SUMMARY
Myeloid derived suppressor cells (MDSCs) and cancer stem cells (CSCs) are important cellular components in the cancer microenvironment, and may affect cancer phenotype and patient outcome. The nature of MDSCs and their interaction with CSCs in ovarian carcinoma are unclear. We examined the interaction between MDSCs and CSCs in patients with ovarian carcinoma and showed MDSCs inhibited T cell activation, enhanced CSC gene expression, sphere formation and cancer metastasis. MDSCs triggered miRNA101 expression in cancer cells. miRNA101 subsequently repressesed the co-repressor gene C-terminal binding protein-2 (CtBP2), and CtBP2 directly targeted stem cell core genes resulting in increased cancer cell stemness, and increasing metastatic and tumorigenic potential. Increased MDSC density and tumor microRNA101 expression, and decreased tumor CtBP2 expression independently predict poor survival. Collectively, the work identifies an immune associated cellular, molecular and clinical network involving MDSCs-microRNA101-CtBP2-stem cell core genes, which extrinsically controls cancer stemness and impacts patient outcome.
Keywords: Myeloid-derived suppressor cell (MDSC), immune suppression, microRNA, CtBP2, stem cell, ovarian cancer and survival
INTRODUCTION
Death rates attributable to ovarian cancer have not changed significantly for decades. Although ovarian cancer patients’ initial response to surgical debulking and chemotherapy is often excellent, relapse with drug-resistant cancer usually occurs and patients succumb to their disease. Given our failure to improve long-term survival from ovarian cancer, there is a need to address the key cellular and molecular mechanisms by which tumor metastasis and chemoresistance occur in these patients, from a novel angle.
The tumor microenvironment is the primary arena where tumor cells and the host immune system interact. The capacity of immunity to modulate cancer phenotype including cancer dormancy has been the subject of intensive investigation (Matsushita et al., 2012). The interaction between tumor cells and host immune system causes immunoediting (Schreiber et al., 2011), fosters tumor immune evasion and ultimately results in tumor dissemination, relapse and metastasis (Pardoll, 2012; Zou, 2005). Tumor infiltrating T cell subsets (Curiel et al., 2004; Galon et al., 2006; Pages et al., 2005; Zhang et al., 2003) have been extensively studied in human cancers. Myeloid derived suppressor cells (MDSCs) are an important immune component in the tumor microenvironment, and are thought to mediate immune suppression in tumor bearing mice and in patients with cancer (Bronte and Zanovello, 2005; Gabrilovich and Nagaraj, 2009; Ma et al., 2011). However, MDSCs are poorly defined in the human cancer microenvironment.
The evolutionarily conserved microRNAs are endogenous ~22 nt non-coding RNAs that fine tune gene expression, regulate cell differentiation and cell-fate determination (Bartel, 2004; Xiao and Rajewsky, 2009). MicroRNA expression and function are linked to cancer development and progression (Korpal et al., 2011; Liu et al., 2011; Ma et al., 2007; Shimono et al., 2009). However, the expression, regulation and potential oncogenic function of any given microRNAs in high grade ovarian serous carcinoma remain unclear.
Identification of cancer stem cells (CSCs) (or cancer-initiating cells) is crucial for advancing cancer biology and therapy. Several markers including aldehyde dehydrogenase (ALDH) have been utilized to identify and investigate human epithelial cancer stem cells (Kryczek et al., 2012; Wicha, 2006). CSCs are thought to contribute to tumor progression, metastasis and therapeutic resistance (Brabletz et al., 2005; Dean et al., 2005; Pardal et al., 2003; Wicha, 2006). However, the factors that determine the invasive, metastatic, and chemoresistant phenotype of tumor cells are not clear. One concept is that the invasive tumor phenotype is defined by cell-autonomous alterations specified by the genomes of cancer cells. Alternatively, metastatic traits may be acquired through interactions between tumor cells and environmental signals within the tumor microenvironment (Karnoub et al., 2007; Kim et al., 2009; Liu et al., 2011; Qian et al., 2011). Among environmental signals affecting tumor cells, the roles of mesenchymal stem cells, macrophages and bone marrow derived progenitors (Dawson et al., 2009; Denardo et al., 2011; Kaplan et al., 2005; Karnoub et al., 2007; Kim et al., 2009; Liu et al., 2011; Qian et al., 2011) have largely been investigated in tumor-bearing mouse models. In this work we focused on the microenvironment in human primary high grade ovarian serous carcinoma, and tested our hypothesis that MDSC, a crucial immune component in the tumor microenvironment, functions as an environmental extrinsic signal, directly targets CSCs, defines tumor phenotype and impact patient outcome. Thus, we have established a cellular, molecular and clinical link among MDSCs, microRNA, CSCs and patients with high grade ovarian serous carcinoma.
RESULTS
Immunological impact of MDSCs in ovarian cancer
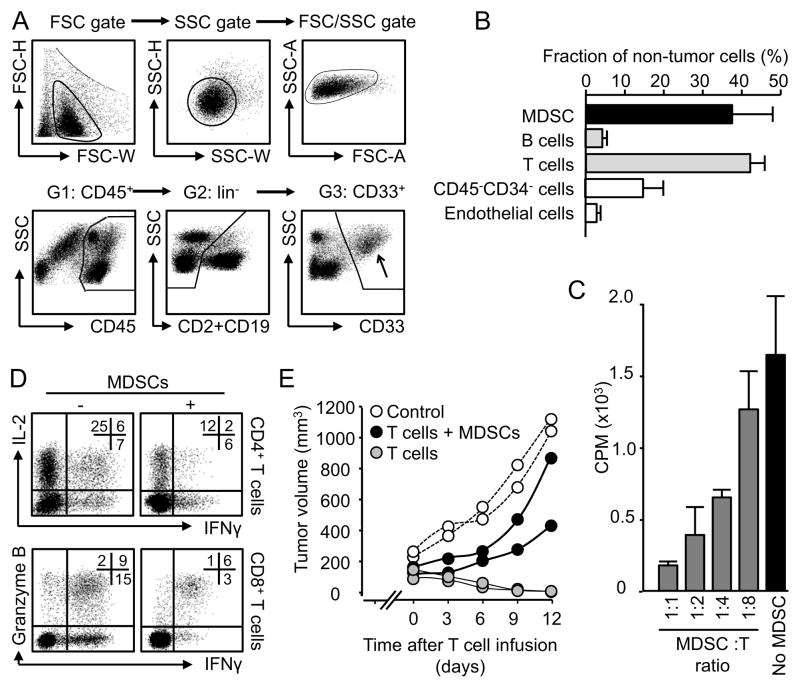
High-grade serous carcinoma is the most common histologic subtype, and most lethal among epithelial ovarian carcinomas. This study included only high grade ovarian serous cancer patients. We first assessed the immunological relevance of MDSCs in ovarian cancer. Polychromatic flow cytometry analysis demonstrated that lin−CD45+CD33+ cells infiltrated ovarian tumors (Figure 1A, Figure S1A, B) and that these cells comprise 37% of non-neoplastic cells in the tumor microenvironment (Figure 1B), and expressed medium levels of HLA-DR (Figure S1A). The majority of these cells were CD15− and CD16−/dim in ovarian cancer (Figure S1A). Lin−CD45+CD33+ cells were also found in peripheral blood (Figure S1C). Sorted tumor-associated (Figure 1C), but not peripheral blood (Figure S1D) lin−CD45+CD33+ cells suppressed T cell proliferation in a dose dependent manner, and inhibited CD4+ and CD8+ T cell effector cytokine interleukin(IL)-2 and interferon (IFN)γ expression and granzyme B expression (Figure 1D). To test the potential direct effect of tumor-associated lin−CD45+CD33+ cells on T cell-mediated anti-tumor immunity, we utilized our xenograft tumor model with primary human ovarian cancer cells (Curiel et al., 2004; Kryczek et al., 2012; Kryczek et al., 2011). We established tumors comprised of primary human ovarian cancer cells into the dorsal flank of female NOD-Shi-scid-IL-2Rγnull (NSG) mice, and treated the mice with tumor associated antigen (TAA)-specific T cells educated or non-educated by tumor-associated lin−CD45+CD33+ cells. We observed that tumor-associated lin−CD45+CD33+ cells weakened T cell-mediated anti-tumor immunity as shown by enhanced tumor volume as compared to tumor alone or tumor plus non-educated T cells (Figure 1E, Figure S1E). Thus, we referred tumor-associated lin−CD45+CD33+ cells as myeloid derived suppressor cells (MDSCs).
Figure 1. MDSCs in patients with ovarian cancer.
Single cells of ovarian cancer tissues were stained for relevant markers. Doublets and apoptotic cells were gated out.
(A) Flow cytometry gate of MDSCs. MDSCs were gated at the basis of their size, localization (FSC/SSC) and phenotypes (CD45+CD2−CD19−CD33+). Arrow pointed to gated MDSCs. One of 40 cancers is shown.
(B) Percentage of non-tumor cells in fresh ovarian cancer tissues. Results are expressed as the mean percentage + SEM. n = 10.
(C, D) MDSCs mediated immune suppression in vitro. Autologous T cells were cultured with MDSCs with different ratios for 3 days. T cell proliferation was determined by thymidine incorporation (C) and cytokine and granzyme B expression were analyzed by FACS (D). Results are expressed as the percent of positive cells in specific T cell subset + SEM. n = 6, P < 0.01, as compared to controls.
(E) MDSCs mediated immune suppression in vivo. Autologous TAA-specific T cells were conditioned with MDSCs and injected into NSG mice bearing established ovarian cancer. Tumor volume was measured. One experiment is shown with 2 mice per group from one patient donor. Similar experiments were done in 3 patient donors. n = 6/group, P < 0.01, as compared to no MDSC and controls.
See also Figure S1
Clinical impact of MDSCs in ovarian cancer
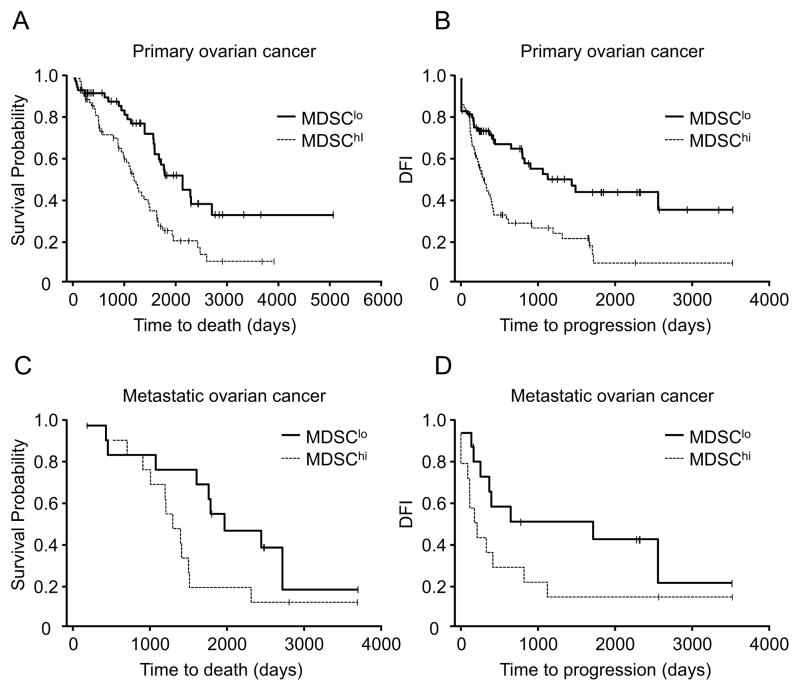
We next examined the clinical relevance of MDSCs in ovarian cancer patients. Multiple reverse gating polychromatic flow cytometry analysis showed that CD33+ cells were confined to lin−CD45+CD33+ MDSCs in fresh ovarian tumor tissue (Figure S1B). Tumor associated lin−CD45+CD33+ MDSCs did not express CD3, CD19, CD34 and CD56 (Figure S1B). Thus, we quantified CD33+ cells with immunohistochemistry (Figure S2) in ovarian tumor tissues from patients whose clinical and pathological information was available (Table S1, S2). Based on the median values of CD33+ MDSC density, we divided patients into “low” and “high” groups.
Age, tumor stage, and debulking were important prognostic factors for survival (Table S1). Overall survival (P = 0.006, n = 137, HR = 1.99, 95% CI: 1.22, 3.25) and disease-free interval (DFI) (P = 0.02, n = 135, HR = 1.75, 95% CI: 1.08, 2.87) were shorter in patients with high MDSC infiltration after adjusting for the important clinical prognostic factors (Figure 2A, B). This relationship between tumor MDSC content and overall survival (P = 0.03, n = 28, HR = 2.87, 95% CI: 1.11, 7.42) (Figure 2C) and DFI (P = 0.02, n = 28, HR = 3.25, 95% CI: 1.21, 9.63) (Figure 2D) remained significant in metastatic cancer. Therefore, increased tumor MDSC numbers are a significant and independent predictor for poor survival in ovarian cancer.
Figure 2. Clinical impact of MDSCs in patients with ovarian cancer.
MDSCs were quantified in primary and metastatic ovarian cancer. Kaplan–Meier estimates of overall survival and progression-free interval were performed according to the median values of CD33+ MDSC density. Overall survival (A, C) and disease-free interval (B, D) in patients with primary cancer (A, B) and metastatic cancer (C, D) were shown.
See also Figure S2, Table S1 and S2.
MDSCs enhance cancer incidence, metastasis and stemness
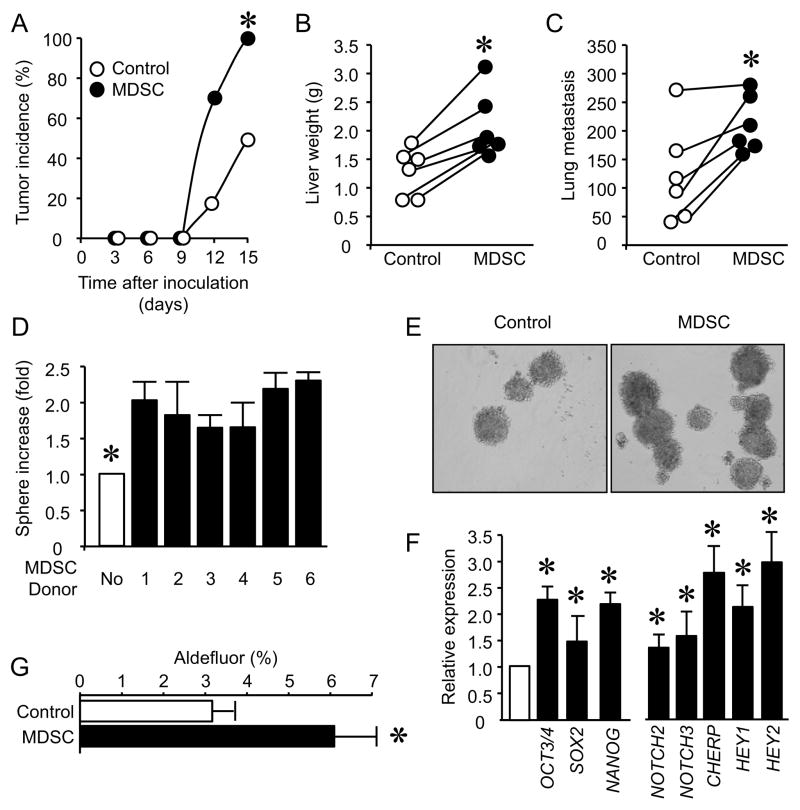
MDSCs may support ovarian cancer progression and metastasis. We tested this hypothesis using our established xenograft tumor model with primary human ovarian cancer cells (Curiel et al., 2004; Kryczek et al., 2012; Kryczek et al., 2011). We injected varying numbers of primary ovarian cancer cells into the NSG mice. We found that ≥ 200,000 primary cancer cells were needed to reach 100% tumor incidence. When primary ovarian cancer cells were cultured (conditioned) with MDSCs, 100,000 MDSC-conditioned primary cancer cells were subcutaneously inoculated into NSG mice, we observed that there was 100% tumor incidence (Figure 3A), and the presence of MDSCs had no effect on tumor volume under this condition (Figure S3A). Similar experiments were performed with titrated tumor cells. MDSCs clearly increased tumor incidence (Figure S3B). These tumor cells were also intravenously injected into NSG mice. There were more metastatic foci of tumor in the liver and lungs of mice that received MDSC-conditioned tumor cells as compared to control animals that were injected with non-conditioned tumor cells (Figure 3B, C, and Figure S3C, D).
Figure 3. MDSCs enhance ovarian cancer incidence, metastasis and stemness.
(A–C) Effects of MDSCs on ovarian cancer incidence and metastasis. MDSC-conditioned primary ovarian cancer cells were subcutaneously (A) or intravenously (B, C) injected into NSG mice. Tumor development was monitored. Results are expressed as the percentage of tumor development (A). Tumor liver and lung metastasis was recorded. Results are expressed as liver weights (B) and the numbers of tumor lung foci (C). n = 6/group. P < 0.05.
(D, E) Effects of MDSCs on cancer spheres formation. Primary ovarian cancer cells and MDSCs were co-cultured in sphere condition. Sphere formation assay was performed. Results are expressed as the fold of increase of sphere numbers ± SD. 6 patients with triplicates. *, P<0.01.
(F) Effects of MDSCs on stem cell associated gene transcripts. Primary ovarian cancer cells were conditioned with MDSCs. Tumor stem cell associated gene transcripts were quantified with real-time PCR and are expressed as the mean values relative to controls ± SD. Five experiments with triplicates, *, P < 0.01.
(G) Effects of MDSCs on ALDH+ cancer stem cells. Primary ovarian cancer cells were cultured with MDSCs for 48 hours. ALDH+ cells are expressed as the mean percentage ± SD, n = 4, derived from 3 different patients. *P <0.05.
See also Figure S3
We hypothesized that MDSCs may affect cancer stem cell properties. In support of this hypothesis, MDSCs promoted tumor sphere formation (Figure 3D, E), and enhanced the expression of multiple stem cell core gene transcripts (Figure 3F). ALDH+ ovarian epithelial cancer cells are enriched with cancer stem cells (Kryczek et al., 2012). We co-cultured MDSCs with primary ovarian cancer cells and found that MDSCs increased ALDH+ cells in the co-culture system (Figure 3G). We performed similar experiments with peripheral blood lin−CD45+CD33+ cells (Figure S3E–G). These blood born cells had no significant effects on tumor incidence (Figure S3E), stem cell-associated gene expression (Figure S3F) and sphere formation (Figure S3G). Thus, tumor associated-MDSCs promote ovarian cancer stemness.
MDSCs stimulate microRNA101 expression in ovarian cancer
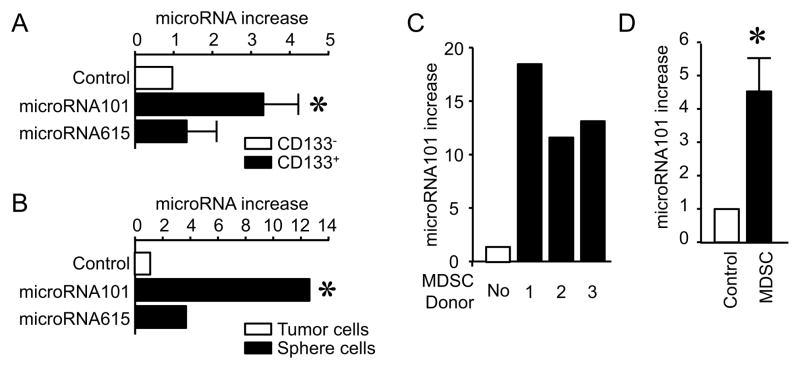
We next investigated the mechanism by which MDSCs enhance cancer stemness. Certain microRNAs affect tumor progression and metastasis (Korpal et al., 2011; Liu et al., 2011; Ma et al., 2007; Shimono et al., 2009). MDSCs may regulate cancer stemness by affecting microRNA expression. To examine this possibility, we conducted microRNA arrays in primary tumor cells conditioned with tumor associated lin−CD45+CD33+ cells (MDSCs), blood lin−CD45+CD33+ cells and medium. The microRNA arrays showed that MDSCs differentially affected the expression of microRNAs as compared to medium and blood controls (Table S3, Figure S4A). Among the increased microRNAs, two microRNAs, microRNA101 and microRNA165 were reliably enhanced more than 4 times by MDSCs as compared to blood lin−CD33+CD45+ cells in different arrays (Table S3, Figure S4A). Therefore, we quantified microRNA101 and microRNA165 in primary CD133+ ovarian cancer stem cells and cancer sphere cells. The levels of microRNA101 and microRNA165 expression were higher in primary CD133+ ovarian cancer stem cells (Figure 4A) and cancer sphere cells (Figure 4B) than primary CD133− ovarian cancer cells (Figure 4A) and non-sphere cancer cells (Figure 4B). However, the levels of microRNA101 were higher than that of microRNA165 in primary CD133+ ovarian cancer stem cells (Figure 4A) and cancer sphere cells (Figure 4B). Based on the data, we assumed that MDSCs might induce microRNA101 expression and in turn regulate cancer stemness. In support of this possibility, we observed that MDSCs stimulated microRNA101 expression in vitro (Figure 4C) and in vivo (Figure 4D) in primary ovarian cancer cells. As additional controls, in line with microRNA arrays (Table S3), MDSCs did not stimulate the expression of microRNA145, microRNA155 and microRNA200 in primary ovarian cancer cells (Figure S4B). Collectively, the findings suggest that microRNA101 induced by MDSCs may be important for maintaining ovarian cancer stemness.
Figure 4. MDSCs stimulate cancer microRNA101 expression.
(A) The expression of microRNA101 and microRNA615 in CD133+ and CD133− cancer cells. CD133+ and CD133− primary cancer cells were sorted from ovarian cancer tissues. microRNAs were quantified by PCR. 3 patients with triplicates are shown. Results are expressed as the mean values relative to controls ± SD. P <0.05 as compared to CD133− cancer cells.
(B) The expression of microRNA101 and microRNA615 in sphere forming cancer cells. microRNAs were quantified by PCR in sphere forming cells and primary cancer cells. One of 5 experiments with triplicates is shown. *, P <0.05.
(C) Effects of MDSCs on cancer microRNA101 expression in vitro. Primary ovarian cancer cells were cultured with MDSCs. Cancer microRNA101 expression was quantified by PCR. 3 patients (donor 1, 2, 3) with triplicates are shown. P <0.05 as compared to controls.
(D) Effects of MDSCs on cancer microRNA101 expression in vivo. Primary ovarian cancer cells and MDSCs were injected into peritoneal cavity of NSG mice. After 24 hours, tumor cells were collected and sorted for microRNA101 detection with quantitative PCR. 3 patients with triplicates are shown. P <0.05 as compared to controls.
MDSCs enhance stemness via microRNA101
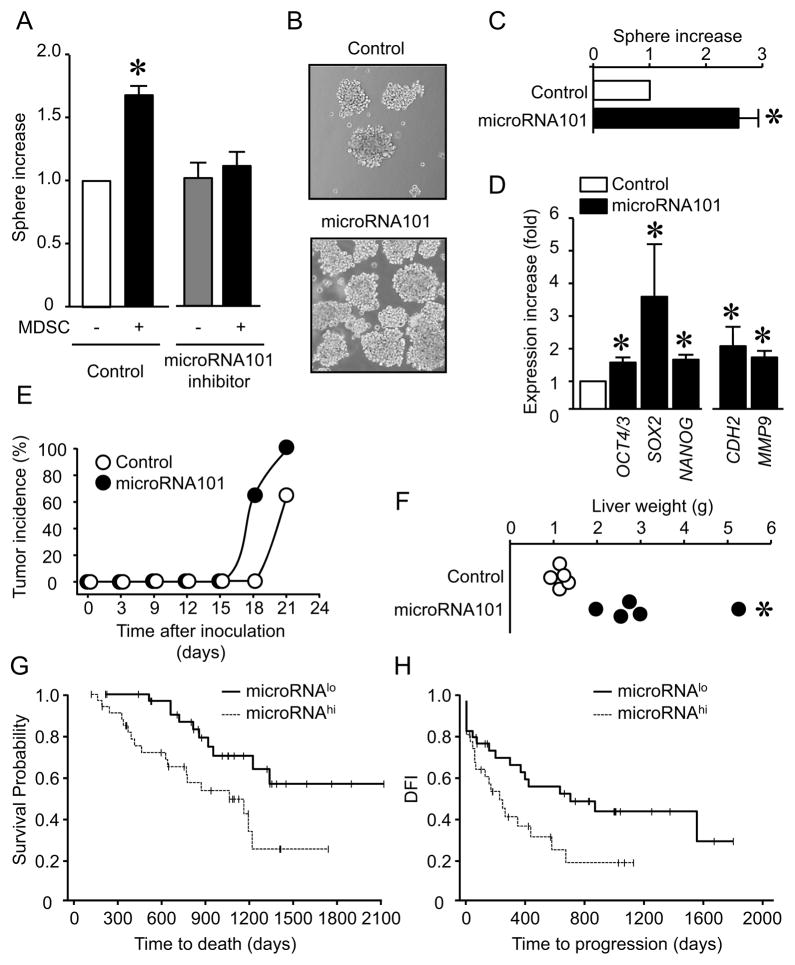
We hypothesized that MDSCs enhance cancer stemness via microRNA101. In support of this hypothesis, a microRNA101 inhibitor blocked MDSC-induced cancer sphere formation (Figure 5A), and microRNA101 overexpression stimulated cancer sphere formation (Figure 5B, C). The increased capacity of cancer sphere formation by microRNA101 was visible in the secondary sphere assay (Figure S5A). Furthermore, microRNA101 overexpression had no effects on cancer cell proliferation (Figure S5B), but enhanced expression of multiple stem cell core genes and genes associated with epithelial to mesenchymal transition (EMT) (Figure 5D), increased tumor incidence (Figure 5E) and liver metastasis (Figure 5F). In further support of the in vivo link among MDSCs, microRNA101 and cancer stemness in patients, we observed positive correlations between CD33 transcripts and microRNA101 (n = 70, P = 0.044, r = 0.26) in snap-frozen primary ovarian cancer tissues. When we dichotomized microRNA101 levels by a median split and divided the patients into high and low groups, high levels of microRNA101 were associated with reduced overall survival (P = 0.041, n = 60, HR = 2.52, 95% CI: 1.07, 6.37)(Figure 5G) and DFI (P = 0.045, n = 65, HR = 1.95, 95% CI: 1.02, 3.75) (Figure 5H). As additional control, we observed that oncogenesis-associated gene FOXO3a and microRNA155 had no impact on patient survival (not shown). These data indicate that MDSCs enhance ovarian cancer stemness by inducing cancer cell microRNA101 expression.
Figure 5. MDSCs promote cancer stemness via microRNA101.
(A) Effect of microRNA101 inhibition on ability of forming spheres by ovarian cancer cells. Sphere assays were performed in the presence of MDSCs and microRNA101 inhibitor. 7 patients with triplicates are shown. Results are expressed as the mean values relative to controls ± SD. *, P<0.01 for testing that the effect of MDSCs depends on microRNA101.
(B, C) Effects of ectopic microRNA101 on sphere formation. Sphere assays were performed in primary ovarian cancer cells transfected with lentiviral vector encoding microRNA101 and control. 3 patients with triplicates are shown. Results are expressed as the mean values relative to controls ± SD. *, P<0.01 as compared to microRNA vector.
(D) Effects of ectopic microRNA101 on stem cell-associated gene transcripts. The relevant genes were quantified in primary ovarian cancer cells transfected with lentiviral vector encoding microRNA101 and control. 3 experiments with triplicates are shown. *, Results are expressed as the mean values relative to controls ± SD. P<0.01 as compared to microRNA vector.
(E, F) Effects of ectopic microRNA101 on ovarian cancer incidence and metastasis. Tumor development was monitored (E). n = 6/group. Tumor liver metastasis was recorded (F). n = 5/group. P < 0.05.
(G, H) Clinical relevance of microRNA101. microRNA101 was quantified by PCR in snap-frozen primary ovarian cancer. Kaplan–Meier estimates of overall survival (G) and progression-free interval (H) were performed according to the median values of microRNA101.
See also Figure S5
microRNA101 taregts CtBP2 and controls stemness
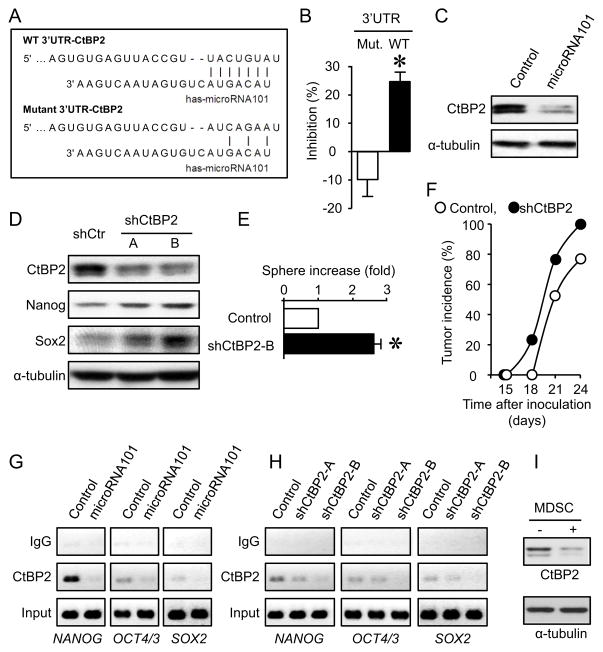
We next investigated how microRNA101 regulates cancer stemness. We searched for the predicted microRNA101 target with potential stemness repressor function (Lewis et al., 2005). There were eight major co-repressor complexes (SWI-SNF, PRC1, NURD, CoREST, NCoR, PRC2, SIN3, TLE) (Perissi et al., 2010). Based on computational analysis with TARGETSCAN software, we found that two co-repressor complexes, CtBP2, the key CoREST complex gene, and Ezh2 and EED, the key PRC2 complex genes were the potential targets of microRNA101. Overexpression of microRNA101 had minimal effects on PRC2 complex gene Ezh2, Suz12 and EED expression (Figure S6A). We further investigated CtBP2. There was a defined target site of microRNA101 at the 3′UTR of CtBP2 (Figure 6A). CtBP2 is involved in normal stem cell regulation (Tarleton and Lemischka, 2010) and prostate cancer development (Thomas et al., 2008). Thus, we hypothesized that microRNA101 targeted CtBP2 and controlled cancer stemness. To test this hypothesis, we cloned the predicted 3′UTR of CtBP2 into a luciferase reporter vector. A mutant containing site mutations at the predicted microRNA101 targeting site was generated as a control (Figure 6A). Overexpression of microRNA101 in primary ovarian cancer cells had no effects on cell proliferation (Figure S5B). Overexpression of microRNA101 decreased the reporter activity containing wild type-3′UTR-CtBP2, but not the mutant (Mut-3′UTR- CtBP2) (Figure 6B). In line with this observation, CtBP2 protein expression levels were decreased in primary ovarian cancer cells overexpressing microRNA101 as compared with the scramble control (Figure 6C). To determine the role of CtBP2 in cancer stemness, CtBP2 expression was genetically knocked down by two specific small hairpin CtBP2 RNAs (shCtBP2-A, shCtBP2-B) in primary ovarian cancer cells. CtBP2 silencing had no effects on cancer cell proliferation (Figure S6B) or tumor growth in vivo (Figure S6C), but resulted in increased stem cell core protein expression (Figure 6D), increased cancer sphere formation (Figure 6E) and tumor incidence (Figure 6F). ChIP analysis confirmed that microRNA101 overexpression (Figure 6G) and knock down of CtBP2 (Figure 6H) resulted in reduced CtBP2 expression and less occupancy on the promoters of NANOG, OCT4/3 and SOX2 in primary ovarian cancer cells. Furthermore, MDSCs increased microRNA101 expression (Figure 4A–C) and reduced CtBP2 protein expression in primary cancer cells (Figure 6I). Thus, microRNA101 targets CtBP2 and controls cancer stemness.
Figure 6. MicroRNA101 targets CtBP2 and controls cancer stemness.
(A) Target of microRNA101 in 3′UTR of CtBP2. The sequence of wild type (WT) and mutant 3′UTR of CtPB2 gene used in luciferase assay was shown.
(B) Effect of microRNA101 on WT-3′UTR-CtBP2 luciferase activity. 3′UTR-CtBP2 luciferase activity was measured in tumor cells transfected with WT-3′UTR and mutant. Results are expressed as the percentage of inhibition of 3′UTR-CtBP2 luciferase activity. 3 experiments with triplicates are shown. Results are expressed as the mean percentage of inhibition ± SEM. *, P<0.01 as compared to mutant-3′UTR- CtBP2.
(C) Effect of microRNA101 on CtBP2 protein expression. Primary ovarian cancer cells were transfected with lentiviral vector encoding microRNA101 and control. Cancer CtBP2 protein was detected by Western blotting. One of 3 experiments is shown.
(D) Effect of CtBP2 on stem cell core protein expression in primary ovarian cancer cells. Primary ovarian cancer cells were transfected with lentiviral vector encoding shCtBP2 and control. Expression of CtBP2, Nanog, Sox2 protein was detected by Western blotting. A, B are two shCtBP2. One of 3 experiments is shown.
(E) Effect of CtBP2 on ovarian cancer sphere formation. Primary ovarian cancer cells were transfected with lentiviral vector encoding shCtBP2. Sphere assay was performed in 3 experiments with triplicates. Results are expressed as the fold increase (mean + SEM). *, P<0.01 as compared to control.
(F) Effect of knock down of CtBP2 expression on tumor incidence. Primary ovarian cancer cells were transfected with lentiviral vector encoding shCtBP2. shCtBP2 cancer cells were injected into NSG mice. Tumor incidence was monitored. Results are expressed as the percentage of tumor development. n = 6 in Control group; n = 5 in sh-CtBP2 group. P < 0.05.
(G, H) Effects of microRNA101 on the binding of CtBP2 to core stem cell gene promoters. ChIP assay was performed in primary ovarian cancer cells expressing microRNA101 (G), shCtBP2 (H) or scramble. One of three experiments is shown.
(I) Effect of MDSCs on CtBP2 protein expression in primary ovarian cancer cells. Primary ovarian cancer cells were cultured with MDSCs. CtBP2 protein expression was detected with Western blot. One of three experiments is shown.
See also Figure S6
MDSC and CtBP2 interaction impacts clinical outcome
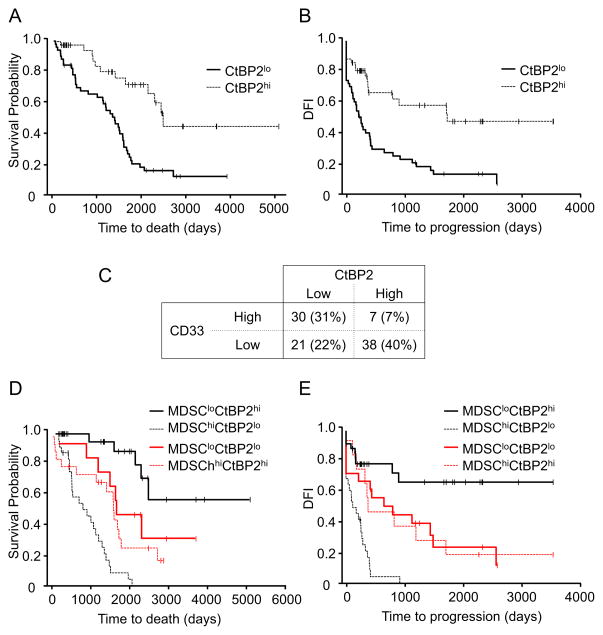
To examine the importance of cancer CtBP2 expression, we quantified the expression of tumor CtBP2 by H-score method (Supplementary experimental procedures and Figure S7), and analyzed its impact on patient survival. Based on the median levels of CtBP2 expression (Figure S7), patients were divided into two groups, low and high CtBP2 expression. High levels of CtBP2 expression in primary tumor cells were associated with increased overall survival (P = 0.006, n = 95, HR = 0.41, 95% CI: 0.21, 0.77) (Figure 7A) and DFI (P = 0.047, n = 93, HR = 0.55, 95% CI: 0.30, 0.98) (Figure 7B).
Figure 7. MDSCs and CtBP2 interaction impacts patient outcome.
(A, B) Impact of CtBP2 on patient survival. CtBP2 expression was quantified in primary ovarian cancer. Kaplan–Meier estimates of overall survival (A) and progression-free interval (B) were performed according to the median levels of CtBP2 expression.
(C) The association and distribution between tumor CtBP2 and CD33+ MDSCs in patients with ovarian carcinoma. Patients were divided into four groups: CD33lowCtBP2high, CD33lowCtBP2low, CD33highCtBP2low and CD33highCtBP2high. Patient distribution in four groups and the association between CtBP2 expression levels and CD33+ cell density are shown.
(D, E) Impact of MDSC and CtBP2 interaction on ovarian cancer overall survival. Kaplan–Meier estimates of overall survival (D) and progression-free interval (E) were performed according to the median levels of CtBP2 expression and MDSC intensity.
See also Figure S7
Finally, we evaluated significance of the two parameters for ovarian cancer survival. High CD33+ MDSC infiltration strongly correlated with low CtBP2 expression in primary tumors (n = 96, r = -0.44, P <0.0001) (Figure 7C), suggesting that MDSCs may impact patient outcome through controlling CtBP2 expression. Thus, we reasoned that the patients with high levels of MDSCs (CD33hi) and low tumor CtBP2 expression (CtBP2lo) would have had a shorter survival than those with a tumor type of CD33loCtBP2hi. Indeed, CD33hiCtBP2lo patients experienced a shorter overall survival (P = 0.0018, HR = 3.8, 95% CI: 1.64, 8.79, n = 96) (Figure 7D) and DFI (P = 0.0049, HR = 3.02, 95% CI: 1.40, 6.53, n = 92) (Figure 7E) than CD33loCtBP2hi patients, after adjusting for important clinical prognostic factors. The combination of CtBP2 and MDSCs may allow for improved prognostic stratification of ovarian cancer overall survival as compared to CD33 or CtBP2. Based on 96 patients with both CD33 and CtBP2 values, we found that the hazard of death for CD33hiCtBP2lo is 3.8 times the hazard of death for CD33loCtBP2hi (P = 0.0018, HR = 3.80, 95% CI: 1.64, 8.79), compared to 1.99 times with CD33 (high vs low: P = 0.02, HR = 1.99, 95% CI: 1.22, 3.25) or 2.47 times with CtBP2 (low vs high: P = 0.006, HR = 2.47, 95% CI: 1.30, 4.71), in addition to age, stage and debulking. Collectively, our data support the functional and clinical importance of the CD33-microRNA101-CtBP2 network in high grade ovarian serous carcinomas.
DISCUSSION
The tumor microenvironment is a main battle ground between tumor cells and the host immune system. The capacity of immunity to control and shape cancer including cancer dormancy has been the subject of intensive investigation (Matsushita et al., 2012). The interaction between tumor cells and host immune system causes immunoediting (Dunn et al., 2002), fosters tumor immune evasion and ultimately results in tumor dissemination, relapse and metastasis (Pardoll, 2012; Zou, 2005). Cancer stem cells are thought to contribute to tumor progression, metastasis and therapeutic resistance (Brabletz et al., 2005; Dean et al., 2005; Pardal et al., 2003). However, it is not well understood what determines the invasive, metastatic, and chemoresistant phenotype of tumor cells. One concept is that the invasive tumor phenotype is defined by cell-autonomous alterations specified by the genomes of cancer cells. Alternatively, metastatic traits may be acquired through interactions between tumor cells and environmental signals within the tumor microenvironment (Karnoub et al., 2007; Kim et al., 2009; Liu et al., 2011; Qian et al., 2011). Among environmental signals affecting tumor cells, the importance of mesenchymal stem cells, macrophages and bone marrow derived progenitors (Dawson et al., 2009; Denardo et al., 2011; Kaplan et al., 2005; Karnoub et al., 2007; Kim et al., 2009; Liu et al., 2011; Qian et al., 2011) have largely been investigated in tumor-bearing mouse models. However, the major immune suppressive components, MDSCs are poorly understood in human cancers. Our current study demonstrates that MDSCs functions as an environmental extrinsic signal, directly targets cancer stem cells, and at least partially shape tumor phenotype. Thus, cancer stemness is partly defined by immune environmental cues including immune suppressive elements.
Here, we isolated viable MDSCs from the microenvironment of primary ovarian tumors for phenotypic, functional, mechanistic and clinical studies. We identified high numbers of immunosuppressive MDSCs in the ovarian tumor microenvironment and showed that MDSCs promote and maintain the ovarian cancer stem cell pool. Dicer and Drosha mRNA expression levels in ovarian cancer have associations with outcomes in patients with ovarian cancer (Merritt et al., 2008). Dicer and Drosha may control microRNA expression. Aberrant microRNA expression can contribute to tumorigenesis, tumor progression and metastasis (Korpal et al., 2011; Liu et al., 2011; Ma et al., 2007; Shimono et al., 2009), but it was previously unknown if tumor environmental immune elements have any impact on microRNA expression and function, and in turn can alter the tumor phenotype and patient outcome. We found that MDSCs stimulate microRNA101 expression in ovarian cancer cells and promote cancer stemness by targeting co-repressor CtBP2. Thus, we have revealed a mechanistic relationship and cross-talk among MDSCs, microRNA101 and CtBP2 at the cellular and molecular levels. We have further demonstrated the clinical relevance of this cross-talk in ovarian cancer patients. Our observations suggest that cancer stemness is partly defined by immune environmental cues. Given the relevance of cancer stem cells in tumor relapse, metastasis and therapy resistance (Brabletz et al., 2005; Dean et al., 2005; Dontu et al., 2003; Pardal et al., 2003; Reya et al., 2001), our observations have important consequences for the way in which we perceive cancer stem cells or MDSCs as independent therapeutic targets, shifting the attention from cancer cells-cancer stem cells or immune suppressive networks (Zou, 2005) to their interaction in the cancer microenvironment. Given that tumor infiltrating effector T cells (Galon et al., 2006; Pages et al., 2005; Zhang et al., 2003) including Th17 cells (Kryczek et al., 2009a) are associated with improved survival in patients with cancer, our data indicate that different immune elements in the tumor microenvironment may play distinct roles in tumor progression, metastasis and therapy, and in turn differentially affect patient outcome. It is reasonable to predict that anti-cancer therapy should simultaneously target host MDSCs and cancer cells-stem cells to improve therapeutic efficiency and reduce therapy resistance.
EXPERIMENTAL PROCEDURES
Ovarian cancer patients and cancer tissue samples
High-grade serous carcinoma is the most common cancer and most lethal type of ovarian carcinoma. Patients diagnosed with high grade serous ovarian carcinomas were recruited for this study. Human subject use in this study was approved by the local Institutional Review Boards. We collected 140 formalin-fixed, paraffin-embedded ovarian tumor tissue blocks, 73 snap-frozen ovarian tumor tissues, and 40 fresh ovarian cancer tissues for this study as described previously (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2006; Zou et al., 2001). After pathological review, a tissue microarray (TMA) was constructed from the most representative area of paraffin-embedded ovarian tumor tissue. For each tumor, a minimum of two representative tumor areas were selected from a hematoxylin- and eosin-stained section of a donor block. Core cylinders (1 mm) were punched from each of these areas and deposited into a recipient paraffin block. Consecutive 6 μm-thick TMA sections were cut and placed on charged Poly-L-Lysine-coated slides for immunohistochemistry analyses. Further information regarding the TMA is described in the supplementary experimental procedures.
In vitro and in vivo T cell immunosuppression
In vitro T cell immunosuppression was tested in coculture system. T cells (2 × 105/ml) were stimulated with 2.5 μg/ml anti-human CD3 and 1.2 μg/ml anti-CD28 (BD) in the presence of different concentrations of peripheral blood lin−CD45+CD33+ cells or MDSCs (2 × 105/ml). Seventy-two hours after coculture, T cell proliferation and cytokine production were evaluated as we described (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2006; Zou et al., 2001). The in vivo T cell suppression was tested with primary ovarian cancer cells in NSG model. Primary ovarian tumor cells (1 × 107) in 200 μl of buffered saline were subcutaneously injected into dorsal tissues of female NSG mice. We established TAA-specific T cells from patients with HLA-A2+ Her2/neu+ ovarian cancer (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2006) (Supplementary method). Autologous TAA-specific T cells (6 × 106) were conditioned with MDSCs (3 × 106) or medium, and were subsequently injected intravenously into mice on day 12 after human tumor inoculation (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2006). Tumor size was measured twice weekly using calipers fitted with a Vernier scale. Tumor volume was calculated based on three perpendicular measurements (Curiel et al., 2004; Curiel et al., 2003; Kryczek et al., 2006; Zou et al., 2001).
Immunohistochemistry staining
Immunohistochemical staining on TMA slides was performed on a DAKO Autostainer (DAKO, Carpinteria, CA) using DAKO LSAB+ and diaminobenzadine (DAB) as the chromogen. Serial sections of de-paraffinized TMA sections were labeled with mouse anti-human CD33 mAb (Clone, NCL-L-CD33, 1:100, Novacastra, Buffalo Grove, IL), or goat anti-human CtBP2 antibody (SC-5966, 1:500, Santa Cruz Biotech, Santa Cruz, CA). Cores from several normal organ tissues were used as tissue controls on each slide. Staining with isotype antibody was used as negative control.
The cores were quantified and analyzed for the expression of CD33 and CtBP2 with an Aperio imaging system (Genetix, San Jose, CA). The specimens were digitalized with an automated platform (Aperio Technologies; Vista, CA) and ScanScope XT and Spectrum Plus using TMA software version 9.1 scanning system. Cores were scored in high resolution of X50. Tissue sections were scored manually on a computer screen, and a mean score for duplicate cores from each individual was calculated. Any discrepancies were resolved by subsequent consultation with diagnostic pathologists. CD33 was localized in the cell membrane/cytoplasm and was scored quantitatively (the numbers of positive cells). The tissues were divided into high (>14) and low (< 14) CD33+ MDSC infiltration based on the median value of CD33+ cells per 10 mm2. CtBP2 was localized in the nuclei, and was scored using the H-score method (Pirker et al., 2012). The tissues were divided into high and low CtBP2 expression based on the median value of CtBP2 expression level per core. The H-score is a method of assessing the extent of nuclear immunoreactivity. The H score takes into account the percentage of positive cells (0–100%) in each intensity category (0–3+) and computes a final score, on a continuous scale between 0 and 300. The score is obtained by the formula: 3 × percentage of strongly staining nuclei + 2 × percentage of moderately staining nuclei + percentage of weakly staining nuclei, giving a range of 0 to 300 (Pirker et al., 2012). The tissues were divided into high and low CtBP2 expression based on the median value of CtBP2 expression level per core. Cores from normal breast, heart, thyroid and tonsil tissues were used as control on each slide. Staining with isotype antibody was used as negative control.
Quantitative real-time PCR to detect microRNA and mRNA expression
Total RNA was isolated from frozen tumor tissue or primary tumor cells with Trizol Reagent (Invitrogen, Carlsbad, CA). MicroRNAs were detected by TaqMan MicroRNA Assay kit (Applied Biosystems, Foster City, CA). Quantitative real-time PCR was carried out to quantify stem cell and EMT associated genes (Kryczek et al., 2011).
Tumor MDSC and cancer stem cell identification and isolation
Fresh ovarian cancer tissues were processed into single cell suspensions and immediately used for cellular phenotyping by a flow cytometry analyzer (LSR II, Becton Dickinson, San Jose, CA) or flow sorting by high-speed sorter (FACSAria, BD) as described (Curiel et al., 2004; Kryczek et al., 2011). Antibodies against CD2, CD3, CD4, CD15, CD16, CD19, CD33, CD45, CD133, and epithelial cell adhesion molecule (EpCAM) were used in the flow experiments (BD Biosciences) (Kryczek et al., 2012). The purity of the sorted cells was >98%.
Ovarian cancer development and metastasis in NSG model
Experimentally manipulated primary ovarian cancer cells were cultured with autologous tumor associated MDSCs then sorted to high purity (>99%) or were stably transfected with specific gene vectors, and were subcutaneously or intravenously injected into female NOD/Shi-scid/IL-2Rcnull (NSG) mice (6 ~ 8 weeks old; Jackson Laboratory, Bar Harbor, ME). The viability of the injected tumor cells was >95%. Subcutaneous tumors were measured every three days using a caliper scale, and tumor volume was calculated based on three perpendicular measurements as described (Kryczek et al., 2012; Kryczek et al., 2011). On day 50 post injection, liver weight and tumor metastatic foci were recorded. Lung metastases were ink stained and counted (Kryczek et al., 2009b). In some cases, primary tumor cells and MDSCs were injected into the peritoneal cavity of mice. After 24 hours, tumor cells were collected and sorted for microRNA101 detection with quantitative PCR. The animal protocol was approved by the University of Michigan Committee on Use and Care of Animals (UCUCA).
Cell culture, MICRORNA aRRAY and in vitro sphere formation
Primary ovarian cancer cells were isolated as described previously (Kryczek et al., 2012). Primary ovarian cancer cells were initially cultured with freshly isolated MDSCs in trans-wells (Corning, One Riverfront Plaza, NY), and were subsequently harvested for microRNA array (Applied Biosystems, Cat# 4470187), quantitative PCR, sphere formation assay (Kryczek et al., 2012) and in vivo experiments.
microRNA array
Primary ovarian cancer cells (5 × 105) were co-cultured with freshly sorted ovarian cancer associated-MDSCs, peripheral blood lin−CD45+CD33+ cells (5 × 105) or medium in a transwell (Corning) for 24 hours. Then, RNA was extracted from the cancer cells by Trizol reagent. The RNAs were applied for microRNA profiling using OpenArray system (Applied Biosystems, Cat# 4470187) (Mestdagh et al., 2008) at University of Michigan Sequencing Core.
Gene transfection and 3′UTR-CtBP2 reporter dual Luciferase assay
Primary ovarian cancer cells stably expressing mciroRNA101 or microRNA scramble were established with lentivirus packaging kit (Invitrogen, Carlsbad, CA) using microRNA101 and control plasmids (Biosettia Inc, San Diego, CA). Primary ovarian cancer cells stably expressing shCtBP2 or scramble control were established by using shCtBP2 or control pGIPZ-GFP plasmids with puromycin selection (Open Biosystem, University of Michigan Vector Core). In some experiments, primary ovarian cancer cells were transiently transfected with microRNA101 inhibitor or control oligonucleotides (100 μM, Thermo Fisher Scientific Dharmacon Products, Lafayette, CO) 24 hours before functional experiments (Vilardo et al., 2010).
The PCR fragment of 3′UTR of CtBP2 (926–1325 bp) containing a predicted microRNA101 targeting site (1109–1115) was cloned into a pMIR luciferase reporter (Applied Biosystems, Foster City, CA) at Spe I and Hid III cloning sites using human genomic DNA template prepared from normal human peripheral blood mononuclear cells. Four site mutations were created at the predicted microRNA101 targeting site using a mutagenesis kit (Promega Biosystems, Sunnyvale, CA). The wild type-(WT)-3′UTR-CtBP2 or mutant (Mut)-3′UTR-CtBP2 reporter plasmids were transiently expressed in primary ovarian cancer cells along with microRNA101 and control plasmids using Lipo2000. pTK-renilla was transfected as an internal control in each transfection. 48 hours after the transfection, cell lysates were applied for dual luciferase activity measurement using the dual luciferase kit (Promega) and a luminometer. RLU represented as CtBP2 3′UTR luciferase activity related to its renilla luciferase activity. The experiments were repeated for three times with triplicates.
Western blot and Chromatin immunoprecipitation (ChIP)
Western blotting was performed using the following primary antibodies CtBP2 (1:1000, 612044, BD), NANOG (1:1000, ab21624, Abcam), SOX2 (1:1000, MAB4343, Millipore) and α-TUBULIN (1:2000, Santa Cruz). Signals were detected by ECL reagent (GE Healthcare, Buckinghamshire, UK).
ChIP was performed as we described (Cui et al., 2008). Briefly, nuclear extracts were prepared from primary ovarian cancer cells. Mouse anti-human CtBP2 (BD) and normal IgG (Santa Cruz) antibodies were used for immunoprecipitation. ChIP primers (Table S4) were designed to detect promoter fragment near transcription start sites on OCT4/3, NANOG and SOX2 genes.
Statistical analysis
Wilcoxon rank-sum tests were used to compare two independent groups, and for paired groups, Wilcoxon signed rank tests were used for the comparison. Correlation coefficients (Spearman correlation, denoted by r, for ordinal data and Pearson correlation, denoted by r, for continuous data), together with a P-value (null hypothesis is that r is in fact zero), were computed to measure the degree of association between biomarkers. ANOVA models were used to evaluate an interaction between MDSC and microRNA101 on cancer sphere formation. Log-rank test was used to compare time to tumor initiation between two groups. Overall patient survival was the defined from date of diagnosis and to disease related death. Disease-free interval was the defined from date of diagnosis and to disease progression or disease related death. Data were censored at the last follow-up for patients who were disease-free or alive at the time of analysis. Survival functions were estimated by Kaplan-Meier methods. Cox’s proportional hazards regression was performed to model survival as a function of MDSC, CtBP2 and microRNA101 (all classified as low and high based on the median value), or the combination of MDSC and CtBP2 (classified as MDSC lowCtBP2high and MDSC highCtBP2low), after adjusting for age, stage and debulking. We assessed the adequacy of the Cox regression model Graphical and numerical methods as described (Lin et al., 1993). All analyses were done using SAS 9.3 software. P < 0.05 considered as significant.
Supplementary Material
Acknowledgments
This work is supported (in part) by the National Institutes of Health grants (CA123088; CA099985; CA156685; CA171306; 5P30CA46592), the Ovarian Cancer Research Fund, and Marsha Rivkin Center for Ovarian Cancer Research. We thank to Deborah Postiff, Michelle Vinco, Ron Craig and Jackline Barikdar in the tissue procurement core for their technical assistance.
Footnotes
Supplementary Information includes Methods, 7 figures and 4 tables.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744–749. doi: 10.1038/nrc1694. [DOI] [PubMed] [Google Scholar]
- Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- Cui TX, Kwok R, Schwartz J. Cooperative regulation of endogenous cAMP-response element binding protein and CCAAT/enhancer-binding protein beta in GH-stimulated c-fos expression. J Endocrinol. 2008;196:89–100. doi: 10.1677/JOE-07-0169. [DOI] [PubMed] [Google Scholar]
- Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, Krzysiek R, Knutson KL, Daniel B, Zimmermann MC, et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat Med. 2003;9:562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
- Dawson MR, Duda DG, Fukumura D, Jain RK. VEGFR1-activity-independent metastasis formation. Nature. 2009;461:E4. doi: 10.1038/nature08254. discussion E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Denardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA, et al. Leukocyte Complexity Predicts Breast Cancer Survival and Functionally Regulates Response to Chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Al-Hajj M, Abdallah WM, Clarke MF, Wicha MS. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003;36(Suppl 1):59–72. doi: 10.1046/j.1365-2184.36.s.1.6.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kim S, Takahashi H, Lin WW, Descargues P, Grivennikov S, Kim Y, Luo JL, Karin M. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 2009;457:102–106. doi: 10.1038/nature07623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celia-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Banerjee M, Cheng P, Vatan L, Szeliga W, Wei S, Huang E, Finlayson E, Simeone D, Welling TH, et al. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood. 2009a;114:1141–1149. doi: 10.1182/blood-2009-03-208249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Liu S, Roh M, Vatan L, Szeliga W, Wei S, Banerjee M, Mao Y, Kotarski J, Wicha MS, et al. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130:29–39. doi: 10.1002/ijc.25967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Wei S, Szeliga W, Vatan L, Zou W. Endogenous IL-17 contributes to reduced tumor growth and metastasis. Blood. 2009b;114:357–359. doi: 10.1182/blood-2008-09-177360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Zhao E, Liu Y, Wang Y, Vatan L, Szeliga W, Moyer J, Klimczak A, Lange A, Zou W. Human TH17 Cells Are Long-Lived Effector Memory Cells. Sci Transl Med. 2011;3:104ra100. doi: 10.1126/scitranslmed.3002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryczek I, Zou L, Rodriguez P, Zhu G, Wei S, Mottram P, Brumlik M, Cheng P, Curiel T, Myers L, et al. B7-H4 expression identifies a novel suppressive macrophage population in human ovarian carcinoma. J Exp Med. 2006;203:871–881. doi: 10.1084/jem.20050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lin DY, WEI LJ, YING Z. Checking the Cox model with cumulative sums of martingale-based residuals. Biometrika. 1993;80:557–572. [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, Pan PY, Eisenstein S, Divino CM, Lowell CA, Takai T, Chen SH. Paired immunoglobin-like receptor-B regulates the suppressive function and fate of myeloid-derived suppressor cells. Immunity. 2011;34:385–395. doi: 10.1016/j.immuni.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36:e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Pirker R, Pereira JR, von Pawel J, Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE, Paz-Ares L, Storkel S, et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012;13:33–42. doi: 10.1016/S1470-2045(11)70318-7. [DOI] [PubMed] [Google Scholar]
- Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton HP, Lemischka IR. Delayed differentiation in embryonic stem cells and mesodermal progenitors in the absence of CtBP2. Mech Dev. 2010;127:107–119. doi: 10.1016/j.mod.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem. 2010;285:18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicha MS. Cancer stem cells and metastasis: lethal seeds. Clin Cancer Res. 2006;12:5606–5607. doi: 10.1158/1078-0432.CCR-06-1537. [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- Zou W, Machelon V, Coulomb-L’Hermin A, Borvak J, Nome F, Isaeva T, Wei S, Krzysiek R, Durand-Gasselin I, Gordon A, et al. Stromal-derived factor-1 in human tumors recruits and alters the function of plasmacytoid precursor dendritic cells. Nat Med. 2001;7:1339–1346. doi: 10.1038/nm1201-1339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.