Progress in microbiology has always been driven by technological advances, ever since Antonie van Leeuwenhoek discovered bacteria by making an improved compound microscope. However, until very recently we have not been able to identify microbes and record their mostly invisible activities, such as nutrient consumption or toxin production on the level of the single cell, not even in the laboratory. This is now changing with the rapid rise of exciting new technologies for single-cell microbiology (1, 2), which enable microbiologists to do what plant and animal ecologists have been doing for a long time: observe who does what, when, where, and next to whom. Single cells taken from the environment can be identified and even their genomes sequenced. Ex situ, their size, elemental, and biochemical composition, as well as other characteristics can be measured with high-throughput and cells sorted accordingly. Even better, individual microbes can be observed in situ with a range of novel microscopic and spectroscopic methods, enabling localization, identification, or functional characterization of cells in a natural sample, combined with detecting uptake of labeled compounds. Alternatively, they can be placed into fabricated microfluidic environments, where they can be positioned, exposed to stimuli, monitored, and their interactions controlled “in microfluido.” By introducing genetically engineered reporter cells into a fabricated landscape or a microcosm taken from nature, their reproductive success or activity can be followed, or their sensing of their local environment recorded.
These novel methods have generated a multitude of fascinating observations and hold great potential for testing ecological and evolutionary theories with rapidly growing microbes under well-controlled conditions. Possibly the most important finding for the future of the discipline of microbiology has been the realization that genetically identical bacterial cells in a well-mixed environment may have individually differing phenotypes. This calls into question the validity of population-level experiments and population models that assume that all individuals are the same.
However, does such individuality of microbes really matter? We argue that the best way to find out is to use individual-based modeling in combination with single-cell and population-level observation in the field or experimentation in the laboratory. By describing relevant characteristics and activities of each individual cell in the population using the data from single-cell observations, the individual-based model will predict what happens at the population level as an emergent property (Fig. 1). Making individual-based models simultaneously reproduce patterns observed at both the individual and population level will make these models structurally realistic so that they deliver independent, testable predictions (3). Individual-based modeling of microbes, adopted from the more established individual-based modeling of larger organisms, has come of age with the development of tools that enable scientists with minimal programming expertise to develop computer simulations for use in their research (e.g., ref. 4).
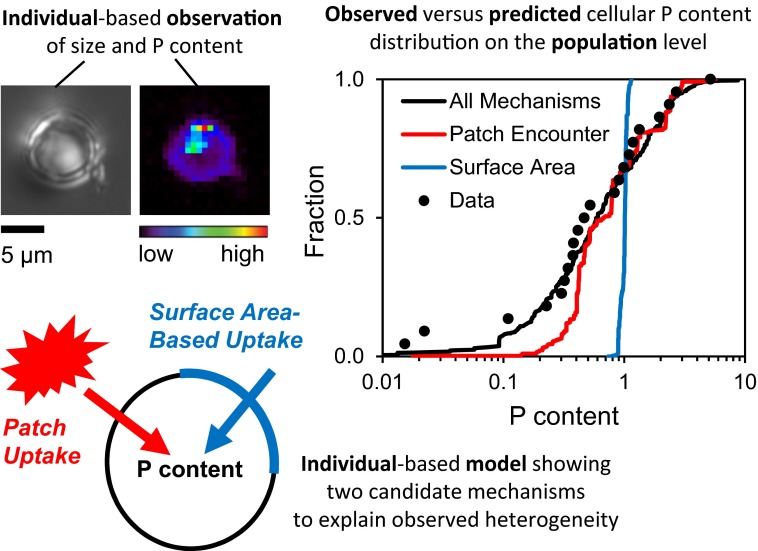
Fig. 1.
Observing and modeling individual heterogeneity in intracellular P content in the freshwater diatom Cyclotella meneghiniana. Some individual-level observations (e.g., size and P content) are used to calibrate the individual-based model, which also implements a range of conceivable mechanisms generating heterogeneity, including microscale patch encounter and surface area-based uptake of P. The model then makes predictions on the population level, which can be compared with population data or other individual-level data (e.g., P content) to identify which of the mechanisms best explains the data. The individual-based model was then used to explore the effect of individual heterogeneity on population growth rate. Variation of P content affects population growth rate because growth rate is a nonlinear function of the internal P content. In this case, population-level growth rate is only 68% of the growth rate a population of cells with average P content would have. (See ref. 6) Diatom image courtesy of Benjamin S. Twining.
The advocated combination of individual-based modeling and experimentation, which we refer to as microbial individual-based ecology (µIBE), may at first only be a marriage of convenience because the models need individual-based data and individual-based models are needed to make the best use of these data. However, a deeper relationship may develop with many mutual benefits. The µIBE agenda is set to mimic a previous success story, namely the development of new systems biology modeling approaches driven by the rise of omics data. Individual-based models can integrate prior knowledge with new data on the individual and population levels. Because these models facilitate the inclusion of mechanistic details, they have the potential to predict observations under changing conditions or in novel environments. These models can also predict concentration gradients of hard-to-detect metabolites, toxins, or chemical signals, given the spatial positions of producers and consumers and the rates of production or consumption.
One key advantage of µIBE, which we believe has tremendous potential, is the ability to incorporate both lower and higher levels of organization into the individual-based framework. First, regarding the lower (i.e., molecular) level, individual-based models lend themselves well to integration with systems biology and synthetic biology (a good example is ref. 5). As individual-based models describe the action of each individual cell, it is straightforward to incorporate models of intracellular dynamics of natural—or synthetic—cells. Such embedded intracellular models will then define the behavior of the individual cell more mechanistically, rather than being based on phenomenological descriptions of the cell’s behavior.
Second, regarding the higher levels of communities and ecosystems, the modeling of diverse communities comprising potentially many different species is also straightforward, as those communities are still made up of individual organisms. Where necessary, species can be aggregated into functional groups to reduce computational complexity. Spatial heterogeneity is taken care of as individuals are localized and interact locally. Furthermore, indirect interactions between individuals of different species (e.g., mediated by diffusible molecules, such as nutrients, toxins, or chemical signals) are an emergent property of the simulation, rather than being interactions that have to be specifically investigated and parameterized. For example, in a community with N species there are 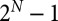 possible species combinations, including intraspecific interactions, such as cannibalism, which makes it infeasible to investigate all possible indirect interactions for even modest values of N.
possible species combinations, including intraspecific interactions, such as cannibalism, which makes it infeasible to investigate all possible indirect interactions for even modest values of N.
The future of µIBE will be bright as experimentalists discover that individual-based models, built using state-of-the-art model development tools, can be used to gain a much deeper understanding of the systems they study than otherwise possible. Modelers will embrace the opportunity to teach the use of the tools, help guide their application, and use the experience to enhance the tools further. Technical obstacles will arise as we move to systems with higher complexity or larger scale, but these obstacles are likely to be overcome and it will be possible to simulate, for example, the human gut, a volume of soil, or part of an ocean as ecosystems teeming with tiny systems biology individuals.
The chance to observe, manipulate, and model from first principles, from molecules via individuals to populations, communities, and ecosystems, goes far beyond the current basis of ecological and evolutionary theory. µIBE will thus lead to insights into ecology and evolution that are simply impossible to obtain with larger organisms. Moreover, µIBE will provide a test-bed for developing the emerging science of agent-based complex systems. The integration of individual-based modeling and experimentation is our best hope toward a future of full understanding of microbial systems, or as Donald Knuth aptly put it (7): “Science is what we understand well enough to explain to a computer. Art is everything else we do.”
Acknowledgments
This work has arisen from the workshop Individual-Based Ecology of Microbes: Observations and Modeling, held at the National Institute for Mathematical and Biological Synthesis in June 2011, sponsored by the National Science Foundation, the US Department of Homeland Security, and the US Department of Agriculture through National Science Foundation Award EF-0832858, with additional support from The University of Tennessee, Knoxville.
Footnotes
Any opinions, findings, conclusions, or recommendations expressed in this work are those of the authors and do not necessarily reflect the views of the National Academy of Sciences.
References
- 1.Musat N, Foster R, Vagner T, Adam B, Kuypers MMM. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol Rev. 2012;36(2):486–511. doi: 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]
- 2.Wessel AK, Hmelo L, Parsek MR, Whiteley M. Going local: Technologies for exploring bacterial microenvironments. Nat Rev Microbiol. 2013;11(5):337–348. doi: 10.1038/nrmicro3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm V, et al. Pattern-oriented modeling of agent-based complex systems: Lessons from ecology. Science. 2005;310(5750):987–991. doi: 10.1126/science.1116681. [DOI] [PubMed] [Google Scholar]
- 4.Lardon LA, et al. iDynoMiCS: Next-generation individual-based modelling of biofilms. Environ Microbiol. 2011;13(9):2416–2434. doi: 10.1111/j.1462-2920.2011.02414.x. [DOI] [PubMed] [Google Scholar]
- 5.Rudge TJ, Steiner PJ, Phillips A, Haseloff J. Computational modeling of synthetic microbial biofilms. ACS Synth Biol. 2012;1(8):345–352. doi: 10.1021/sb300031n. [DOI] [PubMed] [Google Scholar]
- 6.Bucci V, Nunez-Milland D, Twining B, Hellweger F. Microscale patchiness leads to large and important intraspecific internal nutrient heterogeneity in phytoplankton. Aquat Ecol. 2012;46(1):101–118. [Google Scholar]
- 7.Knuth DE. 1996. Foreword. A = B, Petkovsek M, Wilf HS, Zeilberger D (A K Peters/CRC Press, Boca Raton, FL), p vii.