Abstract
To better understand the defense mechanism of Streptococcus thermophilus against superoxide stress, molecular analysis of 10 menadione-sensitive mutants, obtained by insertional mutagenesis, was undertaken. This analysis allowed the identification of 10 genes that, with respect to their putative functions, were classified into five categories: (i) those involved in cell wall metabolism, (ii) those involved in exopolysaccharide translocation, (iii) those involved in RNA modification, (iv) those involved in iron homeostasis, and (v) those whose functions are still unknown. The behavior of the 10 menadione-sensitive mutants exposed to heat shock was investigated. Data from these experiments allowed us to distinguish genes whose action might be specific to oxidative stress defense (tgt, ossF, and ossG) from those whose action may be generalized to other stressful conditions (mreD, rodA, pbp2b, cpsX, and iscU). Among the mutants, two harbored an independently inserted copy of pGh9:ISS1 in two loci close to each other. More precisely, these two loci are homologous to the sufD and iscU genes, which are involved in the biosynthesis of iron-sulfur clusters. This region, called the suf region, was further characterized in S. thermophilus CNRZ368 by sequencing and by construction of ΔsufD and iscU97 nonpolar mutants. The streptonigrin sensitivity levels of both mutants suggest that these two genes are involved in iron metabolism.
Oxygen is an essential element for the viability of most living organisms through its involvement in oxidative phosphorylation that takes place during the aerobic respiration process. However, oxygen can also be harmful due to the production, especially during respiration, of toxic derivatives, namely, reactive oxygen species (ROS) such as hydrogen peroxide, hydroxyl radicals, and superoxide radicals (for a review, see reference 25). The accumulation of these molecules triggers lipid, protein, and nucleic acid oxidation, causing lethal cellular damage (for reviews, see references 7, 25, and 53). In response to such attacks, aerobic microorganisms have evolved antioxidant mechanisms (e.g., catalase, peroxidase, and superoxide dismutase activities) to protect and adapt cells against ROS excess (6, 18, 24, 65, 68).
Although they do not achieve aerobic respiration, anaerobic microorganisms can be subject to ROS. The strict anaerobes Bacteroides fragilis and Clostridium perfringens are able to survive in the presence of a small amount of oxygen due to the induction of an adaptive oxidative stress response that is activated under aerobic conditions (13, 46-49). The peroxide response of B. fragilis shares some characteristics with that of Escherichia coli, such as the involvement of the redox-sensitive transcriptional activator OxyR (46) and some antioxidant activities under its control (47, 49).
The lactic acid bacterium (LAB) Streptococcus thermophilus is a gram-positive bacterium widely used in dairy factories, mainly as a starter of fermentation in yogurt and cheese production. During fermentation processes, S. thermophilus is exposed to oxygen and to its deleterious effects, which can lead to a reduction of cell viability and consequently may have repercussions on the texture, flavor, and safety of the final product.
S. thermophilus is an anaerobic aerotolerant organism that can grow in the presence of oxygen and survive to low concentrations of superoxide and hydroxyl radicals (59). This ability suggests that S. thermophilus displays a defense system that provides protection against the effects of ROS. In the literature, data about the oxidative stress response of this organism are scarce. The existence of at least one ROS defense system and its inducibility by hydrogen peroxide are established for S. thermophilus CNRZ368 (58). This organism possesses a single manganese-containing superoxide dismutase (19) and a glutathione reductase (43). However, no catalase activity was found. In other streptococci, antioxidant enzymes were reported to play roles in defense against oxidative stress (27, 33, 44, 66) and in pathogenesis (45, 67, 69).
The aim of this work was to identify genes involved in the oxidative stress defense of S. thermophilus CNRZ368 without presuming their function. For this purpose, a collection of mutants was created by random insertional mutagenesis and the mutants were screened for their sensitivity to menadione (a superoxide-generating agent). The locus disrupted in 10 S. thermophilus menadione-sensitive clones was identified, and the potential physiological role of the mutated genes in oxidative stress defense is discussed. The implications of these 10 loci during heat shock were also evaluated to assess their specificity to oxidative stress defense. Moreover, the characterization of two of these mutants, disrupted in the sufD and iscU genes, which may participate in [Fe-S] cluster assembly or repair, is more detailed. The results presented in this paper indicate that the inactivation of sufD and iscU genes caused higher sensitivity to superoxide stress and suggest that [Fe-S] cluster repair or assembly could play an important role in the oxidative stress defense of anaerobic bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. thermophilus CNRZ368 was purchased from the Institut National de la Recherche Agronomique-Centre National de Recherches Zootechniques (CNRZ) (Jouy en Josas, France) strain collection. Depending on the experiments, S. thermophilus CNRZ368 and its derivatives were cultivated at 30 or 42°C in milk medium (for precultures), TPPY medium (12), or M17 medium (55) without shaking. Erythromycin was added at 2 μg · ml−1 when required. Streptonigrin sensitivity was monitored by using a TPPYC medium which contained a reduced concentration of metallic dications (including Fe2+). TPPYC was prepared according to the protocol described by Visca et al. (62) and modified as follows: Chelex beads (20 g/liter; Sigma) were added to TPPY medium and stirred at 4°C for 48 h, and the medium was then supplemented with 1 mM MgCl2 and 1 mM CaCl2.
Recombinant plasmids were transformed into E. coli EC101, a TG1 strain [supE hsd-5 thi Δ(lac-proAB) F′ (traD6 proAB+ lacIq lacZΔM15)] containing a chromosomal copy of the pWV01 repA gene (34) and selected at 37°C on Luria-Bertani (LB) (50) agar plates containing 150 μg of erythromycin/ml.
Insertional mutagenesis.
The collection of mutants was obtained by insertional mutagenesis with the plasmid pGh9:ISS1, which carries the Emr gene (35). The mutagenesis procedure and the cloning of the fragments flanking the pGh9:ISS1 insertion locus were carried out as previously described (56).
Selection of mutants impaired in their ability to tolerate menadione.
For the selection of menadione-sensitive clones, two different procedures, differing mainly in the length of the menadione exposure, were assayed. Both assays were preceded by a preculture in milk medium containing 2 μg of erythromycin/ml; precultures of the 2,112 mutants were performed with 96-well microtiter plates.
For long-term menadione exposure, each preculture was transferred onto M17 agar plates containing 2 μg of erythromycin/ml with increasing concentrations of menadione (from 0 to 120 μg/ml). After 20 h of incubation at 42°C, the presence (or absence) of colonies was observed.
For short-term menadione exposure, each preculture was diluted (at a 1/100 ratio) in five different 96-well microtiter plates containing liquid TPPY medium supplemented with 2 μg of erythromycin/ml (in a final volume of 100 μl, resulting in an approximately 1:1 liquid-to-air space ratio). After a 3.5-h growth period at 42°C, five different concentrations of menadione (from 0 to 15 mg/ml) were added to each of the five microtiter plates. Microtiter plates were incubated at 42°C for 3 h. The cultures were then transferred onto TPPY agar plates containing erythromycin (2 μg/ml) by using a hedgehog. The ability of the mutants to survive the menadione stress was assessed after 20 h of incubation at 42°C by observation of the presence or absence of colonies.
For each experiment, clones were considered sensitive when they did not grow on a plate on which more than 90% of the clones were growing. Both procedures were applied twice, and the results were compiled to reveal the mutants exhibiting the more severe phenotype.
DNA manipulations.
The preparation of chromosomal, plasmid, and recombinant DNA and Southern analysis were performed according to standard protocols (50). All cloning junctions and cloned PCR products were checked by sequencing with Dye-Terminator chemistry on an ABI Prism 377 genetic analyzer (PE Biosystems). Sequence data were analyzed by using GenBank, BLAST (2, 3), TMHMM (51), and HMMTOP (61) software.
Construction of the ΔsufD and iscU97 mutants.
Oligonucleotides used to construct the ΔsufD and iscU97 mutants were purchased from MWG Biotech AG (Ebersberg, Germany). To construct the nonpolar truncated ΔsufD mutant, the 5′ and 3′ ends of the sufD gene were independently amplified by PCR. The 5′ end was amplified by using the primers GAGCGTGTTGAATTCCACCG, containing an EcoRI site (in boldface type) and AACTTAACTGCAGCACCGTCT, containing a PstI site (in boldface type). The 3′ end was amplified by using the primers GGTGTTCCAGAATTCAAATAACC, containing an EcoRI site, and TGGTCTGTCATCAGGTCG. The latter primer does not contain a PstI site, but the PCR product generated does contain a PstI restriction site. These two PCR products were digested by PstI and ligated to each other. The resulting fragment was amplified by PCR and then digested by EcoRI and cloned into pGh9. The final construct (pGh9ΔsufD) was used to transform S. thermophilus CNRZ368. For IscU protein inactivation, the strategy consisted of replacing the TGT codon encoding the conserved Cys97 with a TAT codon encoding a Tyr. This was achieved by replacing, in oligonucleotides, the three underlined nucleotides of the sequence AGG TGT ACA, generating an EcoRV restriction site (AGA TAT CCA). Thus, the three amino acids Gly96, Cys97, and Tyr98 were replaced by Arg96, Tyr97, and Thr98, respectively, in the mutated protein. Two internal and complementary primers containing the mutation (GCTGGTGATGATATCACCATTTCAACA and GAAATGGTGATATCATCACCAGCAAAGG) were used with two additional external primers (AAGCGGAATTCAAGCATCCACTCAGGC, containing an EcoRI site, and TGTGACAAGCTTGTAGAGGCAATTCTT, containing an HindIII site), respectively, to amplify the 5′ and 3′ ends of iscU. The two PCR products, of 515 and 262 bp, were digested by EcoRI and EcoRV and by HindIII and EcoRV, respectively, before being ligated to each other. The resulting fragment was cloned into pGh9, which was digested by EcoRI and HindIII. The final construct, pGh9iscU97, was used to transform S. thermophilus CNRZ368. Preparation of S. thermophilus competent cells was performed as previously described by O'Sullivan and Fitzgerald (41). The following gene replacements were done as previously described (8) but in TPPY medium and with 30 and 42°C as the permissive and restrictive temperatures, respectively.
Physiological characterization of S. thermophilus mutant strains.
For short-term methyl viologen exposure, cultures of CNRZ368 and its derivative strains were grown in TPPY medium. At an optical density at 600 nm (OD600) of 0.6, cultures were divided into two subcultures (at a ratio of liquid to air space of approximately 1:1). Methyl viologen (paraquat; Sigma-Aldrich) was added to one of the subcultures at a final concentration of 50 mM. After 3 h of incubation at 42°C, cultures were diluted to appropriate concentrations and spread onto TPPY agar plates. For long-term methyl viologen exposure, strains were grown in TPPY medium until they reached an OD600 of 0.6. Cultures were then diluted to a final theoretical OD600 of 0.001 in TPPY medium (at a ratio of liquid to air space of approximately 5:1) supplemented with 1.7 mM methyl viologen or not supplemented (control experiment). After 18 h of incubation at 42°C, cultures were diluted in TPPY to appropriate concentrations and spread onto TPPY agar plates. In both procedures, methyl viologen sensitivity was estimated by comparing CFU-per-milliliter values obtained for treated cultures with those obtained for nontreated cultures.
Heat shock.
The ability of S. thermophilus and its derivatives to respond to a heat stress was assessed as follows. Each strain was grown at 42°C in TPPY medium until the early log phase (OD600 = 0.3). Then, the culture was divided into two parts. One part was kept at 42°C (control experiment); the other one was immediately transferred to 60°C for 15 min. Appropriate dilutions were spread on TPPY agar plates. The fraction of surviving cells was calculated by dividing the surviving number of CFU per milliliter by the count for the control experiment.
Streptonigrin sensitivity.
Streptonigrin sensitivity was monitored for cells grown overnight in TPPYC medium at 42°C. Cells were harvested by centrifugation, washed twice in TPPYC medium, resuspended in TPPYC, diluted to an OD600 of 0.01, and cultivated in TPPYC for 9 h at 42°C. Then, the cultures were diluted again to an OD600 of 0.005 and divided into four parts, which were all incubated for 21 h at 42°C. One part was supplemented with 0.4% (vol/vol) dimethyl formamide (control experiment), and the other three parts were supplemented with different concentrations of streptonigrin (1, 2, and 4 μg/ml, dissolved in dimethyl formamide). As a control experiment, cultures were also carried out with TPPYC supplemented with 1 mM of 2′2-dipyridyl (an Fe chelator). In each experiment, the OD600 value was used to estimate cellular density after 21 h of incubation. The relative optical density was determined by comparing the cellular densities obtained with and without streptonigrin. All experiments were performed in triplicate.
Nucleotide sequence accession numbers.
For all analyzed sequences, the DNA sequences reported in this paper have been deposited in GenBank under accession numbers AY386239, AY386240, AY386241, AY386242, AY386243, AY386244, and AY386245.
RESULTS
Selection of S. thermophilus CNRZ368 mutants with altered oxidative stress response.
A collection of 2,112 mutants of S. thermophilus CNRZ368 was generated by insertional mutagenesis with the plasmid pGh9:ISS1 (35, 56). This plasmid carries an erythromycin resistance gene for its selection and the insertional element ISS1, which allows random integration of the plasmid into the S. thermophilus genome. The 2,112 mutants obtained were screened to select clones sensitive to menadione (a superoxide-generating compound). Two distinct selection procedures were used to identify mutants susceptible to menadione (see Materials and Methods). The characterization of 10 mutants, disrupted in loci originally named ossA to ossJ (for oxidative stress-sensitive locus), is described in this paper.
Sensitivity of S. thermophilus CNRZ368 mutants to methyl viologen.
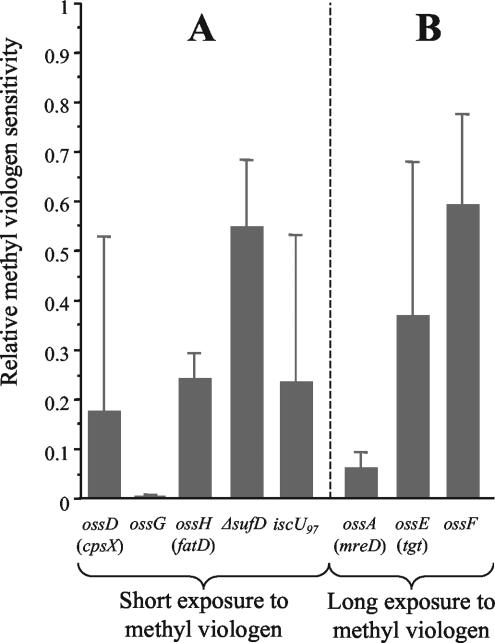
Previously, we demonstrated that ossB and ossC mutants (previously named 6G4 and 18C3, respectively) are compromised in their ability to survive superoxide stress (57). To confirm that the other eight mutants selected were sensitive to superoxide stress, their sensitivity to methyl viologen was examined (Fig. 1). In this experiment, we used the nonpolar mutants ΔsufD and iscU97 constructed in this work (see below) rather than the potential polar clones ossI and ossJ. Short-term methyl viologen exposure revealed the extreme sensitivity of ossG, as well as the four- to fivefold increased sensitivity of the ossD, ossH, and iscU97 mutants (Fig. 1A). Although its sensitivity was less severe, the ΔsufD mutant exhibited about twofold less survival following such stress. Under these conditions, the other three mutants (ossA, ossE, and ossF) did not exhibit any significant sensitivity; however, long-term methyl viologen exposure confirmed their sensitivity. Indeed, long-term methyl viologen exposure (Fig. 1B) showed that the ossA mutant strain displayed a relative survival rate more than 10-fold lower than that of the wild-type strain. The ossE and ossF mutants also showed a twofold lower survival rate than the wild-type strain.
FIG. 1.
Relative methyl viologen sensitivity. (A) Results of short-term methyl viologen exposure. (B) Results of long-term methyl viologen exposure (see Materials and Methods). The relative methyl viologen sensitivity was determined by comparing viabilities with and without treatment. Levels are shown as values relative to the survival level of the wild-type strain. All experiments were performed at least in duplicate. Error bars show two standard deviations of the data.
Survival of the mutants exposed to heat stress.
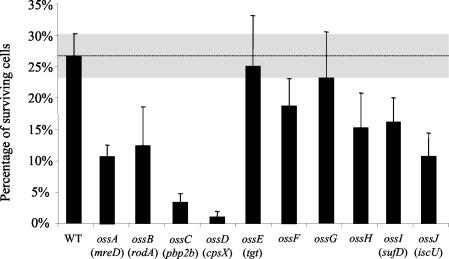
To assess the specificity of these 10 loci to oxidative stress defense, the abilities of the mutants to survive to heat shock were estimated. Five out of the 10 mutants (ossA, ossB, ossC, ossD, and ossJ) isolated as oxidative stress-sensitive clones also showed reduced survival with heat shock compared with the wild type (Fig. 2). These data suggest that the genes impaired in these mutants are not specific to the oxidative stress defense process but are probably involved in a more global response to stresses.
FIG. 2.
Survival after heat shock. The heat shock treatment consisted of 15 min at 60°C. The percentage of surviving cells corresponds to the number of CFU counted on plates after the heat shock exposure divided by the number of CFU before treatment. Error bars show two standard deviations of the data. The dotted line represents the surviving percentage observed for the wild-type (WT) strain, and the gray band comprises two standard deviations of the data.
In contrast, the ossE, ossF, and ossG mutants did not differ significantly from the wild type, indicating that the genes disrupted in these strains did not influence the ability of cells to respond to heat shock. Interestingly, the ossG mutant, which was highly sensitive to methyl viologen, showed no significant decrease of survival when subjected to heat shock compared with that of the wild type, suggesting that the ossG gene, essential for oxidative stress defense, could be specific to such a stress.
Identification of genes implicated in oxidative stress tolerance.
The disrupted locus was identified for each of the 10 mutants by cloning and sequencing the junctions as previously described (56). All of the insertions were mapped within coding sequences. Sequence similarity searches and analysis revealed similar sequences in other bacteria (see Table 1), with an E-value lower than 10−10 for BLASTX searches. In 9 of 10 cases, data about the functions of these genes were available. To facilitate their description, these genes were grouped into five classes (classes I to V) according to their putative functions and renamed when relevant (Table 1).
TABLE 1.
Products of oss genes: database similarities and putative functions
| Gene namea | Accession no. | Gene encoding the most similar productc | Amino acid identityd | Insertion sitee | Putative function | Class |
|---|---|---|---|---|---|---|
| ossA (mreD) | AY386242 | S. mutans mreD (168) | 30 (1-168) | 127 | Cell shape maintenance | I |
| ossB (rodA) | AF399832 | S. mutans rodA (408) | 65 (1-391) | 304 | Peptidoglycan elongation | I |
| ossC (pbp2b)b | AF399833 | S. thermophilus Sfi6 pbp2b (704) | 98 (641-704) | 640 | Peptidoglycan elongation | I |
| ossD (cpsX)b | AY386241 | S. thermophilus cpsX (378) | 98 (212-378) | 264 | Membrane translocation of exopolysaccharide | II |
| ossE (tgt)b | AY386243 | S. pneumoniae tgt (380) | 92 (1-333) | 99 | tRNA-guanine transglycosylase | III |
| ossFb | AY386244 | S. agalactiae gbs0448 (451) | 77 (1-204) | 51 | RNA methyltransferase | III |
| ossG | AY386239 | S. mutans smu.1428c (284) | 67 (1-280) | 170 | Unknown conserved function | IV |
| ossH (fatD) | AY386240 | S. pneumoniae fatD (324) | 55 (22-323) | 119 | Iron ABC transporter | V |
| ossI (sufD) | AY386245 | S. agalactiae sag0142 (420) | 76 (1-420) | 144 | Assembly of Fe/S clusters | V |
| ossJ (iscU) | AY386245 | S. agalactiae gbs0140 (147) | 78 (1-141) | 57 | Assembly of Fe/S clusters | V |
Initial gene name with the definitive gene name in parentheses where applicable.
Disrupted ORF of S. thermophilus CNRZ368 that has been partially sequenced.
Organism, gene name, and (in parentheses) length (in amino acids) of the protein sequence giving the best alignment score.
Percentage of sequence identity with the protein showing the best alignment score with the S. thermophilus Oss protein. In parentheses is the portion of the protein sharing this identity (numbers correspond to amino acid positions).
Deduced amino acid position of the pGh9:ISS1-generated disruption in the full-length protein showing the best alignment score with the Oss protein.
(i) Class I: genes potentially involved in cell wall structure and cell shape determination.
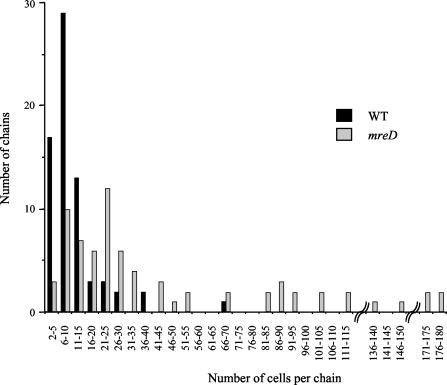
The ossA, ossB, and ossC putative gene products shared sequence identity with MreD, RodA, and Pbp2b, respectively, which putatively direct the elongation of the lateral cell wall in E. coli, conferring on the cell its characteristic rod shape (5, 28, 29). The ossA mutant was defective in a gene encoding a protein with 30% identity to MreD of Streptococcus mutans. Sequence analysis of the surrounding region indicated that the S. thermophilus mreD gene (ossA) was clustered with a gene encoding a putative protein similar to MreC. Moreover, hydropathy profiles of MreD (OssA) determined by using TMHMM and HMMTOP showed five regions of sufficient length and hydrophobicity to be membrane-spanning segments (data not shown), suggesting that this protein is the counterpart of MreD. The relatively low identity (30%) observed between S. thermophilus MreD and S. mutans MreD was in agreement with the general idea that the structure of this protein is more highly conserved than its primary sequence (17). To study the effect of mreD disruption in S. thermophilus, TPPY cultures of the wild-type and mreD mutant strains were observed with light microscopy. In contrast to wild-type cells, mreD mutant cells formed very long chains, with up to 180 cells per chain (Fig. 3). Moreover, a reduction of about 25% of the cell size was observed in the mutant. The growth of the mreD mutant and that of the wild type in TPPY medium were monitored by measuring the OD600. Both strains displayed doubling times of 30 min, but the maximal OD600 of the mreD mutant (1.7 ± 0.1) was lower than that of the wild type (2.4 ± 0.2), showing that mreD mutant cells entered the stationary phase earlier than wild-type cells. The characterization of rodA and pbp2b mutants of S. thermophilus CNRZ368 was described in a previous paper (57).
FIG. 3.
Influence of mreD disruption on chain length. Cell number per chain was determined for 70 chains of the wild-type (WT) and mreD mutant strains grown in TPPY liquid medium.
(ii) Class II: genes potentially involved in exopolysaccharide translocation.
The sequence disrupted in the ossD mutant was partially determined. Its analysis indicated that the C-terminal portion of the predicted encoded protein showed 98% identity to CpsX, a putative membrane protein from S. thermophilus MR-1C. The role of CpsX is still unknown, but the conserved location of cpsX near eps genes of S. thermophilus (14) and the presence of a putative gene related to an ABC transport system in its vicinity argue in favor of CpsX being involved in exopolysaccharide translocation (15). Moreover, the TMHMM and HMMTOP software predictions indicated that CpsX (OssD) from S. thermophilus CNRZ368 would contain highly hydrophobic domains compatible with a location in the cytoplasmic membrane. Although the function of cpsX is not known yet, our work indicates that its disruption is associated with sensitivity not only to superoxide stress but also to heat shock.
(iii) Class III: genes involved in RNA maturation.
In the ossE and ossF mutants, the plasmid inserted into genes whose predicted products share sequence identity with enzymes involved in RNA maturation. The ossE gene encodes a putative tRNA guanine transglycosylase (TGT) which shares 92% identity with the putative Streptococcus pneumoniae TGT. TGT enzyme takes part in the process that removes the guanine residue at position 34 (the wobble position of the anticodon loop) of tRNAHis, tRNAAsp, tRNAAsn, and tRNATyr and replaces it with the modified residue queuosine (39, 40).
ossF potentially encodes a protein that shows 77% identity to a putative RNA methyltransferase of Streptococcus agalactiae (Gbs0448) belonging to the TrmA family. Enzymes of the TrmA family catalyze the formation of the 5-methyluridine in position 54 (localized in the TΨC loop) of tRNA (9). The presence of a TRAM-like domain (RNA binding domain) in the N-terminal part of OssF supports the idea of an interaction between this protein and RNA.
(iv) Class IV: genes of unknown function.
The ossG product shows 67% identity with the product of an S. mutans gene of unknown function (smu.1428c) (Table 1). Although the function of ossG is not known yet, the mutation of this gene is now associated with a phenotype of sensitivity to superoxide stress. The TMHMM and HMMTOP software predictions suggest the existence of two transmembrane domains in the N-terminal region of OssG, making OssG a potential membrane protein.
(v) Class V: genes involved in iron metabolism. (a) Ferric iron transport.
The putative product of ossH shows 55% identity to FatD from S. pneumoniae, a potential membrane-spanning permease of an Fe3+ ABC transporter. Analysis of the adjacent sequence reveals the presence of a gene encoding a putative protein similar to FatC downstream of fatD (ossH) in the same orientation. The fatC gene is also clustered with fatD in the S. pneumoniae genome. The hydropathy profile of FatD (OssH) from S. thermophilus, assessed with TMHMM and HMMTOP, reveals that this protein displays 10 putative-membrane spanning segments compatible with its expected cytoplasmic membrane location.
(b) [Fe-S] cluster biosynthesis.
The sequence affected in the ossI mutant encodes a product that shows 75% identity to Sag0142, a putative protein deduced from the complete genome sequence of S. agalactiae and similar to SufD-like proteins. Analysis of totally sequenced microbial genomes reveals that sufD-like genes are conserved in bacteria and also in some archaea, such as Halobacterium sp. NRC-1, Thermoplasma acidophilum, and Aeropyrum pernix (23). In E. coli and Erwinia chrysanthemi, the sufD gene belongs to the sufABCDSE cluster identified as being involved in iron metabolism and, more precisely, in [Fe-S] cluster biogenesis and/or repair (37, 42). The ossJ gene product shows sequence identity to the gbs0140 product from S. agalactiae (Table 1), probably encoding a IscU protein, which is a NifU family protein. NifU proteins have been proposed to serve as scaffold proteins in [Fe-S] cluster assembly in Synechocystis PCC6803 and would function as a mediator for iron-sulfur cluster delivery (38).
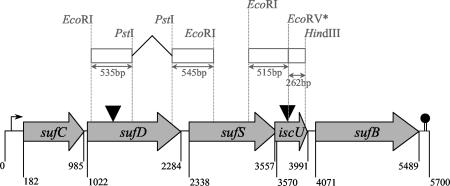
In some genomes, including that of Streptococcus pyogenes, nifU-like genes are clustered with the suf genes (23). To determine whether sufD (ossI) and iscU (ossJ) from S. thermophilus were genetically linked together, the chromosomal DNA regions flanking the pGhost9:ISS1 insertion sites of both mutants were sequenced until they overlapped. Examination of the genes in the neighborhood of sufD and iscU revealed that they both belong to a cluster of five genes all orientated in the same direction. This cluster, named the sufCDS iscU sufB cluster, is shown in Fig. 4. The highest sequence identities found for these gene products are those to putative proteins from S. agalactiae (Table 2). They also share sequence similarity with proteins encoded by E. coli and Erwinia chrysanthemi suf gene clusters. The putative SufC, SufD, SufS, and SufB proteins encoded by the S. thermophilus cluster present, respectively, 72, 48, 65, and 57% similarity to their E. coli counterparts. Moreover, the S. thermophilus IscU presents 47% similarity with IscU from E. coli.
FIG. 4.
Schematic map of the S. thermophilus sufCDS iscU sufB cluster. Genes are indicated by gray arrows, and the positions of putative start and stop codons are given (in base pairs) below the map. The sites of the insertion of the pGh9:ISS1 plasmid into sufD (ossI)- and iscU (ossJ)-disrupted mutants are indicated by filled triangles. The broken arrow indicates the position of the suf cluster putative promoter. The putative rho-independent terminator is represented as hairpin loop. Fragments used for the generation of the nonpolar mutants are represented by open boxes. An asterisk indicates the position of the EcoRV restriction site causing the replacement of Cys97 with a Tyr in the iscU97 mutant.
TABLE 2.
suf cluster gene products of S. thermophilus CNRZ368
| Gene product | Closest similar proteina | Identity, similarity (%) | Function and/or nature of protein |
|---|---|---|---|
| SufC | CAD45782 (S. agalactiae NEM316) | 88, 94 | Putative ABC transporter (ATP-binding protein) |
| SufD | AAM99050 (S. agalactiae 2603V/R) | 76, 87 | Unknown, conserved protein |
| SufS | CAD45784 (S. agalactiae NEM316) | 79, 89 | Similar to SufS |
| IscU | CAD45785 (S. agalactiae NEM316) | 78, 90 | Similar to NifU family proteins |
| SufB | CAD45786 (S. agalactiae NEM316) | 89, 94 | Unknown, conserved protein |
Accession number of the protein giving the best alignment score. The name of the organism is given in parentheses.
Sequences corresponding to promoter regions were searched by inspection of the total sequence of the sufCDS iscU sufB cluster. A unique extended −10 sequence (5′-TTTGTTATAAT-3′) was found upstream of sufC, the first open reading frame (ORF) of the cluster. A potential rho-independent transcriptional terminator was identified 63 bp downstream of sufB, the last ORF of the cluster (ΔG = −10 kcal/mol). These features, together with the short or missing intergenic regions and the same orientation of the genes, suggest that these five ORFs form an operon, as is the case for the Erwinia chrysanthemi suf locus.
Molecular characterization of the sufCDS iscU sufB cluster. (i) Construction of ΔsufD and iscU97 mutants.
Since S. thermophilus suf genes are potentially clustered in an operon, nonpolar ΔsufD and iscU97 mutants were constructed and analyzed. The sufD mutation consists of an internal in-frame deletion leading to the loss of 43% of the whole protein (see Materials and Methods and Fig. 4). Sequence comparison of NifU family proteins from various organisms indicates that the S. thermophilus IscU protein contains the three highly conserved cysteine residues that have been shown to be the assembly site of a transitory [2Fe-2S] center within NifU (70). Thus, the construction of a null iscU mutant was achieved by replacing one of these conserved residues, the Cys97, with a Tyr (see Materials and Methods and Fig. 4). Sequencing of these two constructs confirmed that they did not contain any secondary mutation.
(ii) Role of sufD and iscU in iron metabolism.
To investigate the possibility that ΔsufD and iscU97 mutations impair growth due to the lack of functional [Fe-S] cluster biosynthesis, the mutants were grown in TPPY medium and compared to the wild type. The growth of the ΔsufD and iscU97 mutants showed no significant difference from that of the wild-type strain. Assuming that the high iron concentration of the TPPY medium could allow self-assembly of the [Fe-S] clusters (71) and consequently could be responsible for the absence of a difference in growth rates between strains, the same experiment was conducted in TPPYC, a divalent cation-depleted medium. Under these conditions, the mutants displayed a growth rate similar to that of the wild-type strain, but their maximal OD600 was lower than that of the wild type, showing their premature entry into the stationary phase (Table 3). Moreover, this difference disappeared when the cultures were grown on TPPYC supplemented with 100 μM FeSO4. Taken together, these results suggest (i) that iscU and sufD genes of S. thermophilus are involved in iron metabolism and (ii) that the presence of these genes would be of particular importance under iron-limiting conditions.
TABLE 3.
Growth of wild-type and mutant S. thermophilus strains in TPPYC and in TPPYC supplemented with irona
| Strain | Final OD600 resulting from growth in:
|
|
|---|---|---|
| TPPYC | TPPYC + FeSO4 | |
| Wild type | 0.96 | 1.32 |
| ΔsufD | 0.58 | 1.48 |
| iscU97 | 0.58 | 1.15 |
Cells were grown in milk medium for 15 h at 42°C and diluted in FeSO4-deficient medium (TPPYC medium supplemented with MgCl2 [1 mM] and CaCl2 [1 mM]). After 1 night of growth, cells were washed twice and resuspended either in FeSO4-deficient medium or in medium supplemented with 100 μM FeSO4. The growth of the cells was then followed by the measurement of the OD600. The OD600 was measured after 24 h of incubation at 42°C. This experiment was done in triplicate.
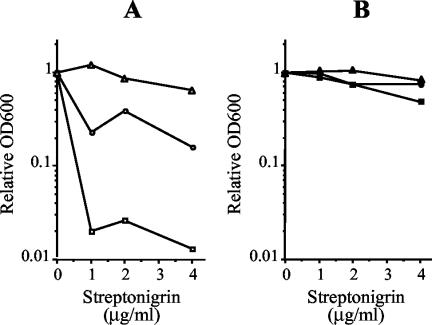
To confirm the involvement of sufD and iscU in iron metabolism, the iron intracellular concentrations of the ΔsufD and iscU97 mutants were checked by measuring the sensitivities of these strains to streptonigrin (an iron-activated antibiotic). As shown in Fig. 5A, the iscU mutant was more sensitive to streptonigrin than the wild-type strain was, whereas the ΔsufD mutant was more resistant. Moreover, the addition of 2,2′-dipyridyl (a ferrous iron chelator) abolished the difference in growth rates between the strains (Fig. 5B), indicating that iron was responsible for the streptonigrin sensitivity observed without 2,2′-dipyridyl. Taken altogether, these results suggest that both iscU and sufD of S. thermophilus play a role in iron metabolism.
FIG. 5.
Streptonigrin tolerance. The tolerance of S. thermophilus strains to streptonigrin was assessed by measuring the OD600 (OD600) after growth in TPPYC medium containing various concentrations of streptonigrin unsupplemented (A) or supplemented with 1 mM of 2,2′-dipyridyl (B) (see Material and Methods for details). The relative OD600 was determined by comparing the OD600s obtained with and without streptonigrin. ○ and •, wild-type strain; □ and ▪, iscU97 mutant; ▵ and ▴, ΔsufD mutant.
DISCUSSION
To investigate the oxidative stress defense system of S. thermophilus, a collection of random insertion mutants was screened to select clones with high sensitivity to menadione. Using this approach, we aimed to identify genes involved in superoxide stress defense without presuming their function. It can be noted that no gene encoding a protein considered a major antioxidant enzyme in other organisms was found by using this approach. Possible explanations for this outcome are (i) that the collection of mutants was nonexhaustive, (ii) that the corresponding mutants could have a weak phenotype, (iii) that such mutations could be lethal, and (iv) that most antioxidant enzymes, except those from SOD, are protective against H2O2 and not O2·−. The genes identified in the present work were classified according to their functions into five groups: genes potentially involved in cell wall metabolism, genes potentially involved in exopolysaccharide translocation, genes potentially involved in tRNA modification, genes potentially involved in iron metabolism, and genes of unknown function.
Perhaps the most striking observation of this study is that 5 out of the 10 sensitive mutants selected (ossH, mreD, rodA, pbp2b, and cpsX) are disrupted in a gene encoding a membrane protein. This finding could reflect that the cell envelope is the main target of the stress applied. Among these mutants, three (mreD, rodA and pbp2b) are impaired in genes potentially encoding proteins described as playing a role in peptidoglycan biosynthesis during cell elongation in E. coli (28, 52, 63). Phenotypic analysis of these three S. thermophilus mutants showed that they all present a reduced cell size associated with a modification of the cell shape, as observed in rodA, pbp2, and mre mutants of E. coli or Salmonella enterica serovar Typhimurium (5, 20, 28, 29, 63, 64). Moreover, the three S. thermophilus mutants also presented increased sensitivity to heat shock compared to the wild-type strain, suggesting that the protective role of MreD, RodA, and PBP2b is not specific to oxidative stress but could be enlarged to a more global stress response. In support of this hypothesis is the existence in Lactococcus lactis of mutations in pbp genes that negatively affect the acid tolerance of a guaA mutant (16).
The major function assigned to the cell wall is the maintenance of cell integrity through the protection of cells not only against osmotic stress but also against other stress conditions. For instance, it has already been shown that in LAB, compromised biosynthesis of peptidoglycan results in increased sensitivity to either UV (22) or acid (11) stress. Additionally, the disruption of mreD, rodA, or pbp2b may cause a modification of the peptidoglycan architecture in S. thermophilus, reducing cell protection against stresses such as oxidative and heat stresses.
tRNA modification is a universal feature of all living organisms, although some described modifications have no assigned function yet. Modifying enzymes affect the tRNA precursor either by chemical modification of an existing residue (in the base or in the sugar) or by nucleotide substitution. These modifications, particularly those present in the anticodon region, can affect translation efficiency and fidelity (for a review, see reference 10). In this study, two clones disrupted in the tgt and ossF loci, encoding proteins showing high identity to tRNA-modifying enzymes, were selected for their high sensitivity to oxidative stress. This sensitivity could suggest that the expression of genes involved in oxidative stress defense may require tRNA maturation or that such modifications could help tRNA stabilization under stress conditions. In agreement with the former hypothesis, Shigella flexneri pathogenicity is regulated by virF mRNA translation efficiency under TGT activity control (21). Additionally, several environmental conditions are suggested to influence tRNA modification. In the case of queuosine (whose synthesis and substitution are carried out by several enzymes, among which is TGT), synthesis is sensitive to the presence of iron (31) and TGT requires oxygen for its full activity (54). Therefore, further investigations on this topic should provide an interesting new regulatory model and could contribute to the emerging observation that translational regulation plays an important role in oxidative stress response, a role ascribed mainly to transcriptional regulation thus far.
Iron is essential for cellular metabolism due to its requirement as a cofactor for a large number of enzymes. However, an excess of iron is toxic because of its involvement in the Fenton reaction, which enhances the formation of ROS. In this work, a gene homologous to fatD, encoding a putative Fe3+ ABC transporter, was found to be involved in the S. thermophilus oxidative stress response. To test whether S. thermophilus FatD is indeed an iron transporter, the sensitivity of the fatD mutant to streptonigrin (an iron-activated antibiotic) was assessed. No significant difference between the mutant and the wild-type strains was detected (data not shown), suggesting that the disruption of fatD would have no effect on intracellular iron concentration. Thus, either FatD is not a major iron transporter under the experimental procedure we used or S. thermophilus fatD encodes an ABC transporter for another metallic cation. A possible cation could be Mn2+, known to have a protective function against oxidative damage either via direct dismutase activity, such as that reported in other organisms (4, 26, 60), or via its role as a cofactor of the only SOD reported in S. thermophilus (19). Thus, the inactivation of an Mn2+ ABC transporter could trigger sensitivity to oxidative stress.
[Fe-S] clusters are oxygen-sensitive prosthetic groups incorporated into proteins via covalent bonds to cysteines. Iron-sulfur proteins, which are found in more than 120 classes of enzymes and proteins (30), are known to play important physiological roles in electron transfer, metabolic reactions, and transcriptional regulation (30, 32). Indeed, because of their intrinsic redox properties, they are likely to sense oxygen or its derivatives, as is the case for SoxR and FNR transcriptional regulators, responding to oxidative stress and oxygen availability (for a review, see reference 32). IscR is also an iron-sulfur transcriptional regulator that is suggested to respond to [Fe-S] cluster-limiting conditions and potentially to hydrogen peroxide in E. coli (32). Although it has been shown that [Fe-S] cluster assembly can occur spontaneously in vitro under anaerobic conditions (36), this process can also be catalyzed enzymatically in vivo. Genes of the suf operon and of the isc locus (iron-sulfur cluster) are conserved and encode [Fe-S] cluster biosynthesis and/or repair pathways (23). More precisely, it has recently been proposed that Isc proteins are necessary for biosynthesis, while Suf proteins are essentially involved in the repair of oxidatively damaged [Fe-S] clusters (37).
Study of ΔsufD and iscU97 nonpolar mutants indicates that SufD and IscU proteins are required for growth in an iron-deficient medium. SufD and IscU inactivation conferred increased resistance and sensitivity, respectively, to streptonigrin. These results indicate that S. thermophilus SufD and IscU are involved in intracellular iron balance. Additionally, the SufD and IscU of S. thermophilus, similar to SufD and NifU family proteins from other microorganisms, are encoded by genes clustered with sufB, sufC, and sufS homologues, also potentially involved in [Fe-S] cluster repair. Hence, the participation of S. thermophilus SufD and IscU proteins in iron intracellular balance is probably the result of their involvement in [Fe-S] cluster repair, as in Erwinia chrysanthemi (37). The NifU family protein function is to provide a scaffold for the assembly of clusters mediated by NifS activity, and such clusters can then be transferred into apo forms of [Fe-S] cluster-containing proteins (1, 70). The role of SufD in [Fe-S] cluster biosynthesis is not well established. However, in S. thermophilus, the increased resistance to streptonigrin of the ΔsufD mutant suggests that its intracellular iron concentration is lower than that of the wild type. At least two alternative hypotheses can be made: (i) SufD could be directly involved in the uptake or assimilation of extracellular iron, or (ii) SufD could inhibit the synthesis and/or repair of the [Fe-S] clusters. In the latter case, SufD inactivation would result in an increase in the synthesis of [Fe-S] clusters and, consequently, in a reduced intracellular free iron concentration.
Additionally, S. thermophilus sufD and iscU mutants displayed increased sensitivity to superoxide radicals compared to the wild type. Thus, SufD and IscU are likely to play a major role in oxidative stress defense, probably resulting from their predicted role in [Fe-S] cluster assembly and/or repair. The involvement of the suf operon in oxidative stress resistance during the early steps of infection by Erwinia chrysanthemi has also been reported (37), as has the induction of the suf gene cluster of E. coli by the hydrogen peroxide response regulator OxyR (72). In spite of a possible spontaneous formation, our data suggest that under oxidative stress, [Fe-S] cluster assembly and/or repair becomes a limiting step in the formation of iron-sulfur proteins and that the cell requires an additional, dedicated system, encoded by the suf gene cluster, to optimize [Fe-S] cluster formation or repair.
Conclusion.
The strategy used in this work allowed the identification of genes involved in the S. thermophilus oxidative stress defense system. Data concerning the functions of some loci are already available in the literature, whereas the roles of other loci are not known. In both cases, the exact roles of these genes in the defense of S. thermophilus against oxidative stress remain to be elucidated.
Acknowledgments
A.T. and A.F. were supported by grants from the Ministère de la Recherche. F.B. was supported by a grant from the Institut National de la Recherche Agronomique.
We are grateful to Paul Hoskinson for his advice regarding the English formulation of the manuscript.
REFERENCES
- 1.Agar, J. N., C. Krebs, J. Frazzon, B. H. Huynh, D. R. Dean, and M. K. Johnson. 2000. IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39:7856-7862. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archibald, F. S., and I. Fridovich. 1981. Manganese, superoxide dismutase, and oxygen tolerance in some lactic acid bacteria. J. Bacteriol. 146:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benov, L. T., and I. Fridovich. 1994. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 269:25310-25314. [PubMed] [Google Scholar]
- 7.Berlett, B. S., and E. R. Stadtman. 1997. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 272:20313-20316. [DOI] [PubMed] [Google Scholar]
- 8.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjork, G. R. 1975. Transductional mapping of gene trmA responsible for the production of 5-methyluridine in transfer ribonucleic acid of Escherichia coli. J. Bacteriol. 124:92-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjork, G. R., J. M. Durand, T. G. Hagervall, R. Leipuviene, H. K. Lundgren, K. Nilsson, P. Chen, Q. Qian, and J. Urbonavicius. 1999. Transfer RNA modification: influence on translational frameshifting and metabolism. FEBS Lett. 452:47-51. [DOI] [PubMed] [Google Scholar]
- 11.Boyd, D. A., D. G. Cvitkovitch, A. S. Bleiweis, M. Y. Kiriukhin, D. V. Debabov, F. C. Neuhaus, and I. R. Hamilton. 2000. Defects in d-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bracquart, P. 1981. An agar medium for the differential enumeration of Streptococcus thermophilus and Lactobacillus bulgaricus in yogurt. J. Appl. Bacteriol. 51:303-305. [Google Scholar]
- 13.Briolat, V., and G. Reysset. 2002. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J. Bacteriol. 184:2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broadbent, J., D. McMahon, C. Oberg, and D. Welker. 2001. Use of exopolysaccharide-producing cultures to improve the functionality of low fat cheese. Int. Dairy J. 11:433-439. [Google Scholar]
- 15.Broadbent, J., D. McMahon, D. Welker, C. Oberg, and S. Moineau. 2003. Biochemistry, genetics, and applications of exopolysaccharide production in Streptococcus thermophilus: a review. J. Dairy Sci. 86:407-423. [DOI] [PubMed] [Google Scholar]
- 16.Budin, A., V. Pichereau, E. Domakova, J. Tremblay, R. Dervyn, E. Auffray, S. D. Ehrlich, S. Kulakauskas, and E. Maguin. 2002. Effect of a guaA mutation on the acid tolerance of L. lactis. Sci. Aliments 22:67-74. [Google Scholar]
- 17.Burger, A., K. Sichler, G. Kelemen, M. Buttner, and W. Wohlleben. 2000. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:1053-1060. [DOI] [PubMed] [Google Scholar]
- 18.Chae, H. Z., K. Robison, L. B. Poole, G. Church, G. Storz, and S. G. Rhee. 1994. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc. Natl. Acad. Sci. USA 91:7017-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang, S. K., and H. M. Hassan. 1997. Characterization of superoxide dismutase in Streptococcus thermophilus. Appl. Environ. Microbiol. 63:3732-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa, C. S., and D. N. Anton. 1993. Round-cell mutants of Salmonella typhimurium produced by transposition mutagenesis: lethality of rodA and mre mutations. Mol. Gen. Genet. 236:387-394. [DOI] [PubMed] [Google Scholar]
- 21.Durand, J. M., B. Dagberg, B. E. Uhlin, and G. R. Bjork. 2000. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol. Microbiol. 35:924-935. [DOI] [PubMed] [Google Scholar]
- 22.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis, K. E., B. Clough, J. W. Saldanha, and R. J. Wilson. 2001. Nifs and Sufs in malaria. Mol. Microbiol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 24.Faguy, D. M., and W. F. Doolittle. 2000. Horizontal transfer of catalase-peroxidase genes between archaea and pathogenic bacteria. Trends Genet. 16:196-197. [DOI] [PubMed] [Google Scholar]
- 25.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fridovich, I. 1983. Superoxide radical: an endogenous toxicant. Annu. Rev. Pharmacol. Toxicol. 23:239-257. [DOI] [PubMed] [Google Scholar]
- 27.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, B. G. Spratt, et al. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 261:7024-7031. [PubMed] [Google Scholar]
- 29.Iwaya, M., R. Goldman, D. J. Tipper, B. Feingold, and J. L. Strominger. 1978. Morphology of an Escherichia coli mutant with a temperature-dependent round cell shape. J. Bacteriol. 136:1143-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, M. K. 1998. Iron-sulfur proteins: new roles for old clusters. Curr. Opin. Chem. Biol. 2:173-181. [DOI] [PubMed] [Google Scholar]
- 31.Kersten, H., and W. Kersten. 1990. Biosynthesis and function of queuine and queuosine tRNAs, p. B69-B108. In C. W. Gehrke and K. C. T. Kuo (ed.), Chromatography and modification of nucleosides. Elsevier, Amsterdam, The Netherlands.
- 32.Kiley, P. J., and H. Beinert. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6:181-185. [DOI] [PubMed] [Google Scholar]
- 33.King, K. Y., J. A. Horenstein, and M. G. Caparon. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 182:5290-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leenhouts, K. 1995. Integration strategies and vectors. Dev. Biol. Stand. 85:523-530. [PubMed] [Google Scholar]
- 35.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merchant, S., and B. W. Dreyfuss. 1998. Metalloprotein and cytochrome assembly in chloroplasts. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:25-51. [DOI] [PubMed] [Google Scholar]
- 37.Nachin, L., M. El Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39:960-972. [DOI] [PubMed] [Google Scholar]
- 38.Nishio, K., and M. Nakai. 2000. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 275:22615-22618. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi, S., Y. Nishimura, Y. Hirota, and S. Nishimura. 1982. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 257:6544-6550. [PubMed] [Google Scholar]
- 40.Okada, N., S. Noguchi, H. Kasai, N. Shindo-Okada, T. Ohgi, T. Goto, and S. Nishimura. 1979. Novel mechanism of posttranscriptional modification of tRNA. Insertion of bases of Q precursors into tRNA by a specific tRNA transglycosylase reaction. J. Biol. Chem. 254:3067-3073. [PubMed] [Google Scholar]
- 41.O'Sullivan, T., and G. Fitzgerald. 1999. Electrotransformation of industrial strains of Streptococcus thermophilus. J. Appl. Microbiol. 86:275-283. [DOI] [PubMed] [Google Scholar]
- 42.Patzer, S. I., and K. Hantke. 1999. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J. Bacteriol. 181:3307-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pébay, M., A. C. Holl, J. M. Simonet, and B. Decaris. 1995. Characterization of the gor gene of the lactic acid bacterium Streptococcus thermophilus CNRZ368. Res. Microbiol. 146:371-383. [DOI] [PubMed] [Google Scholar]
- 44.Poole, L. B., M. Higuchi, M. Shimada, M. L. Calzi, and Y. Kamio. 2000. Streptococcus mutans H2O2-forming NADH oxidase is an alkyl hydroperoxide reductase protein. Free Radic. Biol. Med. 28:108-120. [DOI] [PubMed] [Google Scholar]
- 45.Poyart, C., E. Pellegrini, O. Gaillot, C. Boumaila, M. Baptista, and P. Trieu-Cuot. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocha, E. R., G. Owens, Jr., and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocha, E. R., T. Selby, J. P. Coleman, and C. J. Smith. 1996. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J. Bacteriol. 178:6895-6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rocha, E. R., and C. J. Smith. 1998. Characterization of a peroxide-resistant mutant of the anaerobic bacterium Bacteroides fragilis. J. Bacteriol. 180:5906-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rocha, E. R., and C. J. Smith. 1999. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 181:5701-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Sonnhammer, E. L. L., G. Von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, L. R. D. Sankoff, and C. Sensen (ed.), Proceedings of the 6th International Conference on Intelligent Systems for Molecular Microbiology. AAAI Press, Menlo Park, Calif. [PubMed]
- 52.Spratt, B. G. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62:797-821. [DOI] [PubMed] [Google Scholar]
- 54.Szabo, L., S. Nishimura, and W. R. Farkas. 1988. Possible involvement of queuine in oxidative metabolism. Biofactors 1:241-244. [PubMed] [Google Scholar]
- 55.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thibessard, A., A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2002. Transposition of pGh9:ISS1 is random and efficient in Streptococcus thermophilus CNRZ368. Can. J. Microbiol. 48:473-478. [DOI] [PubMed] [Google Scholar]
- 57.Thibessard, A., A. Fernandez, B. Gintz, N. Leblond-Bourget, and B. Decaris. 2002. Effects of rodA and pbp2b disruption on cell morphology and oxidative stress response of Streptococcus thermophilus CNRZ368. J. Bacteriol. 184:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thibessard, A., A. Fernandez, B. Gintz, N. Leblond-Bourget, and B. Decaris. 2001. Hydrogen peroxide effects on Streptococcus thermophilus CNRZ368 cell viability. Res. Microbiol. 152:593-596. [DOI] [PubMed] [Google Scholar]
- 59.Thibessard, A., N. Leblond-Bourget, A. Fernandez, B. Gintz, and B. Decaris. 2001. Response of Streptococcus thermophilus CNRZ368 and its colonial variants to oxidative stress: evidence for an inducible defence system. Lait 81:311-316. [Google Scholar]
- 60.Tseng, H., Y. Srikhanta, A. McEwan, and M. Jennings. 2001. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Mol. Microbiol. 40:1175-1186. [DOI] [PubMed] [Google Scholar]
- 61.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 62.Visca, P., A. Ciervo, V. Sanfilippo, and N. Orsi. 1993. Iron-regulated salicylate synthesis by Pseudomonas spp. J. Gen. Microbiol. 139:1995-2001. [DOI] [PubMed] [Google Scholar]
- 63.Wachi, M., M. Doi, Y. Okada, and M. Matsuhashi. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171:6511-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wachi, M., M. Doi, S. Tamaki, W. Park, S. Nakajima-Iijima, and M. Matsuhashi. 1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, J., and B. Weiss. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamamoto, Y., M. Higuchi, L. B. Poole, and Y. Kamio. 2000. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 182:3740-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yesilkaya, H., A. Kadioglu, N. Gingles, J. E. Alexander, T. J. Mitchell, and P. W. Andrew. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Youn, H. D., H. Youn, J. W. Lee, Y. I. Yim, J. K. Lee, Y. C. Hah, and S. O. Kang. 1996. Unique isozymes of superoxide dismutase in Streptomyces griseus. Arch. Biochem. Biophys. 334:341-348. [DOI] [PubMed] [Google Scholar]
- 69.Yu, J., A. P. Bryant, A. Marra, M. A. Lonetto, K. A. Ingraham, A. F. Chalker, D. J. Holmes, D. Holden, M. Rosenberg, and D. McDevitt. 2001. Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology 147:431-438. [DOI] [PubMed] [Google Scholar]
- 70.Yuvaniyama, P., J. N. Agar, V. L. Cash, M. K. Johnson, and D. R. Dean. 2000. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. USA 97:599-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, L., R. H. White, V. L. Cash, R. F. Jack, and D. R. Dean. 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng, M., X. Wang, L. J. Templeton, D. R. Smulski, R. A. LaRossa, and G. Storz. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183:4562-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]