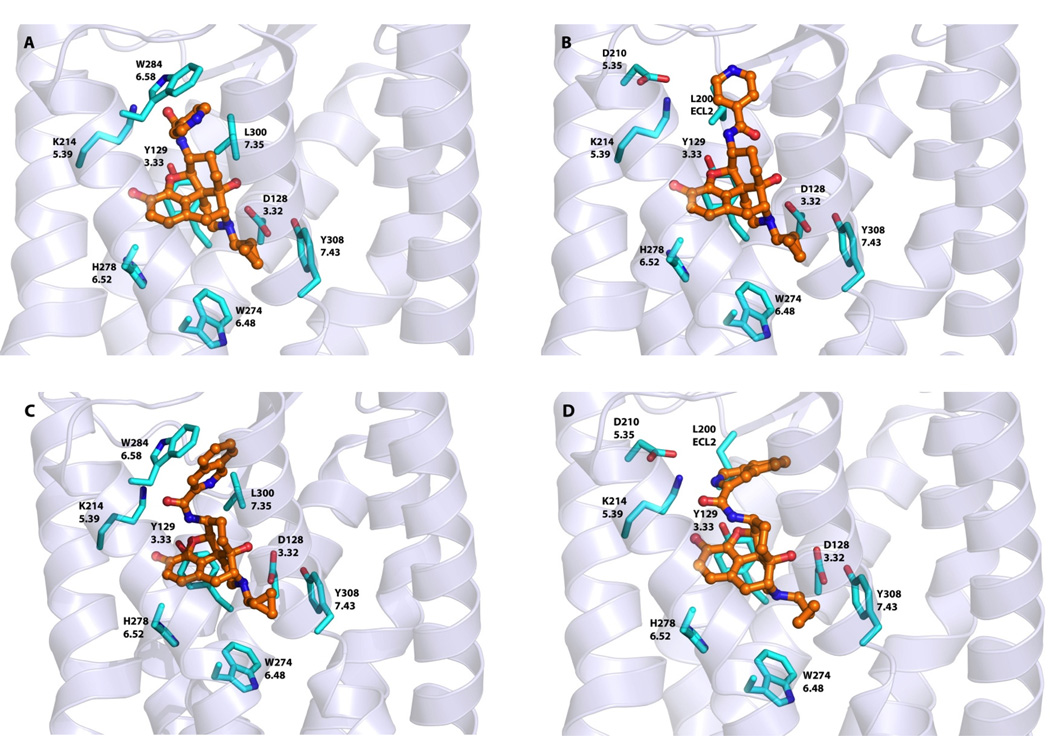
Among a series of novel ligands designed and synthesized to target the “address” domain of the MOR, NAP and NAQ have sub-nanomolar affinity for the MOR and high selectivity over the DOR and the KOR from in vitro radioligand competition binding studies. Both showed only marginal partial agonism in the in vitro GTPγS assay and potent antagonism in an in vivo antinociceptive test against morphine.35 Our initial molecular modeling study suggested that the MOR selectivity for these two ligands could be the result of the previously identified hydrogen bonding between the ligand and the MOR “address” binding locus.35 To further understand how these two lead compounds can have similar affinities and selectivities for the MOR, despite significant chemical differences in their side chain structures, we revisited our docking study by employing the x-ray crystal structure models for the opioid receptors, again using GOLD,45 and the conformations of the co-crystallized ligands as starting points. As with NTX, there is a common core for our ligands and those in the three crystal structures. This was followed by energy minimization of the NAP and NAQ ligand receptor complex models. The results of the docking experiments are presented in Figures 5, 6 and 7.
Figure 5.
Docked poses of NAP and NAQ in the mu opioid receptor. A) NAP in MOR binding Site 1. B) NAP in MOR binding Site 2. C) NAQ in MOR binding Site 1. D) NAQ in MOR binding Site 2. NAP and NAQ atoms: carbon (orange); amino acid residue atoms carbon: (cyan), oxygen (red), nitrogen (blue), sulfur (yellow).
Figure 6.
Docked poses of NAP and NAQ in the kappa opioid receptor. A) NAP in KOR binding Site 1. B) NAP in KOR binding Site 2. C) NAQ in KOR binding Site 1. D) NAQ in KOR binding Site 2.
Figure 7.
Docked poses of NAP and NAQ in the delta opioid receptor. A) NAP in DOR binding Site 1. B) NAP in DOR binding Site 2. C) NAQ in DOR binding Site 1. D) NAQ in DOR binding Site 2.
Analysis of the NAP and NAQ morphinan backbone binding site
As noted above, all three opioid receptors share a very high degree of sequence similarity in the “message” region of ligand-binding pocket. As a result, the morphinan nucleus of NAP and NAQ were both found to be docked within each receptor in a similar fashion as NTX. The composition of the binding pockets is, in fact, practically identical. In other words, the “message” component of the ligands occupied the “message” domain in the receptor. On the other hand, docking results showed that the side chains of the ligands, i.e., the “address” component of the ligands, primarily clustered around two different binding sites in the three opioid receptors: Site 1 located at the top of helix 6 and 7 (including part of ELC3) and Site 2 at the top of helix 5 and ECL2 (see Figures 5–7).
NAP and NAQ in the MOR
Clustering of the docked poses for NAP in the MOR revealed two high scoring pose families related to the two “address” sites suggested above (see Table 3). For one pose, NAP adopted a “syn” conformation at the amide linkage with the pyridinyl substituent pointing towards Site 1, where the side chain could stack with Trp3187.35 with π-π interactions in addition to a possible hydrogen bond (Figure 5A). Also, Lys3036.58 may form a hydrogen bond with the pyridinyl nitrogen of NAP. For the second pose, the ligand indicated its amide linkage to be in the “anti” conformation with the pyridinyl side chain placed in a hydrophobic pocket (Site 2) at the top of helix 5 and near ECL2 (Figure 5B). However, the presence of Glu2295.35, which is close to the pyridinyl side chain, led to a highly negative (i.e. unfavorable) interaction that is reflected by the lower HINT score for this pose (Table 3). Thus, it is likely that NAP prefers interaction with Site 1. NAQ followed a similar pattern in its docking solutions, except that, in both binding sites, the ligand adopted the “syn” conformation. As with NAP in Site 1, the quinolinyl side chain of NAQ appears to interact with Trp3187.35 with π-π stacking, while in Site 2 (Figure 5C), the side chain has an unfavorable interaction with Glu2295.35 (Figure 5D). Meanwhile, Lys3036.58 may also possibly form a hydrogen bond in both binding sites, not with the quinolinyl nitrogen atom of NAQ, but with the spacer portion of the molecule.
NAP and NAQ in the KOR
As in the MOR, docking of NAP in the KOR gave two favorable binding poses. However, in both sites of the KOR, the ligand adopts only the “anti” conformation to achieve high docking scores. In Site 1, although the aromatic side chain may interact with Tyr3127.35 with π-π interactions, the presence of Glu2976.58 in place of Lys3036.58 of the MOR appears to cause deleterious interactions with the ligand side chain (Figure 6A). In Site 2 of the KOR, the Asp2235.35 is one carbon shorter compared to the analogous Glu2295.35 of the MOR, which results in lessened unfavorable interactions with the NAP side chain (Figure 6B). Moreover, the presence of Tyr2195.31 and Ser211 (ECL2) may result in more favorable hydrogen bonding interactions with the nitrogen atom of the side chain. Based on these results, NAP likely adopts Site 2 in the KOR. In contrast, NAQ seems to prefer a “syn” conformation in both its KOR docking poses (Figures 6C, D). The larger hydrophobic group (quinolinyl side chain) of NAQ may result in even more significant hydropathic incompatibility with Glu2976.58, probably negating the thermodynamically favorable aromatic π- stacking interactions of that side chain with Tyr3127.35 at Site 1. Also, HINT scores for NAQ binding in Site 2 (Table 3) indicated no negative interactions with Asp2235.35, while allowing for the possibility of hydrogen bonding between the side chain and Tyr2195.31 and/or Ser211 (ECL2).
NAP and NAQ in the DOR
For docking NAP in the DOR, the ligand adopted a “syn” conformation in Site 1 with the side chain stacked in a hydrophobic pocket formed by Trp2846.58 and Leu3007.35 (Figure 7A), while in Site 2 it was in an “anti” conformation with the pyridinyl side chain placed in a hydrophobic pocket close to the top of helix 5 and ECL2 (Figure 7B). Notably, these putative interactions failed to utilize any of the hydrogen bonding observed in the MOR and the KOR models, which may offer a reason for the lower affinity of NAP for the DOR. NAQ, except for a tendency for the “syn” conformation, adopted nearly identical binding poses as NAP.
Site-directed Mutagenesis
The above results indicated that plausible hydrogen bonding and aromatic stacking interactions between the “address” portions of the two lead compounds and TM7 residues of the MOR may be responsible for their high binding affinity to the MOR vs. the DOR and KOR. In particular, the role of Trp3187.35 seemed to be the most critical. This conclusion is in partial agreement with our earlier homology model-based docking studies for these receptors,35 which suggested possible roles for both Trp3187.35 and Tyr210 (ECL2) in imparting ligand selectivity by recognizing their “address” portion specifically. To further verify the roles of these two residues in the MOR receptor “address” domain for the MOR selectivity of the NAP and NAQ leads, we performed a site-directed mutagenesis study with a transient Chinese hamster ovary (CHO) cell line transfected with wild type and mutated MORs. In these studies, either Trp3187.35 or Tyr210 in MOR was mutated to alanine, and NTX was used as a control. As shown in Table 4, the binding affinities of the NTX control were largely unaffected for either mutated receptor compared to their wild types, which supported the observation from our docking studies that the common “message” portion of the ligands recognized the “message” domain of the receptor to achieve their opioid receptor function. On the other hand, both NAP and NAQ bound to the Y210A mutant with affinities comparable to those of wild type MOR, while their affinities were dramatically decreased for the W318A mutant. These results are in agreement with the docking results described above that indicated the preference of the aromatic NAP and NAQ side chains to π-stack and hydrogen bond with Trp3187.35 of the MOR in Site 1 over Tyr210 (ECL 2) in Site 2. The docking scores of NTX, NAP and NAQ in models of the mutant MOR structures were consistent with experimental results. Together, all of these observations validate our hypothesis that these two leads may recognize an alternative “address” locus in the MOR to confer their selectivity for the mu over the delta and the kappa opioid receptors.
Table 4.
Binding of ligands to site-directed mutated MORs.
| Cmpd | wild type MOR (nM) ± SEM | Y210A MOR (nM) ± SEM | W318A MOR (nM) ± SEM | |||
|---|---|---|---|---|---|---|
| IC50 | Ki | IC50 | Ki | IC50 | Ki | |
| NTX | 3.90 ± 2.96 | 1.85 ± 1.41 | 0.95 ± 0.49 | 0.45 ± 0.23 | 10.35 ± 1.64 | 4.91 ± 0.78 |
| NAP | 2.29 ± 0.15 | 1.09 ± 0.07 | 1.61± 0.17 | 0.77 ± 0.08 | >1000 | NDa |
| NAQ | 5.42 ± 0.70 | 2.57 ± 0.33 | 3.31 ± 1.71 | 1.57 ± 0.81 | >1000 | NDa |
Not determined.
Conclusions
Highly selective opioid receptor antagonists are essential pharmacological probes in opioid receptor structural characterization. We hypothesized that the two novel naltrexamine derivatives NAP and NAQ, previously identified as leads for development as MOR antagonists based on their in vitro and in vivo pharmacological profiles, which included high binding affinity and selectivity for the MOR over the DOR and the KOR, may recognize a unique “address” domain in the MOR that facilitates their selectivity for the receptor. They, along with NTX, were docked in the recently released crystal structures of three antagonist-bound opioid receptors. The modeling results suggest how these novel naltrexamine derivatives may bind to the three opioid receptors to maintain morphinan-like “message” interactions, while their 6-position side chains act as the “address” moiety. This latter component interacts with amino acid residues that are non-conserved among the opioid receptors to confer their selectivity for one receptor over others. Furthermore, these studies also suggest that there may be at least two “address” sites in the receptors, and that the preference of naltrexamine derivatives for these sites depends on the type of opioid receptor. Further site-directed mutagenesis studies of the MOR support the model by proving the importance of Trp3187.35 in the “address” domain. Application of the “message–address” concept, combined with molecular modeling, site-directed mutagenesis and targeted synthesis, indeed helped us in designing and developing more selective ligands for the mu opioid receptor with different pharmacological profiles more recently.49 The same would be expected for other receptor selective ligands design and discovery in the future.
Materials and methods
Sequence analysis and model building
The sequences of human opioid and bovine rhodopsin receptors were obtained from Universal Protein Resource (UniProt) (Entry code: P35372 (MOR), P41145 (KOR), P41143 (DOR) and P02699 (Bovine rhodopsin receptor). The X-ray crystal structures for MOR (4DKL), KOR (4DJH) and DOR (4EJ4) were retrieved from PDB Data Bank at http://www.rcsb.org. Molecular models were built for each receptor in SYBYL-X 2.0; hydrogens were added, Gasteiger–Hückel charges were assigned, and their positions were optimized while holding all heavy atoms fixed as an aggregate with the Tripos force field (TAFF).
Docking studies
Within SYBYL-X 2.0, the chemical structures of the two lead MOR ligands (NAP and NAQ) and the universal ligand NTX were sketched, and their Gasteiger–Hückel charges were assigned before energy minimization (10,000 iterations) with the TAFF. The genetic algorithm docking program GOLD 5.1 was used to perform the docking studies with standard default settings unless otherwise specified. The binding site was defined to include all atoms within 10 Å of the γ-carbon atom of Asp3.32 for the three opioid crystal structures. Automated docking was performed with a distance constraint of 4 Å between the piperidine nitrogen of the ligands’ morphinan nucleus and Asp3.32, and between the ligands’ dihydrofuran oxygen and the phenolic oxygen of Tyr3.33. Based on the fitness scores and the binding orientation of each ligand within the binding cavity, the best GOLD-docked solution was selected and merged into the receptor. The combined receptor–ligand structures were energy-minimized using the parameters described above in order to remove clashes and minimize strain energy, thus optimizing the interactions between ligand and receptor within the binding pocket. These models were then optimized as previously described45,46,47 and subjected to hydropathic analysis with the HINT program.44
Conformational analysis
The NAP and NAQ structures were solvated with a water box. The water box including the ligand was again minimized under conditions similar to those described above. Next, an NVT molecular dynamics simulation was performed in SYBYL-X 2.0 for 100 ns with periodic boundary conditions with a 2 fs time-step. The energy-minimized average for the last 10 ns of the simulation for both ligands is shown.
Site-directed mutagenesis
Single point mutations were introduced into the MOR cDNA, expressed in pcDNA3.1, by mutant strand synthesis reactions through thermal cycling (QuikChange™ Site-Directed Mutagenesis Kit, Stratagene). The mutation was confirmed by DNA sequencing, performed by the DNA Core facility at VCU. The resulting mutant cDNA constructs (in pcDNA 3.1) were transfected into CHO-K1 cells using Lipofectamine. After 48 h, transient transfected cells were harvested and membranes prepared. Receptor expression levels were first determined by [3H]naloxone saturation binding analysis (i.e., with varying concentrations of [3H]naloxone) to determine Bmax values. Cells transfected with the wild-type MOR were adopted as a control. Competition binding assays were conducted following the previously described protocol.35
MOR calcium inhibition assays
MOR-CHO cells were transfected with Gqi5 pcDNA1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommended procedure and maintained in RPMI 1640 supplemented with 10% fetal bovine serum, 100 u/mL penicillin, 100 µg/mL streptomycin, and 1 mg/mL G418 at 37°C and 5% CO2. 48 hours after transfection a total of 2,500,000 cells were spun down and brought back up in 8 mL of 50:1 HBSS:HEPES assay buffer. Cells were then plated at 25,000 cells per well into a clear bottom, black 96-well plate (Greiner Bio-one) and 50 µL of Fluo4 loading buffer (40 µL 2 µM Fluo4-AM (Invitrogen), 100 µL 2.5 mM probenacid, in 5 mL assay buffer) was added to bring the volume up to 130 µL. After incubating for 45 minutes, 50 µL of varying concentrations of ligands and controls were added and the plate was incubated for an additional 15 minutes. Plates were then read on a FlexStation3 microplate reader (Molecular Devices) at 494/516 ex/em for a total of 120 seconds. After 16 seconds of reading, 20 µL of 100 nM DAMGO (NIDA) in assay buffer, or assay buffer alone, was added to the wells to bring the total volume up to 200 µL. The changes in Ca2+ mobilization were monitored and peak height values were obtained using SoftMaxPro software (Molecular Devices). Nonlinear regression curves and IC50’s were generated using GraphPad Prism. All experiments were repeated a total of three times.
Supplementary Material
Table 2.
The non-conserved amino acid residue composition of the two binding sites in the three opioid receptors.
| Receptor | Site 1 | Site 2 |
|---|---|---|
| MOR | K3036.58 and W3187.35 | E2295.35 and T2255.31 |
| KOR | E2976.58 and Y3127.35 | D2235.35 and Y2195.31 |
| DOR | W2846.58 and L3007.35 | D2105.35 and S2065.31 |
Acknowledgements
The work was partially supported by PHS grant DA024022 (Y. Z.) and DA017204 (P. D. M.). The content in this work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available: GOLD docking summary: the top ten scores from the last trial in the automatic docking for all four ligands. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zimmerman DM, Leander JD. Selective opioid receptor agonists and antagonists: research tools and potential therapeutic agents. J. Med. Chem. 1990;33:895–902. doi: 10.1021/jm00165a002. [DOI] [PubMed] [Google Scholar]
- 2.Schmidhammer H. Opioid Receptor Antagonists. Prog. Med. Chem. 1998;35:83–132. [PubMed] [Google Scholar]
- 3.Eguchi M. Recent advances in selective opioid receptor agonists and antagonists. Med. Res. Rev. 2004;24:182–212. doi: 10.1002/med.10059. [DOI] [PubMed] [Google Scholar]
- 4.Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, Befort K, Dierich A, Le Meur M, Dollé P, Tzavara E, Hanoune J, Roques BP, Kieffer BL. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 5.Skoubis PD, Matthes HW, Walwyn WM, Kieffer BL, Maidment NT. Naloxone fails to produce conditioned place aversion in mu-opioid receptor knock-out mice. Neuroscience. 2001;106:757–763. doi: 10.1016/s0306-4522(01)00333-5. [DOI] [PubMed] [Google Scholar]
- 6.Gaveriaux-Ruff C, Kieffer BL. Opioid receptor genes inactivated in mice: the highlights. Neuropeptides. 2002;36:62–71. doi: 10.1054/npep.2002.0900. [DOI] [PubMed] [Google Scholar]
- 7.Substance Abuse and Mental Health Services Administration. NSDUH series H-32. Rockville, MD: Office of Applied Studies; 2007. Results from the 2006 National Survey on Drug Use and Health: national findings. (DHHS publication no. SMA 07-4293.) [Google Scholar]
- 8.Global status report: alcohol policy. Geneva: World Health Organization; 2004. [Google Scholar]
- 9.Updating estimates of the economic costs of alcohol abuse in the United States: estimates, update methods, and data. Bethesda, MD: National Institute on Alcohol Abuse and Alcoholism; 2000. [Google Scholar]
- 10.Contet C, Kieffer BL, Befort K. Mu opioid receptor: a gateway to drug addiction. Curr. Opin. Neurobiol. 2004;14:370–378. doi: 10.1016/j.conb.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Gold MS, Dackis CA, Pottash AL, Sternbach HH, Annitto WJ, Martin D, Dackis MP. Naltrexone, opiate addiction, and endorphins. Med. Res. Rev. 1982;2:211–246. doi: 10.1002/med.2610020302. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez JP, Brogden RN. Naltrexone. A review of its pharmacodynamic and pharmacokinetic properties and therapeutic efficacy in the management of opioid dependence. Drugs. 1988;35:192–213. doi: 10.2165/00003495-198835030-00002. [DOI] [PubMed] [Google Scholar]
- 13.Soyka M, Roesner S. New pharmacological approaches for the treatment of alcoholism. Expert Opin. Pharmacother. 2006;7:2341–2353. doi: 10.1517/14656566.7.17.2341. [DOI] [PubMed] [Google Scholar]
- 14.Oslin DW, Berrettini WH, O'Brien CP. Targeting treatments for alcohol dependence: the pharmacogenetics of naltrexone. Addict Biol. 2006;11:397–403. doi: 10.1111/j.1369-1600.2006.00036.x. [DOI] [PubMed] [Google Scholar]
- 15.Anton RF. Naltrexone for the management of alcohol dependence. N. Engl. J. Med. 2008;359:715–721. doi: 10.1056/NEJMct0801733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto K, McCann M, Basch J, Rawson R, Ling W. Naltrexone and dysphoria: fact or myth? Am. J. Addict. 2002;11:151–160. doi: 10.1080/10550490290087929. [DOI] [PubMed] [Google Scholar]
- 17.Ritter AJ. Naltrexone in the treatment of heroin dependence: relationship with depression and risk of overdose. Aust. N. Z. J. Psychiatry. 2002;36:224–228. doi: 10.1046/j.1440-1614.2002.01012.x. [DOI] [PubMed] [Google Scholar]
- 18.Van Dorp EL, Yassen A, Dahan A. Naloxone treatment in opioid addiction: the risks and benefits. Expert Opin. Drug Saf. 2007;6:125–132. doi: 10.1517/14740338.6.2.125. [DOI] [PubMed] [Google Scholar]
- 19.Baamonde A, Daugé V, Ruiz-Gayo M, Fulga IG, Turcaud S, Fournié-Zaluski MC, Roques BP. Antidepressant-type effects of endogenous enkephalins protected by systemic RB 101 are mediated by opioid delta and dopamine D1 receptor stimulation. Eur. J. Pharmacol. 1992;216:157–166. doi: 10.1016/0014-2999(92)90356-9. [DOI] [PubMed] [Google Scholar]
- 20.Tejedor-Real P, Micó JA, Smadja C, Maldonado R, Roques BP, Gilbert-Rahola J. Involvement of delta-opioid receptors in the effects induced by endogenous enkephalins on learned helplessness model. Eur. J. Pharmacol. 1998;354:1–7. doi: 10.1016/s0014-2999(98)00423-3. [DOI] [PubMed] [Google Scholar]
- 21.Filliol D, Ghozland S, Chluba J, Martin M, Matthes HW, Simonin F, Befort K, Gavériaux-Ruff C, Dierich A, LeMeur M, Valverde O, Maldonado R, Kieffer BL. Mice deficient for delta- and mu-opioid receptors exhibit opposing alterations of emotional responses. Nat. Genet. 2000;25:195–200. doi: 10.1038/76061. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Dergacheva O, Griffioen KJ, Huang ZG, Evans C, Gold A, Bouairi E, Mendelowitz D. Action of kappa and Delta opioid agonists on premotor cardiac vagal neurons in the nucleus ambiguus. Neuroscience. 2004;129:235–241. doi: 10.1016/j.neuroscience.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Schwyzer R. ACTH: a short introductory review. Ann. N. Y. Acad. Sci. 1977;297:3–26. doi: 10.1111/j.1749-6632.1977.tb41843.x. [DOI] [PubMed] [Google Scholar]
- 24.Portoghese PS, Lipkowski AW, Takemori AE. Binaltorphimine and nor-bionaltorphimine, potent and selective kappa opioid receptor antagonists. Life Sci. 1987;40:1287–1292. doi: 10.1016/0024-3205(87)90585-6. [DOI] [PubMed] [Google Scholar]
- 25.Jones RM, Hjorth SA, Schwartz TW, Portoghese PS. Mutational evidence for a common kappa antagonist binding pocket in the wild-type kappa and mutant mu[K303E] opioid receptors. J. Med. Chem. 1998;41:4911–4914. doi: 10.1021/jm9805182. [DOI] [PubMed] [Google Scholar]
- 26.Portoghese PS, Sultana M, Nagase H, Takemori AE. Application of the message-address concept in the design of highly potent and selective non-peptide delta opioid receptor antagonist. J. Med. Chem. 1988;31:281–282. doi: 10.1021/jm00397a001. [DOI] [PubMed] [Google Scholar]
- 27.Schmidhammer H, Burkard WP, Eggstin-Aeppli L, Smith CFC. Synthesis and biological evaluation of 14-alkoxymorphinans. 2. (−)-N-(cyclopropymethyl)-4-14-dimethoxymorphinan-6-one, a selective mu opioid receptor antagonist. J. Med. Chem. 1989;32:418–421. doi: 10.1021/jm00122a021. [DOI] [PubMed] [Google Scholar]
- 28.Marki A, Monory K, Otvos F, Toth G, Krassnig R, Schmidhammer H, Traynor JR, Roques BP, Maldonado R, Borsodi A. Mu-opioid receptor specific antagonist cyprodime: characterization by in vitro radioligand and [35S]GTPgammaS binding assays. Eur J Pharmacol. 1999;383:209–214. doi: 10.1016/s0014-2999(99)00610-x. [DOI] [PubMed] [Google Scholar]
- 29.Portoghese PS, Takemori AE. Affinity labels as probes for opioid receptor types and subtypes. NIDA Res. Monogr. 1986;69:157–168. [PubMed] [Google Scholar]
- 30.Burke TF, Woods JH, Lewis JW, Medzihradsky F. Irreversible opioid antagonist effects of clocinnamox on opioid analgesia and mu receptor binding in mice. J. Pharmacol. Exp. Ther. 1994;271:715–721. [PubMed] [Google Scholar]
- 31.Hawkins KN, Knapp RJ, Lui GK, Gulya K, Kazmierski W, Wan YP, Pelton JT, Hruby VJ, Yamamura HI. [3H]-[H-D-Phe-Cys-Tyr-D-Trp-Orn-Thr-Pen-Thr-NH2] ([3H]CTOP), a potent and highly selective peptide for mu opioid receptors in rat brain. J. Pharmacol. Exp. Ther. 1989;248:73–80. [PubMed] [Google Scholar]
- 32.Kramer TH, Shook JE, Kazmierski W, Ayres EA, Wire WS, Hruby VJ, Burks TF. Novel peptidic mu opioid antagonists: pharmacologic characterization in vitro and in vivo. J. Pharmacol. Exp. Ther. 1989;249:544–551. [PubMed] [Google Scholar]
- 33.Hruby VJ, Toth G, Gehrig CA, Kao LF, Knapp R, Lui GK, Yamamura HI, Kramer TH, Davis P, Burks TF. Topographically designed analogues of [D-Pen,D-Pen5]enkephalin. J. Med. Chem. 1991;34:1823–1830. doi: 10.1021/jm00110a010. [DOI] [PubMed] [Google Scholar]
- 34.Abbruscato TJ, Thomas SA, Hruby VJ, Davis TP. Blood-brain barrier permeability and bioavailability of a highly potent and mu-selective opioid receptor antagonist, CTAP: comparison with morphine. J. Pharmacol. Exp. Ther. 1997;280:402–409. [PubMed] [Google Scholar]
- 35.Li G, Aschenbach LC, Chen J, Cassidy MP, Stevens DL, Gabra BH, Selley DE, Dewey WL, Westkaemper RB, Zhang Y. Design, synthesis, and biological evaluation of 6alpha- and 6beta-N-heterocyclic substituted naltrexamine derivatives as mu opioid receptor selective antagonists. J. Med. Chem. 2009;52:1416–1427. doi: 10.1021/jm801272c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobson KA, Costanzi S. New Insights for Drug Design from the X-Ray Crystallographic Structures of G-Protein-Coupled Receptors. Mol. Pharmacol. 2012;82:361–371. doi: 10.1124/mol.112.079335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Structure of the δ-opioid receptor bound to naltrindole. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seki T, Minami M, Nakagawa T, Ienaga Y, Morisada A, Satoh M. DAMGO recognizes four residues in the extracellular loop to discriminate between µ- and κ-opioid receptors. Eur. Jur. Pharmacol. 1998;350:301–310. doi: 10.1016/s0014-2999(98)00240-4. [DOI] [PubMed] [Google Scholar]
- 41.Ulens C, Baker L, Ratka A, Waumans D, Tytgat J. Morphine-6beta-glucuronide and morphine-3-glucuronide, opioid receptor agonists with different potencies. Biochem. Pharmacol. 2001;62:1273–1282. doi: 10.1016/s0006-2952(01)00761-4. [DOI] [PubMed] [Google Scholar]
- 42.Xu H, Lu Y, Partilla JS, Zheng Q, Wang J, Brine GA, Carroll FI, Rice KC, Chen K, Chi Z, Rothman RB. Opioid peptide receptor studies, 11: involvement of Tyr148, Trp318 and His319 of the rat mu-opioid receptor in binding of mu-selective ligands. Synapse. 1999;32:23–28. doi: 10.1002/(SICI)1098-2396(199904)32:1<23::AID-SYN3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 43.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 44.Kellogg GE, Abraham DJ. Hydrophobicity: Is LogPo/w More than the Sum of its Parts? Eur. J. Med. Chem. 2000;35:651–661. doi: 10.1016/s0223-5234(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 45.Spyrakis F, Amadasi A, Fornabaio M, Abraham DJ, Mozzarelli A, Kellogg GE, Cozzini P. The consequences of scoring docked ligand conformations using free energy correlations. Eur. J. Med. Chem. 2007;42:921–933. doi: 10.1016/j.ejmech.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 46.Kellogg GE, Chen DL. The importance of being exhaustive. Optimization of bridging structural water molecules and water networks in models of biological systems. Chem. Biodivers. 2004;1:98–105. doi: 10.1002/cbdv.200490016. [DOI] [PubMed] [Google Scholar]
- 47.Tripathi A, Fornabaio M, Kellogg GE, Gupton JT, Gewirtz DA, Yeudall WA, Vega NE, Mooberry SL. Docking and hydropathic scoring of polysubstituted pyrrole compounds with antitubulin activity. Biorg. Med. Chem. 2008;16:2235–2242. doi: 10.1016/j.bmc.2007.11.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 49.Yuan Y, Elbegdorj O, Chen J, Akubathini SK, Zhang F, Stevens DL, Beletskaya IO, Scoggins KL, Selley DE, Akbarali HI, Dewey WL, Zhang Y. Design, Synthesis, and Biological Evaluation of 17-Cyclopropylmethyl-3,14β-dihydroxy-4,5α-epoxy-6β-[(4’-pyridyl)carboxamido]morphinan Derivatives as Peripheral Selective Mu Opioid Receptor Antagonists. J. Med. Chem. 2012;55(22):10118–10129. doi: 10.1021/jm301247n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.