Mao et al. (1) published a paper describing a 3D structure of uncleaved, trimeric HIV-1 envelope glycoprotein (Env) at ∼6-Å resolution, following a similar paper last year on the same structure at ∼11-Å resolution (2). Examination of these two papers leads me to doubt that the structures presented are results of a reliable cryo-electron microscopic analysis of purified HIV-1 Env trimers.
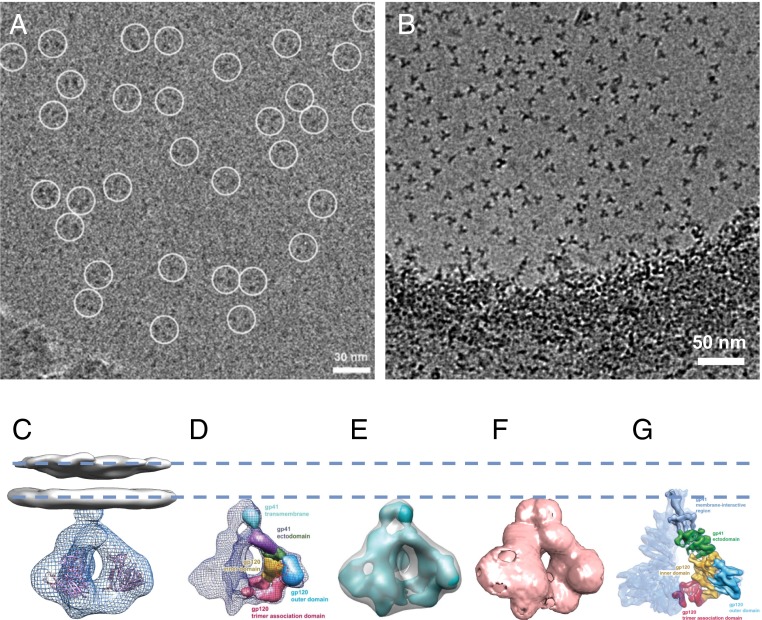
An underlying assumption in structure determination by cryo-electron microscopy is that there is demonstrable evidence that the micrographs contain images of the relevant proteins, although their mere existence in no way guarantees that a structure at 6 Å, or even at 11 Å, can be obtained. The raw electron micrographs presented by Mao et al. (1, 2) do not provide convincing evidence for the presence of molecular images of HIV-1 Env trimers [compare fig. 1A from Mao et al. (1) with fig. 1B from Harris et al. (3), where trimers are clearly visible in the images]. I am therefore concerned that the authors have fallen into the well-known reference-bias trap in image processing by recovering what looks like a real structure starting from images of random noise.
Fig. 1.
HIV-1 envelope glycoprotein structures determined by cryo-electron microscopy. (A) Electron micrograph presented in Mao et al. (1) with putative gp160 trimers circled. (B) For purposes of comparison, see the electron micrograph presented in Harris et al. (3) where the soluble Env trimers are clearly visible. The regions circled in the micrographs presented by Mao et al. (1) have a contrast similar to that of the background, raising doubts about the reliability of the particle selection process, which may have been done using a template. (C) Density map reported for native, cleaved trimeric HIV-1 Env at a resolution of ∼20 Å obtained from cryo-electron tomography of intact virions [from the work of Liu et al. (4)]. The map includes density for the ectodomain of HIV-1 Env (EMD-5019, mesh) and the lipid bilayer (EMD-5022, gray) shown with fitted coordinates for the gp120 protomer (Protein Data Bank ID code 3DNN). (D) Density map reported for uncleaved, detergent-solubilized trimeric HIV-1 Env at a resolution of ∼11 Å obtained by cryo-electron microscopy [from the work of Mao et al. (2)]. (E) Superposition of the map in D with the ectodomain of the map of native trimeric Env in C indicates that the region identified by Mao et al. (2) to be the transmembrane region corresponds to the base of the ectodomain region of the native Env reported by Liu et al. (4). (F) Inversion of the densities in EMD-5418 results in an outline that closely matches that of EMD-5019 shown in C. (G) Density map reported for uncleaved, detergent-solubilized trimeric HIV-1 Env at a resolution of ∼6 Å obtained by cryo-electron microscopy [from the work of Mao et al. (1)]. The dashed lines are drawn to provide a visual guide to the location of the lipid bilayer. A is reproduced from figure S1A of Mao et al. (1), and B is reproduced from figure 1C of Harris et al. (3). D and E are reprinted from Macmillan Publishers Ltd: Nature Structural & Molecular Biology (2), copyright 2012, and G is reproduced from figure 1 of Mao et al. (1).
Support for this concern comes from inspection of the density maps deposited in the Electron Microscopy Data Bank (EMD-5447 and EMD-5418 from refs. 1 and 2, respectively). EMD-5418 closely matches a related map determined earlier using cryo-electron tomography (ref. 4, EMD-5019) that may have been used as a mask during image processing (Fig. 1 C–F). Further, the claim that the structure of the uncleaved trimeric HIV-1 Env as presented in the work of Mao et al. (1, 2) is in agreement with the structure of native, cleaved trimeric HIV-1 Env presented in Liu et al. (4) is incorrect. EMD-5019 represents only the ectodomain of the Env trimer; it does not include the transmembrane region, which was deposited separately (EMD-5022). Assignment of the end regions of the maps presented by Mao et al. to the gp41 transmembrane helices (1, 2) (Fig. 1 D and G) thus contradicts the earlier cryo-electron tomography results (4).
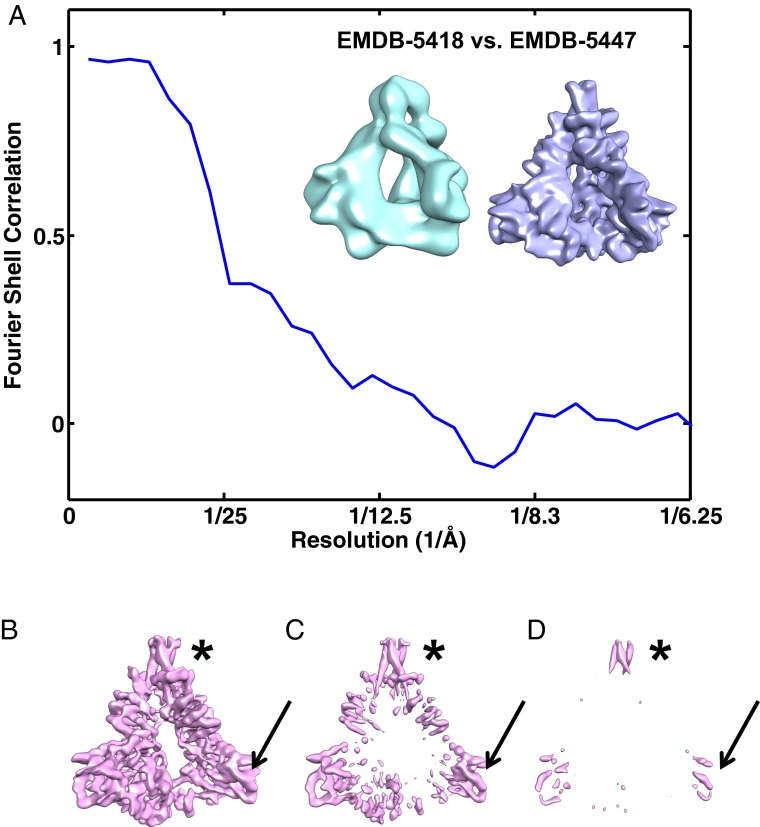
Quantitative comparison of the maps deposited by Mao et al. at resolutions of ∼11 (2) and ∼6 Å (1) shows that the Fourier shell coefficient falls to zero at ∼11 Å (Fig. 2A). If the latter map was derived from the former, as the authors write, the correlation at ∼11 Å would be nearly 100% and not zero. Further, filtering the 6-Å map to a resolution of 11 Å yields a map that is markedly different from EMD-5418 (Fig. 2A, Inset). The most ordered regions of EMD-5447 are the gp41 transmembrane helices, which stand out clearly against the background, and the V4 loop, which is disordered in crystal structures of monomeric gp120 (5) (Fig. 2 B–D). A plausible explanation for these surprising map features is that the authors used a molecular model to select particles from their micrographs; in this case, the refinement process would be circular, resulting in a final map that recapitulates the starting template.
Fig. 2.
Comparison of the two maps reported by Mao et al. (1, 2). (A) Fourier shell correlation plot between density maps EMD-5418 (reported resolution ∼11 Å) and EMD-5447 (reported resolution ∼6 Å) indicating that the two maps are essentially uncorrelated beyond resolutions of ∼20 Å. (Inset) Comparison of EMD-5447 (filtered to a resolution of 11 Å) with EMD-5418 (reported to have a resolution of 11 Å) indicates that features in these two maps do not match. (B–D) Isosurface contours of EMD-5447 rendered using UCSF Chimera at three different thresholds (0.0074, 0.018, and 0.34 respectively) to determine the most ordered regions of the map. The asterisks and arrows point to the regions identified in Mao et al. (1) to correspond to the location of the gp41 transmembrane region and the V4 variable loop of gp120, respectively.
It will not be possible to determine the validity of the results of Mao et al. (1, 2) unless they make publicly available their original micrographs, list of locations where particles were selected, and a complete description of the protocols used for image processing.
Note Added in Proof.
This letter is accompanied by a related Letter from Marin van Heel (6) and Perspective article by Richard Henderson (7).
Footnotes
The author declares no conflict of interest.
References
- 1.Mao Y, et al. Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc Natl Acad Sci USA. 2013;110(30):12438–12443. doi: 10.1073/pnas.1307382110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao Y, et al. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nat Struct Mol Biol. 2012;19(9):893–899. doi: 10.1038/nsmb.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris A, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci USA. 2011;108(28):11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455(7209):109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong PD, et al. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8(12):1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 6.van Heel M. Finding trimeric HIV-1 envelope glycoproteins in random noise. Proc Natl Acad Sci USA. 2013;110:E4175. doi: 10.1073/pnas.1314353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson R. Avoiding the pitfalls of single particle cryo-electron microscopy: Einstein from noise. Proc Natl Acad Sci USA. 2013;110:18037–18041. doi: 10.1073/pnas.1314449110. [DOI] [PMC free article] [PubMed] [Google Scholar]