Abstract
The distribution of tfdAα and cadA, genes encoding 2,4-dichlorophenoxyacetate (2,4-D)-degrading proteins which are characteristic of the 2,4-D-degrading Bradyrhizobium sp. isolated from pristine environments, was examined by PCR and Southern hybridization in several Bradyrhizobium strains including type strains of Bradyrhizobium japonicum USDA110 and Bradyrhizobium elkanii USDA94, in phylogenetically closely related Agromonas oligotrophica and Rhodopseudomonas palustris, and in 2,4-D-degrading Sphingomonas strains. All strains showed positive signals for tfdAα, and its phylogenetic tree was congruent with that of 16S rRNA genes in α-Proteobacteria, indicating evolution of tfdAα without horizontal gene transfer. The nucleotide sequence identities between tfdAα and canonical tfdA in β- and γ-Proteobacteria were 46 to 57%, and the deduced amino acid sequence of TfdAα revealed conserved residues characteristic of the active site of α-ketoglutarate-dependent dioxygenases. On the other hand, cadA showed limited distribution in 2,4-D-degrading Bradyrhizobium sp. and Sphingomonas sp. and some strains of non-2,4-D-degrading B. elkanii. The cadA genes were phylogenetically separated between 2,4-D-degrading and nondegrading strains, and the cadA genes of 2,4-D degrading strains were further separated between Bradyrhizobium sp. and Sphingomonas sp., indicating the incongruency of cadA with 16S rRNA genes. The nucleotide sequence identities between cadA and tftA of 2,4,5-trichlorophenoxyacetate-degrading Burkholderia cepacia AC1100 were 46 to 53%. Although all root nodule Bradyrhizobium strains were unable to degrade 2,4-D, three strains carrying cadA homologs degraded 4-chlorophenoxyacetate with the accumulation of 4-chlorophenol as an intermediate, suggesting the involvement of cadA homologs in the cleavage of the aryl ether linkage. Based on codon usage patterns and GC content, it was suggested that the cadA genes of 2,4-D-degrading and nondegrading Bradyrhizobium spp. have different origins and that the genes would be obtained in the former through horizontal gene transfer.
2,4-dichlorophenoxyacetic acid (2,4-D) has been used as a herbicide since the 1940s, and a number of its degrading bacteria have been isolated worldwide from a variety of environments and extensively examined on a molecular basis (1, 2, 4, 17, 20, 24, 27, 34, 38, 39, 41). The most extensive studies have been done with Ralstonia eutrophus JMP134, which carries a 2,4-D-degrading gene cluster on the transmissible plasmid pJP4 (4, 24, 32). The genes and enzymes involved in the 2,4-D degradation pathway have been characterized in several isolates, and comparative studies of their phylogenetic positions, nucleotide and amino acid sequences, and gene organization and rearrangement have allowed us to understand adaptive and evolutionary processes for degrading this xenobiotic compound (2, 11, 24, 28, 41, 42, 43, 45).
2,4-D-degrading bacteria are considered to be categorized into three groups on evolutionary and physiological bases (20). The first group consists of copiotrophic bacteria belonging to the β and γ subdivisions of Proteobacteria, which include genera such as Achromobacter, Burkholderia, Delftia, Halomonas, Pseudomonas, Ralstonia, Rhodoferax, and Variovorax (2, 11, 28, 43). These bacteria have been isolated from agricultural and industrial soils and sediments and have tfd genes in common (2, 11, 28, 43). 2,4-D is transformed into 2,4-Dichlorophenol (2,4-DCP) by an α-ketoglutarate (α-KG)-dependent 2,4-D dioxygenase encoded by tfdA (9, 10), and 2,4-DCP is subsequently hydroxylated to 3,5-dichlorocatechol by 2,4-DCP hydroxylase encoded by tfdB (8, 26, 34). 3,5-Dichlorocatechol is further degraded through a modified ortho-cleavage pathway encoded by tfdCDEF (24, 34). Nucleotide sequence similarities of tfdA, tfdB, and tfdC in this group are more than 76, 34, and 55%, respectively (28, 43), and the tfd genes are usually present on a transmissible plasmid (2, 4, 28). 2,4-D degraders in this group are supposed to obtain the 2,4-D-catabolizing ability by independent tfd gene recruitment through horizontal gene transfer (11, 18, 19, 32, 42, 44). The incongruency of the phylogeny between the tfd genes and 16S rRNA genes supports this suggestion (28, 43).
The second group consists of α-Proteobacteria which are closely related to a Bradyrhizobium sp. based on their 16S rRNA gene sequences (16, 20). They are oligotrophic and have been isolated at first from pristine environments in Hawaii, Canada, and Chili (20) and subsequently also from arable soil in Japan (16). Although 2,4-D-degrading genes in this group had not been characterized until recently, remarkable findings have been made. CadAB genes, which have a nucleotide sequence similar to that of tftAB encoding 2,4,5-trichlorophenoxyacetic acid (2,4,5-T) oxygenase of Burkholderia cepacia AC1100 (3), were found in Bradyrhizobium sp. strain HW13 isolated from Hawaiian soil (23). In addition, a tfdA-like gene (tfdAα) was found in strain RD5-C2 isolated in Japan (16, 17). The CadAB genes were transformed into Sinorhizobium meliloti, and the transformant acquired the degrading activities for 2,4-D and its related compounds. The gene homologs were also found in other Hawaiian and Canadian isolates by Southern hybridization (23). The tfdAα gene showed approximately 60% identity to canonical tfdA, and the gene was expressed in Escherichia coli as a MalE-TfdAα fusion showing α-KG-dependent 2,4-D dioxygenase activity. The protein exhibited about 10 times greater activity for phenoxyacetate (PAA) than 2,4-D. tfdAα was also found in the other 2,4-D-degrading strains isolated from pristine environments (17).
Members of the third group belong to a species of the genus Sphingomonas, which consists of copiotrophic α-Proteobacteria. They have been isolated from agricultural and industrial samples in the same way as the first group (28, 39, 43). Of the genes involved in 2,4-D degradation in this group, tfdB and tfdC show similarity to those of the first group, but the initial step of 2,4-D oxidation is not likely performed by α-KG-dependent 2,4-D dioxygenase but by another enzyme system (11, 28, 39, 43). Recently, the presence of cadA gene homologs was suggested by Southern hybridization that used a cadA gene fragment as a probe of the 2,4-D-degrading Bradyrhizobium strain HW13 (23).
The 2,4-D-degrading Bradyrhizobium strains have conspicuous characteristics, because they were isolated from pristine environments with no exposure to 2,4-D, and they have two different systems for degrading 2,4-D. Therefore, it is supposed that they could provide useful information on the origin and evolutionary relationship of the 2,4-D catabolic genes. As both tfdAα and cadAB are carried independently of the presence of 2,4-D in this group of 2,4-D degraders, the enzyme systems should maintain their original functions in addition to degrading 2,4-D, and it is expected that the phylogenetically related Bradyrhizobium spp. also have tfdAα and cadAB.
In this paper, we report that tfdAα genes are present in all Bradyrhizobium strains tested, which include type strains of Bradyrhizobium japonicum USDA110 and Bradyrhizobium elkanii USDA94 and those isolated from nodules of American and Australian native legumes. None of the strains showed 2,4-D-degrading activity; however, some strains showed activity toward less chlorinated phenoxyacetate derivatives. In addition, tfdA-like genes were found in 2,4-D-degrading Sphingomonas strains. On the other hand, cadA was found only in the Bradyrhizobium and Sphingomonas strains, which can degrade the phenoxy compounds. We also suggest that cadA was acquired via lateral gene transfer in Bradyrhizobium sp. and Sphingomonas sp. based on their GC content and codon usage frequencies.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used are listed in Table 1. Bradyrhizobium strains, Agromonas oligotrophica JCM1494, and Rhodopseudomonas palustris JCM2524 were grown at 25°C on modified HM medium (29). Sphingomonas strains were grown at 25°C on peptone-yeast extract-tryptone-glucose medium (20).
TABLE 1.
Strains used in this study
| Strain | tfdAαa | cadAa | 16Sb | 2,4-Dc | 4-CPAc | PAAc | Relevant characteristic(s) | Reference |
|---|---|---|---|---|---|---|---|---|
| Bradyrhizobium spp. | ||||||||
| RD5-C2 | + | + | e | + | + | + | 2,4-D degrader; Japanese isolate | 16 |
| HW13 | + | + | e | + | + | + | 2,4-D degrader; Hawaiian isolate | 20 |
| HWK12 | + | + | e | + | + | + | 2,4-D degrader; Hawaiian isolate | 20 |
| BTH | + | + | e | + | + | + | 2,4-D degrader; Canadian isolate | 20 |
| USDA 110 | + | − | j | − | − | − | B. japonicum type strain | 30 |
| USDA 94 | + | + | e | − | + | + | B. elkanii type strain | 30 |
| jwc91-2 | + | + | e | − | + | − | U.S. isolate from Amphicarpaea bracteata | 33 |
| th-b2 | + | + | e | − | − | − | U.S. isolate from Amphicarpaea bracteata | 33 |
| DesT1 | + | + | e | − | + | − | U.S. isolate from Desmodium glutinosum | 33 |
| DesB1 | + | + | e | − | − | − | U.S. isolate from Desmodium glutinosum | 33 |
| ApB16 | + | − | j | − | − | − | U.S. isolate from Apios americana | 33 |
| BDV5111 | + | − | e | − | − | − | Australian isolate from native shrubby legumes | 25 |
| BDV5329 | + | − | j | − | − | − | Australian isolate from native shrubby legumes | 25 |
| BDV5419 | + | − | e | − | − | − | Australian isolate from Hardenbergia violacea | 25 |
| BDV5680 | + | − | j | − | − | − | Australian isolate from Bossiaea ensata | 25 |
| Sphingomonas sp. | ||||||||
| B6-5 | + | − | − | − | − | Originally a 2,4-D degrader and then lost the ability; Canadian isolate | 11 | |
| B6-10 | + | + | + | − | − | 2,4-D degrader; Canadian isolate | 11 | |
| TFD26 | + | + | + | + | + | 2,4-D degrader; U.S. isolate | 41 | |
| TFD44 | + | + | + | + | + | 2,4-D degrader; U.S. isolate | 11 | |
| A. oligotrophica JCM1494 | (+) | − | − | − | − | 36 | ||
| R. palustris JCM2524 | (+) | − | − | + | − | 36 |
+, PCR amplified and sequenced; (+), detected by Southern hybridization; −, not detected.
e, 16S rRNA sequence belongs to the cluster of B. elkanii; j, 16S rRNA sequence belongs to the cluster of B. japonicum.
+, degrading ability; −, no degrading ability.
DNA preparation and analysis of tfdAα and cadA.
Total DNA samples were prepared as described previously (14, 17). A partial tfdAα fragment was amplified by using primers designed for the conserved regions of the tfdAα genes of Bradyrhizobium 2,4-D-degrading strains (17). A partial cadA fragment was amplified with primers designed from the cadA sequence of strain HW13 (23): 5′-AAGCTGCA(A/G)TTTGA(A/G)AA(C/T)GG-3′ and 5′-(A/C)GGATTGAAGAAATCCTG(A/G)TA-3′. The amplified gene fragments were sequenced directly or after cloning into a pT7Blue T-Vector (Novagen) with an ABI 377 DNA sequencing system (Perkin-Elmer Japan) by using the same primers or the M13 primers M4 and RV (Takara). DNA sequences around the PCR-amplified fragments were determined by using a PCR in vitro cloning kit (Takara). To examine the distribution of tfdAα and cadA gene homologs among the bacterial strains used, the amplified tfdAα and cadA gene fragments of strain RD5-C2 were labeled by a DIG DNA labeling kit (Roche Molecular Biochemicals) and used as probes for Southern hybridization analysis. The DNA samples were digested by SalI and blotted onto Hybond N nylon membranes (Amersham) and then hybridized with each labeled probe under low-stringency conditions as described previously (23). All procedures described here were conducted according to the suppliers' instructions.
The tfdAα and cadA sequences were aligned by using the CLUSTAL W (version 1.8) program (40), and the sites where nucleotides were not resolved for all sequences were excluded from the alignments. Phylogenetic analysis was performed with the CLUSTAL W program using the neighbor-joining (NJ) method (37), and bootstrap analysis based on 1,000 replicates was used to place confidence estimates on the trees.
Degrading ability for 2,4-D and its derivatives.
Cells grown on the designated medium were aseptically harvested by centrifugation (6,000 × g for 10 min) and resuspended in the same medium containing 2,4-D, 4-chlorophenoxyacetic acid (4-CPA), or PAA (each at a concentration of 10 mg/liter) to give a final turbidity of 0.25 at 600 nm. Cell suspensions were incubated at 25°C for 1 week, and the amount of remaining substrate was measured with a high-performance liquid chromatograph (LC-10AT; Shimadzu, Kyoto, Japan), equipped with a SUMIPAX ODS-A212 column (interior diameter, 6 mm; length, 150 mm) (Sumika Chemical Analysis Service, Osaka, Japan). The mobile phase was a mixture of acetonitrile-0.1% acetic acid (60/40) with a flow rate of 1.0 ml/min. 2,4-D, 4-CPA, and PAA (and their corresponding phenol derivatives) were detected by a Shimadzu SP10-10A UV-visible light detector at 284, 279, and 269 nm, respectively, and identified based on their retention times. Chemicals used were purchased from Wako Pure Chemical Industries (Osaka, Japan).
Comparison of codon usage frequency and GC content.
Codon usage patterns and the GC content of the tfdAα and cadA genes of the 2,4-D-degrading and non-2,4-D-degrading Bradyrhizobium strains were compared with those of the nodulation genes (nod), NifA-regulated genes, and housekeeping genes of Bradyrhizobium spp. (31, 35, and www.kazusa.or.jp/codon) to elucidate the difference in origin of tfdAα and cadA in this genus. Nineteen genes of B. japonicum (35) and the dnaK and gyrB genes of B. elkanii were used as the housekeeping genes of Bradyrhizobium spp. Patterns of synonymous codon frequency except for stop codons and codons for methionine and tryptophan were grouped by cluster analysis (Ward linkage using squared Euclidean distance) by using the software SPSS for Windows (version 11.0.1 J, SPSS Inc., Chicago, Ill.). The comparison was also made for tfdA-like and cadA genes of the Sphingomonas strains. The genes bglI, ces10, and ugpG were used as the housekeeping genes of Sphingomonas spp.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the DDBJ, EMBL, and GenBank databases under the accession numbers AB119224 to AB119237 for tfdAα and AB119238 to AB119248 for cadA.
RESULTS
Phylogenetic analysis of tfdAα and cadA genes in Bradyrhizobium strains and Sphingomonas 2,4-D degraders.
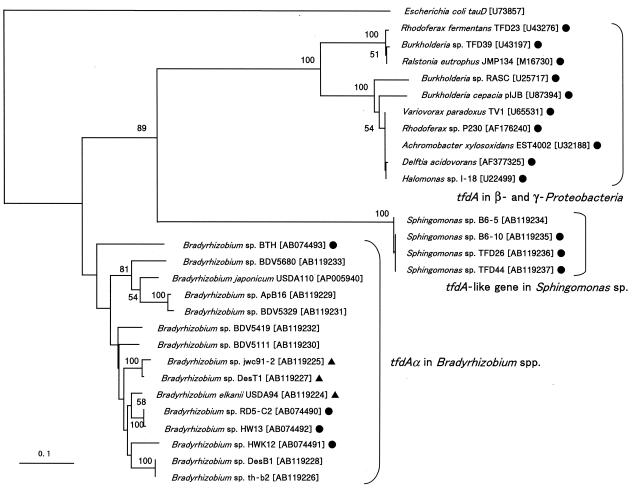
tfdAα gene fragments were amplified by PCR from all the Bradyrhizobium and Sphingomonas strains used in this experiment (Table 1) but not from A. oligotrophica JCM1494 or R. palustris JCM2524. Based on the nucleotide sequences, a phylogenetic tree of tfdAα was constructed together with canonical tfdA gene sequences from β- and γ-Proteobacteria (Fig. 1). E. coli tauD, encoding taurine/α-KG dioxygenase (6), was used as an outgroup. The tfdAα and tfdA genes were clearly separated, and the tfdA-like genes of Sphingomonas sp. were positioned beside the tfdA cluster. The nucleotide sequence identities between tfdAα and tfdA were 51 to 57%, and those of Sphingomonas sp. with tfdAα and tfdA were 48 to 51% and 46 to 49%, respectively.
FIG. 1.
Phylogenetic position of tfdAα in Bradyrhizobium spp. and the tfdA-like gene of Sphingomonas sp. beside canonical tfdA in β- and γ-Proteobacteria. E. coli tauD encodes taurine/α-KG dioxygenase. 2,4-D-degraders (•) and 4-CPA- and/or PAA-degraders (▴) are indicated. Designations in brackets are DDBJ, EMBL, and GenBank database accession numbers. The phylogenetic tree was constructed based on common partial sequences (307 bp) by using the NJ method. Bootstrap values above 50% are shown at nodes. The scale bar indicates substitutions per site.
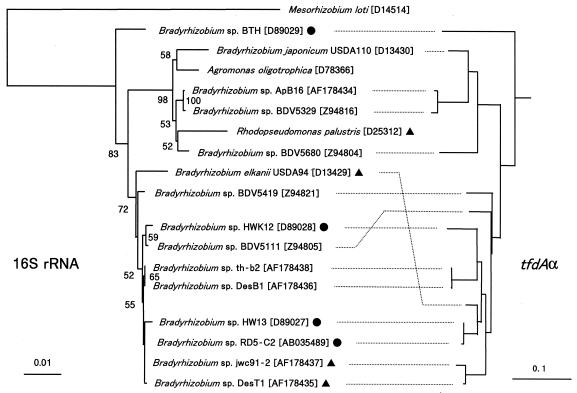
To examine the evolutionary characteristics of tfdAα in Bradyrhizobium spp., phylogenetic trees of tfdAα and the 16S rRNA genes were compared. As shown in Fig. 2, Bradyrhizobium strains were separated into two groups based on the 16S rRNA genes, resulting in clusters including B. japonicum or B. elkanii. The strains belonging to the cluster of B. japonicum in 16S rRNA genes clustered also in tfdAα; the same was true for the strains belonging to the cluster of B. elkanii, indicating congruency in the phylogenetic trees between tfdAα and the 16S rRNA genes. The nucleotide sequence identities of tfdAα in B. japonicum and B. elkanii were 81 to 87%.
FIG. 2.
Phylogenetic relationship of Bradyrhizobium strains based on 16S rRNA and tfdAα sequences. 2,4-D-degraders (•) and 4-CPA- and/or PAA-degraders (▴) are indicated. Designations in brackets are DDBJ, EMBL, and GenBank database accession numbers. The phylogenetic tree was constructed based on common partial sequences (1300 bp) by using the NJ method. Bootstrap values above 50% are shown at nodes. The scale bar indicates substitutions per site.
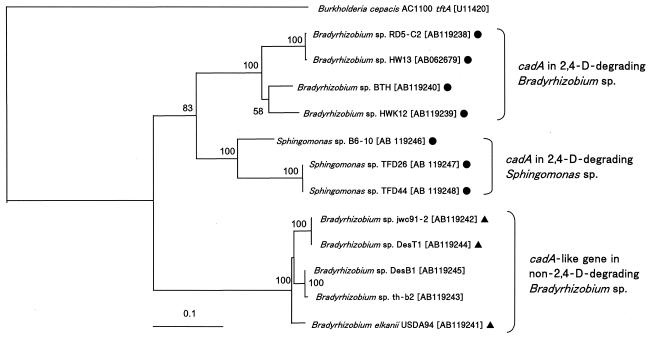
In contrast to the pattern of distribution of tfdAα, cadA was amplified by PCR only in the 2,4-D-degrading Bradyrhizobium and Sphingomonas strains and in the non-2,4-D-degrading Bradyrhizobium strains USDA94, jwc91-2, th-b2, DesT1, and DesB1. The phylogenetic tree of cadA is shown in Fig. 3 with tftA of 2,4,5-T-degrading Burkholderia cepacia AC1100 as an outgroup. The nucleotide sequence identities of cadA and tftA were 46 to 53%. In the α-Proteobacteria, cadA genes were phylogenetically separated between 2,4-D-degrading and nondegrading strains, and the separation between the 2,4-D-degrading Bradyrhizobium sp. and Sphingomonas sp. was apparent with 74 to 78% of the nucleotide sequence identities. As a result, congruency of cadA with 16S rRNA genes was not observed between Bradyrhizobium sp. and Sphingomonas sp.
FIG. 3.
Phylogenetic tree of cadA from 2,4-D-degrading α-Proteobacteria. Burkholderia cepacia AC1100 tftA encodes 2,4,5-T oxygenase. 2,4-D-degraders (•) and 4-CPA- and/or PAA-degraders (▴) are indicated. Designations in brackets are DDBJ, EMBL, and GenBank database accession numbers. The phylogenetic tree of cadA was constructed on the basis of common partial sequences (375 bp) by using the NJ method. Bootstrap values above 50% are shown at nodes. The scale bar indicates substitutions per site.
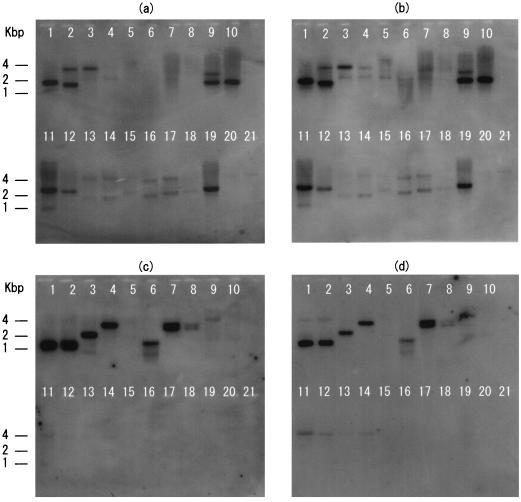
Southern hybridization.
Positive signals by a tfdAα probe of strain RD5-C2 were detected in all the Bradyrhizobium strains and phylogenetically related strains of A. oligotrophica JCM1494 and R. palustris JCM2524 in which no tfdAα fragment was amplified by PCR, suggesting the inefficiency of the primers used (Fig. 4a). No tfdAα homologs were detected in the 2,4-D-degrading Sphingomonas strains even under low-stringency conditions; however, positive signals were observed when a PCR-amplified tfdA-like gene fragment of B6-5 was used as a probe (Fig. 4b). When cadA was used to probe RD5-C2, it hybridized with DNA of the 2,4-D-degrading Bradyrhizobium and Sphingomonas strains, and weak signals were also observed in Bradyrhizobium strains USDA94, jwc91-2, th-b2, DesT1, and DesB1 (Fig. 4c). Stronger signals were detected in the Bradyrhizobium strains when a cadA-like probe of USDA94 was used (Fig. 4d).
FIG. 4.
Southern hybridization by the tfdAα (a) and cadA (c) probes from 2,4-D-degrading strain RD5-C2 in Bradyrhizobium sp., by the tfdA-like gene (b) from Sphingomonas sp. strain B6-5, and by the cadA-like gene (d) from Bradyrhizobium sp. strain USDA94. Results for the following strains are shown: 2,4-D-degrading strains RD5-C2 (lane 1), HW13 (lane 2), HWK12 (lane 3), and BTH (lane 4) in Bradyrhizobium sp.; 2,4-D-degrading strains B6-5 (2,4-D-degrading ability lost) (lane 5), B6-10 (lane 6), TFD26 (lane 7), and TFD44 (lane 8) in Sphingomonas sp.; B. elkanii USDA94 (lane 9); B. japonicum USDA110 (lane 10); Bradyrhizobium sp. strains jwc91-2 (lane 11), DesT1 (lane 12), DesB1 (lane 13), th-b2 (lane 14), ApB16 (lane 15), BDV5111 (lane 16), BDV5329 (lane 17), BDV5419 (lane 18), and BDV5680 (lane 19); Agromonas oligotrophica JCM1494 (lane 20); and Rhodopseudomonas palustris JCM2524 (lane 21).
Degradation of 2,4-D and its related compounds.
The abilities of the strains used in the present study to degrade 2,4-D and its related compounds are indicated in Table 1 and in Fig. 1 to 3. The 2,4-D-degrading Bradyrhizobium and Sphingomonas strains degraded 4-CPA and PAA equally well except for Sphingomonas sp. strain B6-10, which did not degrade 4-CPA and PAA. Strains BTH, TFD26, and TFD44 did not degrade phenol, which accumulated during the degradation of PAA. None of the other strains degraded 2,4-D; however, strains USDA94, jwc91-2, DesT1, and R. palustris JCM2524 degraded 4-CPA. They did not degrade 4-chlorophenol beyond what accumulated during the degradation of 4-CPA. PAA was not degraded by any strains other than the 2,4-D degraders and USDA94. The degrading abilities were repeatedly observed and were not lost during successive cultivation under nonselective conditions on 2,4-D.
Codon usage frequency and GC content.
The genes of the Bradyrhizobium spp. examined were separated into two groups based on their GC content as summarized in Table 2. Distinctions made between the two groups on the basis of the GC content and codon usage patterns were in good agreement (data not shown). The first group, which had a lower total GC content (55.7 to 59.4%), consisted of cadA of 2,4-D-degrading Bradyrhizobium sp., nod, and the NifA-regulated genes of Bradyrhizobium spp. (31, 35, and www.kazusa.or.jp/codon). The second group was made up of the cadA gene of non-2,4-D-degrading Bradyrhizobium sp., the tfdAα gene of both 2,4-D-degrading and nondegrading Bradyrhizobium spp., and the housekeeping genes of Bradyrhizobium spp. (35) and showed a higher total GC content (63.9 to 65.7%). The difference between the two groups was distinctive when the GC bias at the third codon position was considered.
TABLE 2.
GC content of tfdAα and cadA genes of Bradyrhizobium spp. and Sphingomonas spp.
| Gene | No. of genes | GC content (mol%)
|
|||
|---|---|---|---|---|---|
| Total | 1st codon position | 2nd codon position | 3rd codon position | ||
| tfdAα of 2,4-D-degrading Bradyrhizobium sp. | 4 | 64.4 | 65.3 | 39.2 | 89.0 |
| tfdAα of non-2,4-D-degrading Bradyrhizobium spp | 11 | 63.0 | 64.9 | 37.3 | 86.9 |
| cadA of 2,4-D-degrading Bradyrhizobium sp. | 4 | 55.7 | 59.1 | 43.8 | 64.5 |
| cadA of non-2,4-D-degrading Bradyrhizobium sp. | 5 | 65.7 | 64.1 | 48.4 | 84.6 |
| nod genes of Bradyrhizobium japonicuma | 12 | 57.8 | Nab | Na | 64.4 |
| NifA-regulated genes of B. japonicuma | 14 | 57.7 | Na | Na | 69.4 |
| Housekeeping genes of B. japonicuma | 19 | 64.9 | Na | Na | 86.4 |
| nod genes of B. elkaniic | 18 | 59.4 | 63.2 | 45.6 | 69.4 |
| Housekeeping genes of B. elkaniid | 2 | 64.3 | 66.3 | 38.3 | 88.1 |
| tfdA-like genes of 2,4-D-degrading Sphingomonas sp. | 4 | 64.8 | 67.3 | 45.2 | 81.7 |
| cadA genes of 2,4-D-degrading Sphingomonas sp. | 3 | 55.9 | 59.1 | 41.4 | 67.3 |
| Housekeeping genes of Sphingomonas spp.e | 3 | 68.2 | 71.9 | 47.6 | 85.3 |
In Sphingomonas sp., tfdA-like genes were similar to the housekeeping genes on the basis of a higher GC content (64.8 and 68.2%, respectively) and distinguished from cadA with a lower GC content (55.9%).
DISCUSSION
A number of 2,4-D degraders that belong to a variety of genera have been isolated worldwide, and it has been revealed that there are at least two catabolic systems for 2,4-D degradation. 2,4-D degraders in β- and γ-Proteobacteria use α-KG-dependent 2,4-D dioxygenase encoded by tfdA (11, 28, 44), whereas 2,4-D degrading α-Proteobacteria use the enzyme that is similar to 2,4,5-T oxygenase, encoded by cadAB (23). Besides these, tfdAα genes have been found in 2,4-D-degrading Bradyrhizobium strains in α-Proteobacteria that were isolated from pristine environments (17). However, the evolutionary origin of the genes still remains unknown because there are very few similar genes and amino acid sequences in databases. As cadAB and tfdAα genes have been found in microorganisms isolated from pristine environments where lava was deposited 4,800 years ago (20), we speculate that ancestral cadAB and tfdAα genes in this group of bacteria may serve as the origin of the tftAB and tfdA genes, respectively.
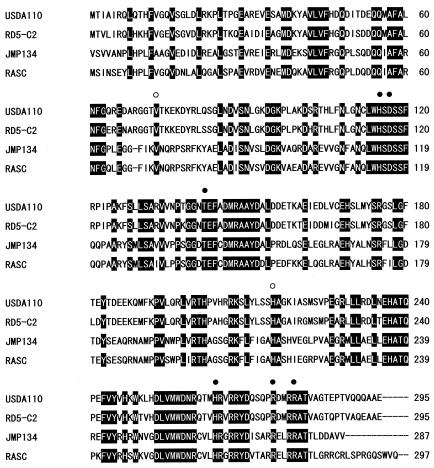
In this study, we revealed that all of the Bradyrhizobium strains including type strains of B. japonicum USDA110 and B. elkanii USDA94 have the gene tfdAα. The gene seems to be ubiquitous among Bradyrhizobium spp. independent of their 2,4-D-degrading ability. Therefore, TfdAα should have an original substrate or function, which should have no relation to xenobiotic compounds. Naturally occurring aryl ethers and cinnamic acid are proposed as candidate substrates (5), as well as naturally produced organohalogen compounds (13). Very recently, the complete genomic sequence of B. japonicum USDA110 has been determined (22). Gene arrangements around tfdAα were almost identical between strains RD5-C2 and USDA110 (our unpublished observations), and there are no related tfd genes (tfdB and tfdCDEF) that have been found in the 2,4-D-degrading plasmid pJP4. As genes for d-β-hydroxybutyrate dehydrogenase and oxidoreductase are present just upstream of tfdAα, the function of TfdAα might be related to fatty acid metabolism. As shown in Fig. 5, alignment of the deduced amino acid sequence of B. japonicum USDA110 TfdAα with the other α-KG-dependent 2,4-D dioxygenase indicates that the predicted active-site residues are well conserved among them as follows: two His residues and one Asp residue, which are suspected of being iron-binding ligands; a Thr and an Arg residue, suspected of forming a salt bridge with the C-5 carboxylate of α-KG; and a His and an Asp residue, suspected of forming a salt bridge with the 2,4-D carboxylate (7, 15). Therefore, it is supposed that TfdAα in Bradyrhizobium spp. would belong to the α-KG-dependent dioxygenase family (15).
FIG. 5.
Multiple alignment of the deduced amino acid sequences of TfdAα of Bradyrhizobium strains RD5-C2 and USDA110 and TfdA of Ralstonia eutropha JMP134 and Burkholderia sp. strain RASC. Conserved residues are boxed. The predicted iron- and α-KG-binding ligands are marked with filled circles. The predicted 2,4-D-binding ligands in TfdA are marked with open circles.
The phylogenetic trees of the tfdAα gene and the corresponding 16S rRNA genes were almost congruent, indicating the evolution of the genes without horizontal gene transfer in Bradyrhizobium spp. As tfdAα was also detected in phylogenetically closely related A. oligotrophica and R. palustris, the distribution of tfdAα seems not to be limited within Bradyrhizobium spp. However, no genes similar to tfdAα could be detected in Rhizobium loti and Sinorhizobium meliloti, both of which are symbiotic nitrogen-fixing bacteria belonging to α-Proteobacteria (12, 21).
The 2,4-D-degrading Sphingomonas sp. has been believed to have no tfdA gene homologs based on the negative results of PCR amplification and hybridization experiments (11, 28, 43, 44). In this study, however, tfdA-like gene fragments were amplified by PCR and detected by Southern hybridization in Sphingomonas strains. The phylogenetically distinct position of Sphingomonas tfdA-like genes suggests that the evolution occurred without recent horizontal gene transfer.
In addition to the previous evidence that 2,4-D-degrading Bradyrhizobium and Sphingomonas strains have cadA homologs (23), the genes were also detected in the non-2,4-D-degrading Bradyrhizobium strains (Table 1 and Fig. 3). In contrast to tfdAα, however, the distribution was limited to strains USDA94, jwc91-2, th-b2, DesT1, and DesB1, which belong to the group of B. elkanii, based on the 16S rRNA gene sequences, and have been isolated in the United States (33). A worldwide survey of the geographical distribution of cadA in 2,4-D-degrading Bradyrhizobium spp. and non-2,4-D-degrading Bradyrhizobium spp. could provide information on the endemicity of the gene.
The phylogenetic tree of cadA suggests the separate evolution of the gene in 2,4-D-degraders versus non-2,4-D-degraders (Fig. 3). The former cluster was made up of two distinct clusters of Bradyrhizobium sp. and Sphingomonas sp., suggesting the independent evolution in the genera from a common ancestral gene. The incongruency of cadA with the 16S rRNA genes would be an indication of the horizontal transfer of cadA between the genera, and the distinct separation of the tree between 2,4-D-degrading Bradyrhizobium sp. and Sphingomonas sp. suggests that they could be separated at an early stage of evolution.
The apparent differences in the codon usage patterns and the GC content of cadA between 2,4-D-degrading and non-2,4-D-degrading Bradyrhizobium spp. (Table 2) also suggests a different origin of cadA in 2,4-D-degraders versus nondegraders. Ramseier and Gottfert reported that B. japonicum nodulation and the NifA-regulated genes have different codon usage and GC content from the housekeeping genes in B. japonicum and suggested that nodulation and the NifA-regulated genes were acquired relatively late in evolution via lateral gene transfer from other microorganisms (35). The presence of the DNA region of lower GC content (680 kb), in which the symbiotic island region (410 kb) was included, was demonstrated in the complete genomic sequence of B. japonicum USDA110 (22). The codon usage patterns and the GC content of cadA of non-2,4-D-degrading Bradyrhizobium sp. were similar to those of the housekeeping genes, whereas for cadA of 2,4-D-degrading Bradyrhizobium sp., they were similar to those of the symbiotic genes, suggesting the possibility that the cadA gene of 2,4-D-degrading Bradyrhizobium sp. was obtained through horizontal gene transfer. The same assumption could be made for cadA of Sphingomonas sp., in which the GC content was distinctively distinguished from that of the housekeeping genes.
Among the non-2,4-D-degrading Bradyrhizobium strains having cadA homologs, USDA94, jwc91-2, and DesT1 degraded 4-CPA. They degrade 4-CPA with the accumulation of 4-chlorophenol as an intermediate metabolite, suggesting that the cadA homologs are involved in the cleavage of the aryl ether linkage. The cadA gene with 2,4-D-degrading ability might have emerged from cadA homologs, which had been distributed among the limited group of Bradyrhizobium spp. in the form of naturally occurring aryl ether compounds as substrates. As 2,4-D-degrading Bradyrhizobium strains were isolated from pristine environments, cadA should have evolved from natural compounds and the degradation of 2,4-D would be coincidental. The inability to degrade 4-chlorophenol also suggests the unrelated presence of the gene with the 2,4-D-degrading pathway. The limited distribution of cadA homologs in Bradyrhizobium spp. indicates that the genes are not essential for their proliferation.
It has been revealed that two systems for 2,4-D degradation, TfdA and CadAB, have evolved in Bradyrhizobium spp. and Sphingomonas sp. in α-Proteobacteria. Although the α-KG-dependent 2,4-D dioxygenase activity was demonstrated in TfdAα (17), a good agreement between the presence of cadA and the ability to degrade phenoxy compounds suggests that the CadAB system might have a greater responsibility for the degradation. The distinct separation of cadA between Bradyrhizobium 2,4-D degraders and nondegraders in phylogeny and GC content suggests their evolution from different origins. A comparison of their genetic organization around cadA would provide useful information on the evolution of 2,4-D degraders.
Acknowledgments
We thank M. A. Parker (State University of New York) and CSIRO Plant Industry for providing Bradyrhizobium strains.
REFERENCES
- 1.Amy, P. S., J. W. Schulke, L. M. Frazier, and R. J. Seidler. 1985. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl. Environ. Microbiol. 49:1237-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalca, L., A. Hartmann, N. Rouard, and G. Soulas. 1999. Diversity of tfdC genes: distribution and polymorphism among 2,4-dichlorophenoxyacetic acid degrading soil bacteria. FEMS Microb. Ecol. 29:45-58. [Google Scholar]
- 3.Danganan, C. E., R. W. Ye, D. L. Daubaras, L. Xun, and A. M. Chakrabarty. 1994. Nucleotide sequence and functional analysis of the genes encoding 2,4,5-trichlorophenoxyacetic acid oxygenase in Pseudomonas cepacia AC1100. Appl. Environ. Microbiol. 60:4100-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunning Hotopp, J. C., and R. P. Hausinger. 2001. Alternative substrates for 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J. Mol. Catal. B Enzym. 15:155-162. [Google Scholar]
- 6.Eichhorn, E., J. R. van der Ploeg, M. A. Kertesz, and T. Leisinger. 1997. Characterization of α-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem. 272:23031-23036. [DOI] [PubMed] [Google Scholar]
- 7.Elkins, J. M., M. J. Ryle, I. J. Clifton, J. C. Dunning Hotopp, J. S. Lloyd, N. I. Burzlaff, J. E. Baldwin, R. P. Hausinger, and P. L. Roach. 2002. X-ray crystal structure of Escherichia coli taurine/α-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry 41:5158-5192. [DOI] [PubMed] [Google Scholar]
- 8.Farhana, L., and P. B. New. 1997. The 2,4-dichlorophenol hydroxylase of Alcaligenes eutrophus JMP134 is a homotetramer. Can. J. Microbiol. 43:202-205. [DOI] [PubMed] [Google Scholar]
- 9.Fukumori, F., and R. P. Hausinger. 1993. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J. Biol. Chem. 268:24311-24317. [PubMed] [Google Scholar]
- 10.Fukumori, F., and R. P. Hausinger. 1993. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulthorpe, R. R., C. McGowan, O. V. Maltseva, W. H. Holben, and J. M. Tiedje. 1995. 2,4-Dichlorophenoxyacetic acid-degrading bacteria contain mosaics of catabolic genes. Appl. Environ. Microbiol. 61:3274-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. B. Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. H. Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 13.Gribble, G. W. 2003. The diversity of naturally produced organohalogens. Chemosphere 52:289-297. [DOI] [PubMed] [Google Scholar]
- 14.Hiraishi, A. 1992. Direct automated sequencing of 16S rRNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett. Appl. Microbiol. 15:21-213. [DOI] [PubMed] [Google Scholar]
- 15.Hogan, D. A., S. R. Smith, E. A. Saari, J. McCracken, and R. P. Hausinger. 2000. Site-directed mutagenesis of 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase. Identification of residues involved in metallocenter formation and substrate binding. J. Biol. Chem. 275:12400-12409. [DOI] [PubMed] [Google Scholar]
- 16.Itoh, K., R. Kanda, Y. Momoda, Y. Sumita, Y. Kamagata, K. Suyama, and H. Yamamoto. 2000. Presence of 2,4-D-catabolizing bacteria in a Japanese arable soil that belong to BANA (Bradyrhizobium-Agromonas-Nitrobacter-Afipia) cluster in α-Proteobacteria. Microbes Environ. 15:113-117. [Google Scholar]
- 17.Itoh, K., R. Kanda, Y. Sumita, H. Kim, Y. Kamagata, K. Suyama, H. Yamamoto, R. P. Hausinger, and J. M. Tiedje. 2002. tfdA-like genes in 2,4-dichlorophenoxyacetic acid-degrading bacteria belonging to the Bradyrhizobium-Agromonas-Nitrobacter-Afipia cluster in α-Proteobacteria. Appl. Environ. Microbiol. 68:3449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ka, J. O., and J. M. Tiedje. 1994. Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergenetic transfer. J. Bacteriol. 176:5284-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Genetic and phenotypic diversity of 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacteria isolated from 2,4-D-treated field soils. Appl. Environ. Microbiol. 60:1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima. M. Kohara. M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa, W., S. Takami, K. Miyauchi, E. Masai, Y. Kamagata, J. M. Tiedje, and M. Fukuda. 2002. Novel 2,4-dichlorophenoxyacetic acid degradation genes from oligotrophic Bradyrhizobium sp. strain HW13 isolated from a pristine environment. J. Bacteriol. 184:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, C. M., J. H. J. Leveau, A. J. B. Zehnder, and J. R. van der Meer. 2000. Characterization of a second tfd gene cluster for chlorophenol and chlorocatechol metabolism on plasmid pJP4 in Ralstonia eutropha JMP134(pJP4). J. Bacteriol. 182:4165-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lafay, B., and J. J. Burdon. 1998. Molecular diversity of rhizobia occurring on native shrubby legumes in southeastern Australia. Appl. Environ. Microbiol. 64:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, T., and P. J. Chapman. 1984. Purification and properties of a plasmid-encoded 2,4-dichlorophenol hydroxylase. FEBS Lett. 173:314-318. [DOI] [PubMed] [Google Scholar]
- 27.Maltseva, O., C. McGowan, R. Fulthorpe, and P. Oriel. 1996. Degradation of 2,4-dichlorophenoxyacetic acid by haloalkaliphilic bacteria. Microbiology 142:1115-1122. [DOI] [PubMed] [Google Scholar]
- 28.McGowan, C., R. Fulthorpe, A. Wright, and J. M. Tiedje. 1998. Evidence for interspecies gene transfer in the evolution of 2,4-dichlorophenoxyacetic acid degraders. Appl. Environ. Microbiol. 64:4089-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minamisawa, K., T. Isawa, Y. Nakatsuka, and N. Ichikawa. 1998. New Bradyrhizobium japonicum strains that possess high copy numbers of the repeated sequence RSα. Appl. Environ. Microbiol. 64:1845-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minamisawa, K., M. Itakura, M. Suzuki, K. Ichige, T. Isawa, K. Yuhashi, and H. Mitsui. 2002. Horizontal transfer of nodulation genes in soils and microcosms from Bradyrhizobium japonicum to B. elkanii. Microbes Environ. 17:82-90. [Google Scholar]
- 31.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neilson, J. W., K. L. Josephson, I. L. Pepper, R. B. Arnold, G. D. DiGiovanni, and N. A. Sinclair. 1994. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl. Environ. Microbiol. 60:4053-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parker, M. A. 1999. Relationship of bradyrhizobia from the legumes Apios americana and Desmodium glutionosum. Appl. Environ. Microbiol. 65:4914-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perkins, E. J., M. P. Gordon, O. Caceres, and P. F. Lurquin. 1990. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J. Bacteriol. 172:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramseier, T. M., and M. Göttfert. 1991. Codon usage and G+C content in Bradyrhizobium japonicum genes are not uniform. Arch. Microbiol. 156:270-276. [DOI] [PubMed] [Google Scholar]
- 36.Saito, A., H. Mitsui, R. Hattori, K. Manamisawa, and T. Hattori. 1998. Slow-growing and oligotrophic bacteria phylogenetically close to Bradyrhizobium japonicum. FEMS Microb. Ecol. 25:277-286. [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Streber, W. R., K. N. Timmis, and M. H. Zenk. 1987. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J. Bacteriol. 169:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suwa, Y., A. D. Wright, F. Fukumori, K. A. Nummy, R. P. Hausinger, W. E. Holben, and L. J. Forney. 1996. Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid/α-ketoglutarate dioxygenase from Burkholderia sp. strain RASC. Appl. Environ. Microbiol. 62:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonso, N. L., V. G. Matheson, and W. E. Holben. 1995. Polyphasic characterization of a suite of bacterial isolates capable of degrading 2,4-D. Microb. Ecol. 30:3-24. [DOI] [PubMed] [Google Scholar]
- 42.Top, E. M., W. E. Holben, and L. J. Forney. 1995. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl. Environ. Microbiol. 61:1691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vallaeys, T., L. Courde, C. McGowan, A. D. Wright, and R. R. Fulthorpe. 1999. Phylogenetic analyses indicate independent recruitment of diverse gene cassettes during assemblage of the 2,4-D catabolic pathway. FEMS Microb. Ecol. 28:373-382. [Google Scholar]
- 44.Vallaeys, T., R. R. Fulthorpe, A. M. Wright, and G. Soulas. 1996. The metabolic pathway of 2,4-dichlorophenoxyacetic acid degradation involves different families of tfdA and tfdB genes according to PCR-RFLP analysis. FEMS Microb. Ecol. 20:163-172. [Google Scholar]
- 45.van der Meer, J. R., W. M. de Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]