Significance
Trimeric forms of HIV-1 envelope glycoproteins are being used for structural and vaccine studies. The most common way to make these proteins is to eliminate the cleavage site between the glycoprotein (gp)120 and gp41 subunits. We show that doing so creates trimers that adopt irregular, nonnative configurations. Cleaved, stabilized trimers, in contrast, resemble the native spikes on the HIV-1 virus. Our findings will help structural and vaccine programs by showing how to make native-like trimers. The rationale for vaccine trials based on the use of uncleaved gp140 trimers should be reevaluated.
Abstract
We compare the antigenicity and conformation of soluble, cleaved vs. uncleaved envelope glycoprotein (Env gp)140 trimers from the subtype A HIV type 1 (HIV-1) strain BG505. The impact of gp120–gp41 cleavage on trimer structure, in the presence or absence of trimer-stabilizing modifications (i.e., a gp120–gp41 disulfide bond and an I559P gp41 change, together designated SOSIP), was assessed. Without SOSIP changes, cleaved trimers disintegrate into their gp120 and gp41-ectodomain (gp41ECTO) components; when only the disulfide bond is present, they dissociate into gp140 monomers. Uncleaved gp140s remain trimeric whether SOSIP substitutions are present or not. However, negative-stain electron microscopy reveals that only cleaved trimers form homogeneous structures resembling native Env spikes on virus particles. In contrast, uncleaved trimers are highly heterogeneous, adopting a variety of irregular shapes, many of which appear to be gp120 subunits dangling from a central core that is presumably a trimeric form of gp41ECTO. Antigenicity studies with neutralizing and nonneutralizing antibodies are consistent with the EM images; cleaved, SOSIP-stabilized trimers express quaternary structure-dependent epitopes, whereas uncleaved trimers expose nonneutralizing gp120 and gp41ECTO epitopes that are occluded on cleaved trimers. These findings have adverse implications for using soluble, uncleaved trimers for structural studies, and the rationale for testing uncleaved trimers as vaccine candidates also needs to be reevaluated.
Trimeric envelope glycoprotein (Env gp) spikes on the HIV type 1 (HIV-1) surface mediate entry of the viral genome into the target cell (1, 2). When spikes interact with their cell-surface receptors, a series of conformational changes within the Env culminates in virus–cell membrane fusion. Neutralizing antibodies (NAbs) against various Env epitopes antagonize these events (2, 3). Hence, Env glycoproteins are a focus of vaccine design programs intended to induce NAbs and thereby prevent HIV-1 transmission (3, 4). Env trimers are composed of three gp120 surface glycoprotein subunits and three gp41 transmembrane glycoproteins, the six subunits all associated via noncovalent interactions (5, 6). A critical event in trimer assembly is proteolytic cleavage of the gp160 precursor into its gp120 and gp41 components, a process essential for HIV-1 entry not least because it liberates the fusion peptide (FP) at the gp41 N terminus (5, 6).
Trimer-based vaccine strategies involve expressing soluble, recombinant versions of the virion-associated (i.e., native) spikes. To facilitate production and purification, the membrane-spanning and cytoplasmic domains that anchor spikes to the virion, but that are not NAb targets, are eliminated (7–12). However, the resulting proteins, known as gp140s, are highly unstable and disintegrate into their gp120 and gp41-ectodomain (gp41ECTO) components, making them useless as immunogens. Two fundamentally different protein-engineering strategies have been used to create gp140s that can be produced and purified without falling apart (3, 4, 7–17). The most common method involves eliminating the cleavage site between gp120 and gp41ECTO, creating uncleaved gp140s (gp140UNC) where the two subunits remain covalently linked (7–12). Additional trimerization motifs are often added to the gp41ECTO C terminus (10–12). Our alternative approach is based on the premise that cleavage is a fundamental feature of Env structure and involves stabilizing fully cleaved gp140s. The critical changes are an appropriately positioned disulfide bond (referred to as “SOS”) to link gp120 to gp41ECTO covalently, and an Ile/Pro (IP) substitution at residue 559 to strengthen inter-gp41ECTO interactions (13–17). The resulting cleaved trimers are designated SOSIP gp140s (14). Additional modifications have improved their stability, homogeneity, and antigenicity (15–17). Our current design, based on the BG505 subtype A env gene, yields SOSIP.664 trimers that mimic native, virion-associated Env spikes antigenically and when viewed by negative-stain electron microscopy (EM) (17–19).
Here we show that cleavage is essential for producing stable, soluble gp140 trimers that resemble native Env spikes. EM studies reveal that purified, trimeric gp140UNC proteins are heterogeneous and that the irregularly shaped images rarely resemble a native spike; we refer to them as “aberrant configurations” (ACs). In contrast, cleaved SOSIP gp140 trimers are homogeneous and mimic native spikes; we designate them native-like (NL) trimers. The antigenic properties of the cleaved (NL) and uncleaved (AC) trimers, assessed by surface plasmon resonance (SPR) and enzyme-linked immunoabsorbance assays (ELISA), are consistent with the EM images. Nonneutralizing gp120 and gp41ECTO epitopes are exposed on gp140UNC trimers but occluded on cleaved ones, whereas quaternary structure-dependent epitopes indicative of proper folding are present only on cleaved trimers. Our findings have substantial implications, because uncleaved trimers are being studied structurally and developed as vaccine candidates (3, 9, 10, 12, 20).
Results
Cleavage and Trimer-Formation Properties of gp140 Mutants.
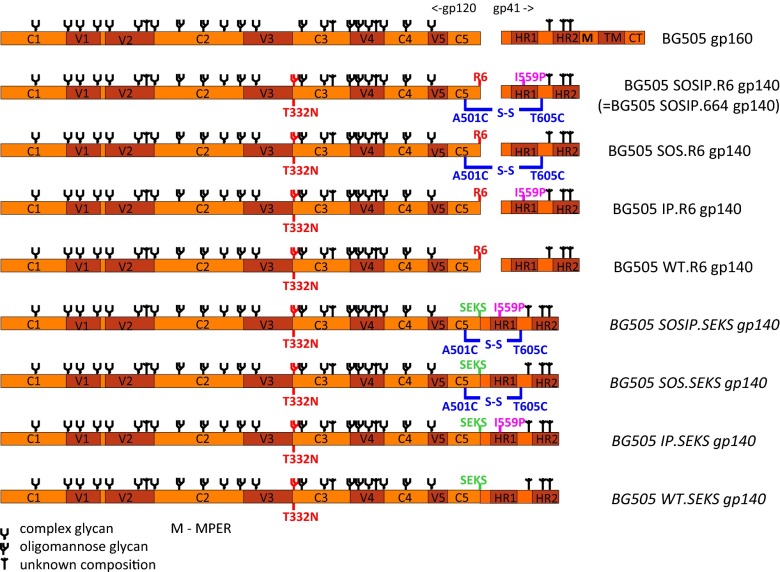
Eight constructs based on BG505 env were used to explore the influence of cleavage and other stabilizing changes on trimer stability and conformation. The constructs are grouped into two subcategories: those that can (hexa-Arg or R6 cleavage site) or cannot (Ser-Glu-Lys-Ser or SEKS cleavage site) be cleaved. Mutant Env proteins also either contained or lacked the disulfide bond (SOS) between gp120 and gp41ECTO and/or the I559P change in gp41ECTO. In all eight constructs, gp41ECTO was terminated at residue 664, eliminating most of the membrane-proximal external region (MPER), the transmembrane domain, and the cytoplasmic domain (Fig. 1).
Fig. 1.
Design of cleaved and uncleaved gp140s. Linear representation of BG505 gp160 and the SOSIP.R6, SOS.R6, IP.R6, WT.R6, SOSIP.SEKS, SOS.SEKS, IP.SEKS, and WT.SEKS gp140 constructs. Modifications in red are described in Results and Materials and Methods. Env subdomains are as follows: five conserved domains (C1–C5); five variable domains (V1–V5); heptad repeats 1 and 2 (HR1 and HR2); the MPER; transmembrane domain (TM); and cytoplasmic tail (CT). Glycan assignments are as previously reported (17).
The eight soluble gp140s were expressed transiently in HEK293T cells; the Furin protease was coexpressed with the four cleavable variants. The IP.R6 and WT.R6 variants were cleaved efficiently, but were too unstable to form gp140 trimers (Table 1) and were not studied further. The other six Env proteins were purified on a 2G12 mAb affinity column and analyzed by reducing SDS/PAGE (Fig. 2A). As expected, SDS/PAGE confirmed that the two Env proteins containing the R6 site were fully (>95%) cleaved, whereas the four with the SEKS site were not (<5%) (Fig. 2A). Low-level cleavage of SEKS variants may be via a secondary-site eight residues upstream of the primary Furin site (21). Nonreducing SDS/PAGE showed that, unlike SOSIP.R6 and SOS.R6, the four gp140UNC preparations contained oligomers that survived heating with SDS (Fig. 2B) and that are probably covalently linked via aberrant, intersubunit disulfide bonds (10, 11).
Table 1.
Properties of cleaved and uncleaved gp140s
| Mutant | % cleavage* | % trimers† | Presence of native-like trimers‡ |
| SOSIP.R6 | >95 | ∼60 | ++++ |
| SOS.R6 | >95 | ∼20 | ++ |
| IP.R6 | >95 | <5 | N/A |
| WT.R6 | >95 | <5 | N/A |
| SOSIP.SEKS | <5 | ∼60 | ++ |
| SOS.SEKS | <5 | ∼50 | − |
| IP.SEKS | <5 | ∼60 | + |
| WT.SEKS | <5 | ∼50 | − |
| SOSIP.R6-MPER | >95 | ∼60 | ++++ |
| SOSIP.SEKS-MPER | <5 | ∼60 | ++ |
| WT.SEKS-MPER | <5 | ∼50 | − |
| CN54 gp140 | N/A | N/A | − |
| UG37 gp140 | N/A | N/A | − |
N/A, not analyzed; but see data sheets at www.polymun.at.
gp140 and gp120 bands on reducing SDS/PAGE were quantified using ImageJ (National Institutes of Health). The percentage of total Env protein that was cleaved to gp120 and gp41ECTO was calculated.
The gp140 trimer, gp140 dimer, gp140 monomer, gp120 monomer, and Env-aggregate bands on BN/PAGE of 2G12-purified Env proteins were quantified using ImageJ. The percentage of the total Env protein in the form of gp140 trimers was calculated.
The formation of NL trimers was semiquantitatively assessed by negative-stain EM as described in Materials and Methods, and categorized as follows: −, <2%; +, 2–10%; ++, 10–50%; +++, 50–100%; ++++, ∼100%.
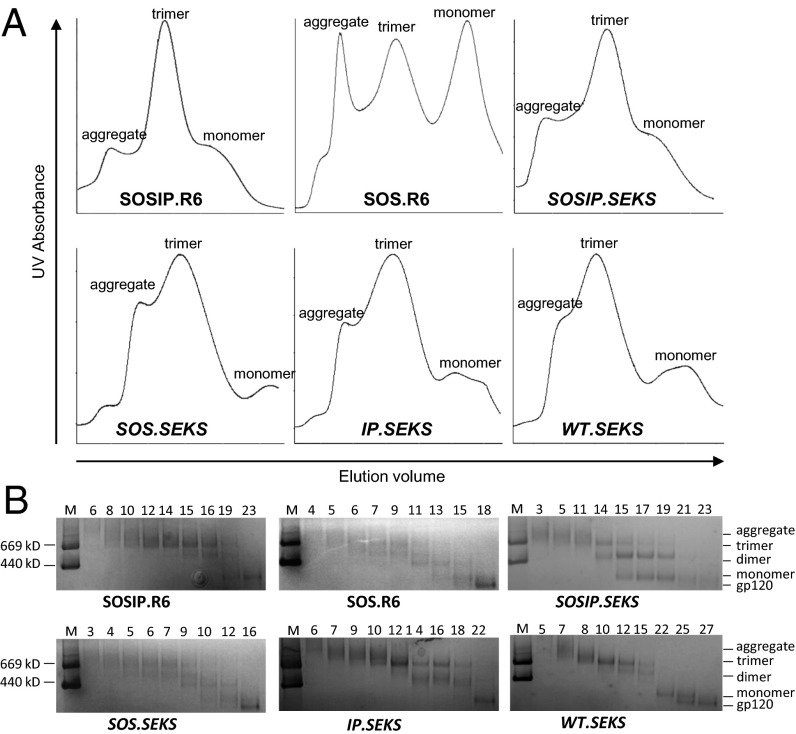
Fig. 2.
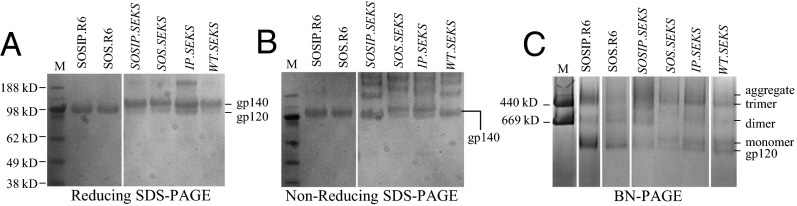
Biochemical characterization of cleaved and uncleaved gp140s. Reducing SDS/PAGE (A), nonreducing SDS/PAGE (B), and BN/PAGE (C) analysis followed by Coomassie blue staining of 2G12-purified Env proteins. Molecular weights of marker (M) proteins (thyroglobulin and ferritin) are indicated.
Blue native (BN)/PAGE analysis of 2G12-purified Env proteins confirmed that the SOSIP.R6 construct produced trimers efficiently; the overall yield was ∼50–60% (Fig. 2C and Table 1). The SOS.R6 variant produced some trimers (∼20%), but the predominant Env species was monomeric gp140, that is, one gp120 linked to gp41ECTO via the SOS bond. As noted, the IP.R6 and WT.R6 constructs formed trimers inefficiently (<5%). Thus, the only cleaved gp140 for which trimers predominated was SOSIP.R6, consistent with previous reports that SOS and I559P changes are both needed to stabilize cleaved trimers (13, 14). However, the SOS bond alone (SOS.R6) was sufficient to allow some cleaved trimers to be expressed from BG505 env. All four 2G12-purified gp140UNC preparations contained trimers, but were accompanied by Env dimers and aggregates (Fig. 2C). Nontrimeric Env species are commonly seen before trimer purification on sizing columns (14, 22). Some monomeric gp120 was also present in gp140UNC preparations, probably due to low-level cleavage at the secondary site (21). The presence or absence of SOS and I559P changes had no effect on how gp140UNC proteins migrated, the lack of cleavage being sufficient for trimer formation.
The six 2G12-purified Env preparations were analyzed by size-exclusion chromatography (SEC) (Fig. 3A), with the eluted fractions assessed by BN/PAGE (Fig. 3B). As previously found, the BG505 SOSIP.R6 construct formed trimers efficiently, with only minor amounts of aggregates, dimers, and monomers visible (17). For SOS.R6, some trimers were produced but the greater amounts of aggregates and monomers eluted from the SEC column are consistent with the BN/PAGE study (Figs. 2C and 3B). Significant amounts of SOS.R6 gp120 monomers were also visible. The contrast between the SEC profiles for SOS.R6 and SOSIP.R6 (gp120 monomers present vs. absent) suggests that the gp120–gp41ECTO disulfide bond forms more efficiently when the I559P substitution is present. All four uncleaved gp140s yielded a major trimer peak as well as aggregates and gp120 monomers.
Fig. 3.
SEC analysis of cleaved and uncleaved gp140s. (A) SEC profiles for 2G12-purified Env proteins on a Superdex 200 26/60 column. (B) Selected fractions were analyzed by BN/PAGE and Coomassie blue staining. Molecular weights of marker (M) proteins (thyroglobulin and ferritin) are indicated.
We conclude that only gp140 constructs with a covalent link between gp120 and gp41ECTO form Env trimers efficiently. That link can be either a peptide (between residues 511 and 512 for gp140UNC) or a disulfide bond (between residues 501 and 605 for cleaved SOS or SOSIP gp140).
Electron Microscopy of Cleaved and Uncleaved gp140 Trimers.
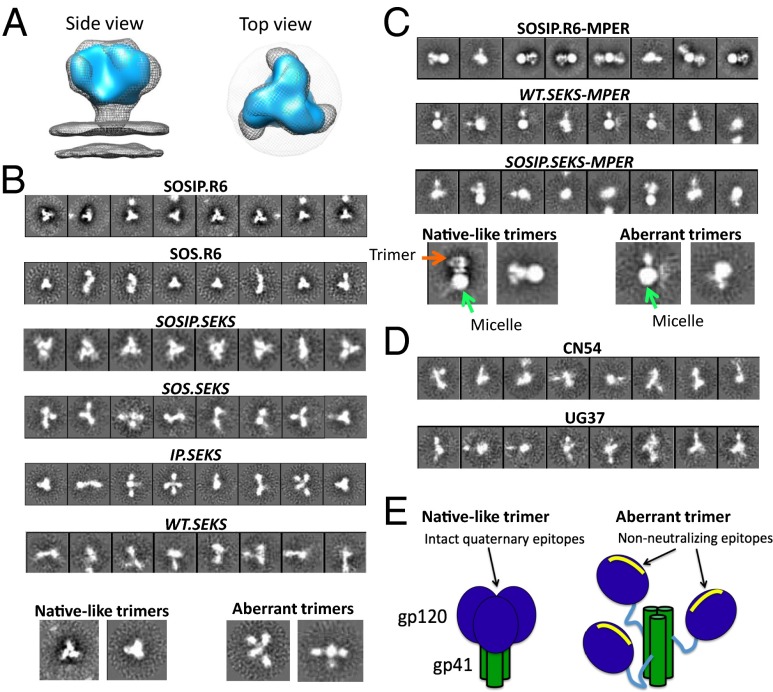
The above six 2G12- and SEC column-purified trimers were viewed by negative-stain EM, and the reference-free 2D class averages were inspected to determine their overall morphology. The cleaved SOSIP.R6 trimers were regular and homogeneous (Fig. 4 A and B), consistent with earlier reports of the same construct (there designated BG505 SOSIP.664 gp140) (17–19). A semiquantitative analysis of the class averages showed that ∼100% of the SOSIP.R6 trimer images resembled virion-associated, native trimers from HIV-1 BaL (EMDB-5019) (Fig. 4B, Table 1, and Fig. S1) (6). Hence, we refer to SOSIP.R6 gp140 trimers as “native-like.”
Fig. 4.
Negative-stain EM of cleaved and uncleaved trimers. (A) Comparison of the reconstruction of the soluble BG505 SOSIP.R6 (NL) trimer (blue) (17) with the structure of the membrane-associated native Env spike of the subtype B BaL strain (mesh) (6). (B) Reference-free 2D class averages for various 2G12/SEC-purified, MPER-deleted trimers. (C) Reference-free 2D class averages for 2G12/SEC-purified SOSIP.R6-MPER, SOSIP.SEKS-MPER, and WT.SEKS-MPER trimers. (D) Reference-free 2D class averages for CN54 (subtype C) and UG37 (subtype A) uncleaved gp140s. (E) Cartoon showing the NL and AC conformations of the Env trimer (see also refs. 13 and 31).
About a third of the purified SOS.R6 trimers had an NL configuration, the rest adopting a range of unusual shapes (Fig. 4B, Table 1, and Fig. S1). Noting also the complete absence of trimers from the WT.R6 and IP.R6 constructs, the gp120–gp41ECTO SOS bond is necessary and, to an extent, sufficient to yield NL gp140 trimers, at least from the BG505 gene. However, as most SOS.R6 trimers were of the irregular AC form, the I559P change is also important for making cleaved, NL gp140 trimers quantitatively.
Uncleaved WT.SEKS gp140 trimers (i.e., the standard gp140UNC design) were highly heterogeneous, with multiple different shapes visible and no predominant conformation (Fig. 4B). These AC trimers often resemble three gp120 subunits dangling off a central core, which is presumably a trimeric form of gp41ECTO. If so, the gp120 moieties must remain attached to gp41ECTO solely via the uncleaved link between the C5 region and the FP. Overall, only a very small fraction (<2%) of WT.SEKS gp140 trimers had the NL configuration (Table 1).
The SOS.SEKS gp140UNC trimers, the standard gp140UNC with an added SOS bond, were also mostly ACs, with <2% in NL form (Fig. 4B, Table 1, and Fig. S1). This low percentage was the same as with WT.SEKS, so the subunit-linking SOS bond does not facilitate NL trimer formation when cleavage is prevented.
The IP.SEKS trimers were also mostly ACs, although now approximately a tenth of the images resembled NL trimers (Fig. 4B, Table 1, and Fig. S1). The trimer-stabilizing I559P change does therefore allow a minor subset of gp140UNC trimers to adopt the NL configuration. However, the lack of cleavage still dominates the outcome, as >90% of IP.SEKS trimers were of the AC type.
Approximately two-thirds of the uncleaved SOSIP.SEKS trimers that contained both the SOS and I559P changes adopted the NL configuration (Fig. 4B, Table 1, and Fig. S1). Thus, adding both the SOS and I559P changes partially rescues the NL phenotype in the absence of cleavage, albeit not completely. We have noted, however, that SOSIP.SEKS NL trimers seem quite fragile, in that the proportion in this form is much lower in some preparations and may be dependent on incubation or storage conditions.
MPER Deletion Does Not Explain the Rarity of Native-Like gp140UNC Trimers.
The above experiments used gp140s from which the MPER was deleted to increase trimer homogeneity (23, 24). To assess whether the absence of the MPER influenced trimer configuration, we made SOSIP.R6-MPER, SOSIP.SEKS-MPER, and WT.SEKS-MPER constructs that are identical to the corresponding variants described above except for truncation at residue 681, not residue 664. The EM images show a large spherical blob of density that masks the base of gp41ECTO in the MPER-containing trimers (Fig. 4C and Fig. S1). This density is far larger than can be accounted for by the additional 17 amino acids, but resembles a micelle that associated with similar KNH1144.681 trimers when a small amount of detergent was added to prevent aggregate formation (23, 24). Although there was some aggregation of the MPER-containing BG505 trimers, the extent was much less than with the KNH1144 versions. The MPER-containing BG505 trimers may have acquired lipids or other cellular amphiphiles during production, creating a micelle around the hydrophobic MPER that survives the purification process. Overall, the EM images of the three MPER-containing trimers are generally consistent with their MPER-deleted counterparts (Fig. 4C). Thus, a clear majority of 2D class averages from the visible SOSIP.R6-MPER trimers had the NL configuration, whereas few if any WT.SEKS-MPER NL trimers were seen (Fig. 4C, Table 1, and Fig. S1). We noted that, similar to SOSIP.SEKS, a substantial fraction of SOSIP.SEKS-MPER trimers adopted the NL form (Fig. 4C, Table 1, and Fig. S1), but again AC forms were also present. Finally, we studied two uncleaved, MPER-containing gp140 trimers based on other env genes: CN54 (subtype C) and UG37 (subtype A) (12, 25). They were overwhelmingly in AC form, with almost no NL images observed (Fig. 4D, Table 1, and Fig. S2). We conclude that the presence or absence of the MPER does not determine whether a soluble gp140 trimer adopts the NL or AC configuration.
Antigenic Structure of Cleaved and Uncleaved gp140 Trimers.
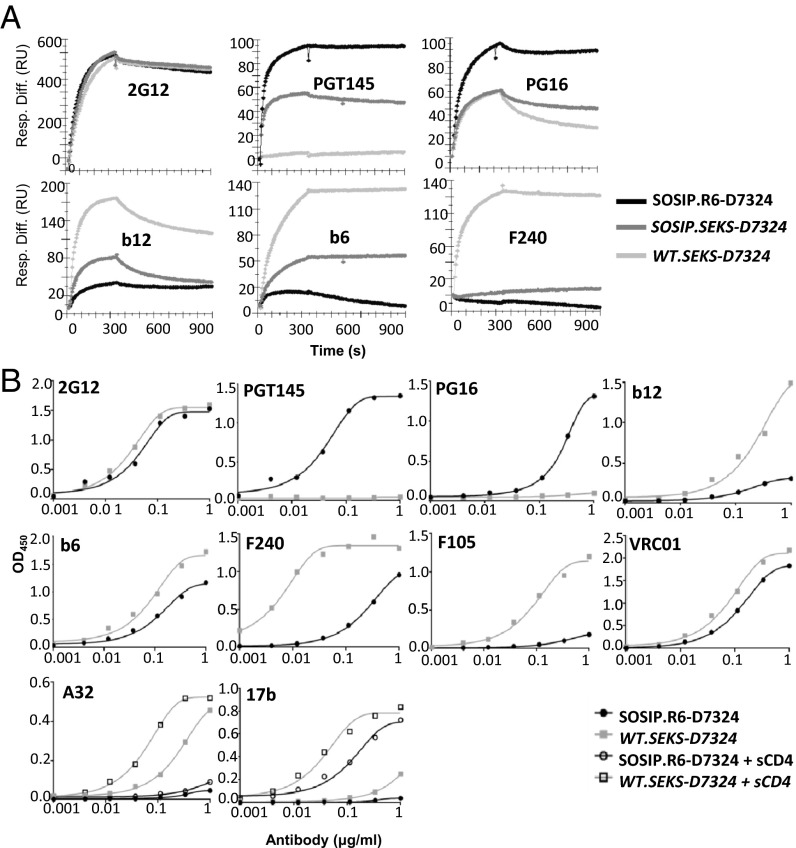
Cleaved and uncleaved, soluble or membrane-bound trimers express NAb and non-NAb epitopes very differently (13, 26–28). We therefore assessed the antigenicity of purified, D7324-tagged versions of SOSIP.R6, SOSIP.SEKS, and WT.SEKS gp140 trimers. An SPR study showed that NAb and non-NAb epitopes were expressed differently on the two versions of trimers (Fig. 5A). The cleaved, SOSIP.R6-D7324 NL trimers bound the quaternary epitope-dependent NAbs PGT145 and PG16 strongly, whereas the uncleaved WT.SEKS-D7324 AC trimers did so to a lesser extent (PG16) or not at all (PGT145). Note that PG16, but not PGT145, binds monomeric BG505 gp120 to a limited degree, which would account for PG16 reactivity with the AC trimers (17). In marked contrast, uncleaved WT.SEKS-D7324 AC trimers exposed the epitopes for non-NAbs F240 (gp41, cluster I) and b6 and b12 [gp120, CD4 binding site (CD4bs)] far more than cleaved, SOSIP.R6-D7324 NL trimers [NAb b12 is nonneutralizing for HIV-1 BG505 (17)]. The two trimer forms bound broadly neutralizing antibody (bNAb) 2G12 equivalently; this nonquaternary epitope is on the exposed surface of gp120. The antigenic properties of the SOSIP.SEKS-D7324 trimers were generally intermediate between SOSIP.R6-D7324 and WT.SEKS-D7324 (Fig. 5A), which is consistent with the mixture of NL and AC forms visible in the EM images of the SOSIP.SEKS trimers (Fig. 4B and Fig. S1).
Fig. 5.
Antigenicity of cleaved and uncleaved trimers. (A) SPR sensorgrams (one of two or three similar replicates) for the indicated NAbs and non-NAbs, injected at 500 nM over purified, cleaved SOSIP.R6-D7324 and uncleaved WT.SEKS-D7324 or SOSIP.SEKS-D7324 gp140 trimers captured by D7324 onto CM5 chips. Response units (RU) are given on the y axis as a function of time (s) after injection on the x axis (5-min association, 10-min dissociation). (B) ELISA binding curves (from two or three replicates) for the indicated NAbs and non-NAbs and purified, cleaved SOSIP.R6-D7324 and uncleaved WT.SEKS-D7324 gp140 trimers. The curves for A32 and 17b were derived with and without sCD4 (10 µg/mL), as indicated.
ELISA data were consistent with the SPR analysis. Cleaved SOSIP.R6-D7324 trimers again bound bNAbs PG16 and PGT145 far more strongly than uncleaved WT.SEKS-D7324 trimers, whereas the converse applied to non-NAbs F240, b6, b12, and F105 (also to the CD4bs). Like 2G12, the CD4bs bNAb VRC01 (which neutralizes BG505) bound the two trimer forms comparably (Fig. 5B). The A32 epitope was not exposed on cleaved trimers whether soluble CD4 (sCD4) was present or not, but this non-NAb bound well to uncleaved trimers. The 17b non-NAb also bound more strongly to uncleaved trimers, but sCD4 induced its epitope on cleaved trimers (Fig. 5B).
Comparative Protease Sensitivity of Cleaved and Uncleaved Trimers.
We assessed the sensitivity of SOSIP.R6 or WT.SEKS gp140 trimers to trypsin, chymotrypsin, and proteinase K. The cleaved SOSIP.R6 trimers were completely resistant to trypsin and chymotrypsin (1 µg/mL), whereas the uncleaved WT.SEKS trimers were degraded at this concentration. Similarly, the uncleaved trimers were markedly more sensitive to proteinase K (Fig. S3).
Discussion
Our principal conclusion is that Env cleavage is a critical determinant of the configuration of soluble gp140 trimers. Thus, cleaved, SOSIP-stabilized BG505 gp140 trimers [SOSIP.R6; referred to elsewhere as “SOSIP.664” (17–19)] are highly homogeneous and their configurations resemble native Env spikes, as judged by negative-stain EM (17–19) (Fig. 4 A and B). The antigenicity of BG505 SOSIP.664 trimers correlates strongly with neutralization of the corresponding virus (17). In the context of the BG505 genotype, the gp120–gp41ECTO SOS bond is necessary, and to a limited extent sufficient, to produce these NL trimers. Thus, the cleaved SOS.R6 construct (i.e., no I559P change) yielded some trimers (approximately one-third of the total) that appeared indistinguishable from SOSIP.R6. However, the overall SOS.R6 trimer yield was also reduced (by approximately threefold), with gp140 monomers held together by the intersubunit disulfide bond predominating, and most SOS.R6 trimers were of the AC form, that is, highly heterogeneous and not at all like native spikes. The I559P substitution is, therefore, not only necessary to yield cleaved trimers efficiently (14) but also helps keep them in the NL configuration. The cleaved gp140 constructs lacking the SOS bond, IP.R6 and WT.R6, failed to form trimers, the gp120 components presumably dissociating rapidly from a gp41ECTO monomer or trimer.
We propose that the engineered SOS bond anchors the gp120 subunit to gp41ECTO, not by creating a new site of interaction but by supporting the weaker, noncovalent linkages that form naturally and which may be stabilized to some extent by the membrane-spanning domain. Membrane-associated, full-length (i.e., gp160) SOS trimers are still fusion-competent, provided the disulfide bond is reduced after receptor engagement; the SOS bond does not compromise trimer structure and function (29). In contrast, the I559P change is incompatible with fusion, presumably because it impedes the transition(s) from the prefusion form to the fusion intermediate and/or postfusion forms of gp41 (14, 30). A loop-to-helix transition around residue 559 that is blocked by a helix-breaking proline may be involved in those conformational changes (14, 30).
In marked contrast to cleaved SOSIP gp140 trimers, uncleaved trimers adopted multiple, irregular conformations that only very rarely (<2%) resembled native Env spikes. The simplest interpretation of the gp140UNC trimer EM images is that the three gp120 moieties dangle from a central gp41ECTO core (now in the postfusion, six-helix bundle form), to which they remain tethered by the still-intact linkage between the gp120 C5 domain and the FP region of gp41ECTO (Fig. 4E) (13, 31). The greater sensitivity of the WT.SEKS gp140UNC trimers to protease digestion, compared with the cleaved SOSIP.R6 versions, is consistent with uncleaved trimers adopting a splayed-out conformation that is more accessible to the digesting enzymes (Fig. S3). The same conclusions about the overall configuration of gp140UNC trimers of several genotypes were recently drawn from independent, biophysical analyses (31).
We propose that, for gp140UNC trimers, the normal association between gp120 and gp41ECTO either never forms or is too unstable to persist. Introducing the SOS bond into gp140UNC to form SOS.SEKS does not create NL trimers, whereas the I559P change is partially beneficial (5–10% in NL form). Introducing both the SOS and I559P changes rescued a larger percentage of SOSIP.SEKS gp140UNC NL trimers (approximately two-thirds), the rest being in AC form. The antigenicity properties of SOSIP.SEKS trimers are also consistent with their being a mixture of NL and AC forms. These NL trimers also appear to be less stable, and more sensitive to incubation/storage conditions, than SOSIP.R6. The stark contrast between gp140UNC proteins and cleaved SOSIP.R6 trimers (∼100% NL) clearly shows the importance of cleavage for creating stable, soluble trimers that resemble Env spikes seen on virus particles. Although uncleaved gp140 trimers contain an appropriate number of gp120 subunits (i.e., three), from the perspective of creating mimics of native Env spikes we suggest they are trimers in name only (TINOs).
Why do uncleaved gp140 trimers very seldom adopt NL structures? Do such conformations rarely form, or are they too unstable to survive? Although both factors may be relevant, we think that instability is the more important, namely that the noncovalent bonds between gp120 and gp41ECTO are too weak for soluble gp140 trimers to persist in the prefusion form for very long after they are secreted. Without the engineered SOS bond, cleaved gp140 trimers are also highly unstable and disintegrate, and for many genotypes this also applies to cleaved trimers lacking the I559P change (14, 16). In that catastrophic process, the gp120 and/or gp41ECTO moieties physically separate but, when the same event takes place for their gp140UNC counterparts, the dissociating subunits still remain attached via the uncleaved peptide link between C5 and the FP. The same conclusion has recently been drawn independently (31). The resulting Env structures are “trimeric” in that three gp120 and gp41ECTO subunits are all physically associated, but as noted earlier, they are TINOs. These conclusions are not dependent on the presence or absence of the MPER, as the EM images for uncleaved WT.SEKS and WT.SEKS-MPER trimers showed that NL trimers were rarely seen in either case. MPER-containing, uncleaved gp140s of several genotypes also did not form NL trimers when studied by EM (here) or using H/D-exchange mass spectrometry, which is sensitive to local structural order (31).
Why do the stabilizing SOS and I559P changes have different quantitative and qualitative effects on cleaved and uncleaved trimers, yielding SOSIP.R6 and SOSIP.SEKS NL structures at approximate respective frequencies of 100% vs. 70% (less in some preparations and/or storage conditions)? We propose that gp140 cleavage itself has a modest stabilizing effect on the trimer. More specifically, we suggest that the normal gp120–gp41ECTO association is more likely to form, or to persist longer, after cleavage occurs, increasing the probability that an NL structure will survive long enough to be further stabilized by the engineered SOS bond. One hypothesis is that cleavage-dependent release of the gp120 C5 region from the FP liberates gp120 and/or gp41ECTO residues necessary for formation of the complete intersubunit binding site; the C5 region is certainly involved in gp120–gp41 association (32).
The antigenicity data are consistent with how cleaved and uncleaved trimers appear by EM. Both SPR and ELISA show that cleaved SOSIP.R6 trimers express epitopes for the quaternary structure-dependent NAbs PGT145 and PG16 far better than the WT.SEKS gp140UNC trimers. In contrast, CD4bs non-NAbs (for HIV-1 BG505) b12 and F105 bind uncleaved trimers far more strongly than cleaved ones, and the same trend is seen with b6. The VRC01 CD4bs bNAb, which does neutralize BG505, binds cleaved and uncleaved trimers comparably in ELISA. The A32 non-NAb bound only to the gp140UNC trimers. It is argued that this epitope is important for antibody-dependent cellular cytotoxicity (ADCC) activity (33), but we note that Env on the surface of virus-infected cells, the most likely ADCC targets in vivo, is in the form of nascent virions on which Env is predominantly cleaved (34). Cleaved SOSIP.R6 trimers also expose the cluster I gp41ECTO epitope for the non-NAb F240 to a far lesser extent than uncleaved WT.SEKS trimers. This gp41ECTO region is, therefore, inaccessible on the compact, cleaved trimers, buried under the properly sited gp120 components, but well-exposed on gp140UNC trimers, where the gp120 subunits are physically separated from the six-helix bundle configuration of gp41ECTO (31).
Here we have only studied how cleavage affects soluble gp140 trimers, but many studies show that cleaved and uncleaved, membrane-associated gp160 trimers differ antigenically in quite substantial ways. For example, multiple non-NAbs are much more reactive with various gp120 epitopes on uncleaved trimers, as are ones to the six-helix bundle form of gp41 (13, 26–28). Thus, we believe that the aberrant soluble gp140 structures that we have visualized by EM will have their counterparts on the surface of cells expressing uncleaved Env proteins. Perhaps additional stability imparted by the transmembrane region will reduce the extent to which membrane-associated trimers decay into AC forms, but a wealth of antigenicity data strongly argues that such structures are commonplace on the surface of cells expressing uncleaved gp160s (13, 26–28).
This study used a set of mutants based on the BG505 sequence, but the EM images of two other gp140UNC trimers of different genotypes are essentially identical to those based on BG505. Biophysical studies of gp140UNC proteins from several genotypes from various genetic subtypes yielded the same conclusion about their nonnative conformation, and the addition of nominally stabilizing changes such as a trimerizing foldon sequence does not overcome such defects (31). Thus, the problems we have identified with the heterogeneity and structural aberrance of uncleaved BG505 gp140 trimers do appear generalizable, which could be verified experimentally. As gp140UNC trimers are vaccine candidates, there are strategic implications for the production of homogeneous antigens and how they behave as immunogens (3, 9, 10, 12, 20). Although gp140UNC trimers are modestly superior to the corresponding monomeric gp120s at inducing NAbs in animals (10–12, 20), any such benefit now seems very unlikely to be based on their mimicry of native Env structures. One possibility is that the well-exposed gp41ECTO moiety might confer an immunogenicity benefit, if any antibodies to this subunit register as NAbs in assays such as the one based on A3R5 cells (12). The greater size, and perhaps multivalency, of uncleaved trimers might also confer an immunological advantage over a gp120 monomer, particularly if Env aggregates form during immunization. Whatever the case, hypotheses that underlie clinical trials of uncleaved trimers should be revisited. Moreover, the EM and biophysical methods described here and elsewhere (31) should be used to evaluate all Env trimers being considered for clinical trials.
We conclude that cleavage of the gp120–gp41ECTO junction is essential for a trimer to adopt a stable native-like structure, probably because that event imparts additional robustness to intersubunit interactions. Uncleaved, soluble gp140 trimers have aberrant, nonnative configurations when viewed by negative-stain EM, and their antigenicity profile is consistent with their appearance. Cleavage is not, however, sufficient for a cleaved, soluble trimer to be usefully stable, as these entities rapidly disintegrate unless they also include the stabilizing SOS and I559P changes. The resulting soluble, cleaved, SOSIP-stabilized trimers are highly homogeneous, resemble native Env spikes, and are useful for structural (17–19) and immunogenicity studies. The cryo-EM and X-ray diffraction structures of the BG505 SOSIP.664 gp140 trimers at ∼5 Å resolution show they adopt the prefusion form of Env (35, 36).
Materials and Methods
Soluble BG505 gp140s.
The subtype A BG505 SOSIP.664 gp140 construct, here designated SOSIP.R6, is described elsewhere (17). It incorporates the following sequence changes (HxB2 numbering): T332N in gp120 (restoration of Asn332 glycan-dependent epitopes); A501C and T605C (SOS; gp120–gp41 disulfide bond); I559P (IP; trimer-stabilizing); REKR to RRRRRR (R6) in gp120 (cleavage enhancement); and the stop codon at residue 664 to delete the MPER (improved homogeneity and solubility). The following mutants were made (Table 1): SOS.R6 lacks the I559P change; IP.R6 lacks the gp120–gp41ECTO SOS bond; WT.R6 lacks both I559P and SOS; WT.SEKS, the standard gp140UNC construct with the REKR cleavage site changed to noncleavable SEKS; SOS.SEKS, gp140UNC with the SOS bond added; IP.SEKS, gp140UNC with the I559P change added; and SOSIP.SEKS, gp140UNC with both the SOS and I559P changes added. The four italicized constructs cannot be cleaved at the primary site between gp120 and gp41ECTO. Variants bearing a D7324 epitope tag at the gp41ECTO C terminus were purified in the same way, and designated by appending “-D7324” to the gp140 descriptor (e.g., SOSIP.R6-D7324, WT.SEKS-D7324). MPER-containing SOSIP.R6-MPER, WT.SEKS-MPER, and SOSIP.SEKS-MPER constructs were based on the SOSIP.681 gene (23, 24). See also SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to D. R. Burton, J. Mascola, and P. D. Kwong for reagents. This work was supported by National Institutes of Health (NIH) Grants P01 AI82362, R37 AI36082, and R01 AI84817; Aids Fonds Netherlands Grant 2011032; and the International AIDS Vaccine Initiative. J.-P.J. has a CIH Research Fellowship. R.W.S. has a Netherlands Organization for Scientific Research Vidi Grant and a Starting Investigator Grant (ERC-StG-2011–280829-SHEV). EM data were collected at the National Resource for Automated Molecular Microscopy at The Scripps Research Institute, supported by NIH Grant P41 RR017573.
Footnotes
Conflict of interest statement: R.W.S., A.C., J.-P.J., I.A.W., P.J.K., A.B.W., and J.P.M. are listed on a patent application relating to the design and use of soluble, cleaved envelope glycoprotein trimers.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314351110/-/DCSupplemental.
References
- 1.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: The HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klasse PJ. The molecular basis of HIV entry. Cell Microbiol. 2012;14(8):1183–1192. doi: 10.1111/j.1462-5822.2012.01812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zwick MB, Burton DR. HIV-1 neutralization: Mechanisms and relevance to vaccine design. Curr HIV Res. 2007;5(6):608–624. doi: 10.2174/157016207782418443. [DOI] [PubMed] [Google Scholar]
- 4.Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: Current approaches and future directions. Curr Opin Immunol. 2010;22(3):358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410(4):582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earl PL, et al. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J Virol. 1994;68(5):3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao F, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group M consensus envelope glycoprotein. J Virol. 2005;79(2):1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spearman P, et al. HIV Vaccine Trials Network of NIAID A trimeric, V2-deleted HIV-1 envelope glycoprotein vaccine elicits potent neutralizing antibodies but limited breadth of neutralization in human volunteers. J Infect Dis. 2011;203(8):1165–1173. doi: 10.1093/infdis/jiq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Wyatt R, Sodroski J. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol. 2001;75(3):1165–1171. doi: 10.1128/JVI.75.3.1165-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, et al. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76(9):4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovacs JM, et al. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci USA. 2012;109(30):12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binley JM, et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74(2):627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders RW, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002;76(17):8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Binley JM, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76(6):2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beddows S, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology. 2007;360(2):329–340. doi: 10.1016/j.virol.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Sanders RW, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9):e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci USA. 2013;110(11):4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien JP, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9(5):e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders RW. Clinical evaluation of a soluble trimeric HIV-1 envelope glycoprotein vaccine. Expert Rev Vaccines. 2011;10(8):1117–1120. doi: 10.1586/erv.11.97. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa Y, Barsov E, Jones I. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J Virol. 1993;67(6):3601–3604. doi: 10.1128/jvi.67.6.3601-3604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Center RJ, Lebowitz J, Leapman RD, Moss B. Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J Virol. 2004;78(5):2265–2276. doi: 10.1128/JVI.78.5.2265-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klasse PJ, et al. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol. 2013;87(17):9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khayat R, et al. Structural characterization of cleaved, soluble HIV-1 envelope glycoprotein trimers. J Virol. 2013;87(17):9865–9872. doi: 10.1128/JVI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiffner T, et al. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. J Virol. 2013;87(18):10163–10172. doi: 10.1128/JVI.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si Z, Phan N, Kiprilov E, Sodroski J. Effects of HIV type 1 envelope glycoprotein proteolytic processing on antigenicity. AIDS Res Hum Retroviruses. 2003;19(3):217–226. doi: 10.1089/088922203763315722. [DOI] [PubMed] [Google Scholar]
- 27.Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332(1):145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 28.Tong T, Osawa K, Robinson JE, Crooks ET, Binley JM. Topological analysis of HIV-1 glycoproteins expressed in situ on virus surfaces reveals tighter packing but greater conformational flexibility than for soluble gp120. J Virol. 2013;87(16):9233–9249. doi: 10.1128/JVI.01145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binley JM, et al. Redox-triggered infection by disulfide-shackled human immunodeficiency virus type 1 pseudovirions. J Virol. 2003;77(10):5678–5684. doi: 10.1128/JVI.77.10.5678-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders RW, Busser E, Moore JP, Lu M, Berkhout B. Evolutionary repair of HIV type 1 gp41 with a kink in the N-terminal helix leads to restoration of the six-helix bundle structure. AIDS Res Hum Retroviruses. 2004;20(7):742–749. doi: 10.1089/0889222041524544. [DOI] [PubMed] [Google Scholar]
- 31.Guttman M, Lee KK. A functional gp41-gp120 interaction is observed in monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J Virol. August 21, 2013;87(21):11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65(4):2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari G, et al. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol. 2011;85(14):7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parren PW, et al. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J Virol. 1998;72(5):3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 Env trimer. Science. 2013 doi: 10.1126/science.1245627. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer in complex with a glycan-dependent broadly neutralizing antibody. Science. 2013 doi: 10.1126/science.1245625. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.