Abstract
Nonselected and natural populations of Escherichia coli from 12 animal sources and humans were examined for the presence and types of 14 tetracycline resistance determinants. Of 1,263 unique E. coli isolates from humans, pigs, chickens, turkeys, sheep, cows, goats, cats, dogs, horses, geese, ducks, and deer, 31% were highly resistant to tetracycline. More than 78, 47, and 41% of the E. coli isolates from pigs, chickens, and turkeys were resistant or highly resistant to tetracycline, respectively. Tetracycline MICs for 61, 29, and 29% of E. coli isolates from pig, chickens, and turkeys, respectively, were ≥233 μg/ml. Muliplex PCR analyses indicated that 97% of these strains contained at least 1 of 14 tetracycline resistance genes [tetA, tetB, tetC, tetD, tetE, tetG, tetK, tetL, tetM, tetO, tetS, tetA(P), tetQ, and tetX] examined. While the most common genes found in these isolates were tetB (63%) and tetA (35%), tetC, tetD, and tetM were also found. E. coli isolates from pigs and chickens were the only strains to have tetM. To our knowledge, this represents the first report of tetM in E. coli.
Problems associated with the presence of antibiotic-resistant bacteria have reached epidemic proportions in recent years, with cost estimates exceeding $4 billion in the United States alone (6, 12). The spread of antibiotic-resistant bacteria in the environment is dependent on the presence and transfer of resistance genes among microorganisms, mutations, and selection pressure to keep these genes in a population. Selection pressure has been neatly provided by the approximately 50 million pounds of antibiotics that are produced and used each year in the United States (14). Only half of these antibiotics are used for humans, while the remainder are administered to animals or other organisms (8). The causes and effects of antibiotic overuse are varied. One of the most controversial applications of antibiotics, however, is for growth promotion in livestock, and this application has raised concerns about its contribution to the presence of resistant bacteria in humans (1, 25).
Tetracyclines have become the drugs of choice to treat Mycoplasma- and Chlamydia-induced pneumonia (13) and have been used to treat other atypical pneumonias, rickettsial infections, Lyme disease, ehrlichiosis, and other diseases and cancers (23). The clinically useful chlortetracycline was introduced in 1948 (24). Only a year later, it was shown that young chickens fed tetracyclines had enhanced growth characteristics (10). However, by 1953, it was reported that Shigella dysenteriae had developed resistance to tetracycline antibiotics, and by 1955, a Shigella sp. strain had developed multidrug resistance (20). Because of that history and the broad clinical use of tetracycline, this antibiotic was chosen, along with commensal strains of Escherichia coli, to provide a prototypical view of the use of antibiotics and their effects on bacterial populations (21). Tetracycline is a broad-spectrum antibiotic that inhibits bacterial protein synthesis by preventing aminoacyl-tRNA from binding to the bacterial ribosome (20). Resistance to the antibiotic is conferred by 1 or more of the 36 currently described tet genes, which encode one of three mechanisms of resistance: an efflux pump, a method of ribosomal protection, or direct enzymatic inactivation of the drug (7). Efflux mechanisms appear to be more abundant among gram-negative microorganisms, while ribosomal protection mechanisms are more common among gram-positive organisms (7). Generally speaking, the rapid spread of tetracycline resistance among bacteria is due to the localization of tet genes on plasmids, transposons, and integrons (7, 15, 21).
While several studies have examined tetracycline resistance among bacteria, most have employed clinically isolated bacteria (4, 11, 17) or populations specifically isolated for their ability to grow in the presence of tetracyclines (5, 22). These studies, while useful, do not give an unbiased appraisal of the presence and types of tet genes that are present in natural (nonclinical), nonselected populations of bacteria in the environment.
Only a limited number of studies have examined tetracycline resistance determinants in bacteria isolated from a large variety of animal species with different histories of exposure to tetracyclines or in environmental samples (11). While Sengeløv and coworkers (22) examined 100 E. coli isolates for the presence of five tet resistance determinants and Blake et al. (5) used PCR to examine 200 tetracycline-resistant E. coli strains for seven tet genes, few have examined a large number of tet determinants in nonclinical E. coli isolates from a variety of animal species. To better understand the distribution of resistance genes in the environment and to provide insight into selection pressures involved with the use of antibiotics in animal feed, we investigated tetracycline resistance among natural and unselected populations of E. coli from 12 animal sources and humans and determined which resistance genes were present in this population.
Isolates and determination of MIC.
In order to characterize tetracycline resistance in natural, nonclinical E. coli strains from both human and animal sources, 1,263 unique isolates were obtained from humans, cats, cows, deer, turkeys, ducks, sheep, geese, dogs, pigs, horses, chickens, and goats (Table 1). Fecal materials were collected by swabbing the rectal or cloacal region of individual wild and domesticated animals located throughout Minnesota and western Wisconsin as previously described (9). Fecal samples were kept at 4°C and analyzed within 6 h of swabbing. Fecal material was streaked onto mFC agar plates (Difco, BD Diagnostic Systems, Sparks, Md.) and incubated at 44.5°C for 24 h, and six blue colonies from the mFC agar plates were picked and evaluated by using selective and differential growth media as previously described (9). Only isolates giving growth and color responses on all media that were typical for E. coli were used in these studies. Three E. coli colonies from each individual fecal sample were used for DNA fingerprinting. All isolates were subjected to DNA fingerprint analysis using rep-PCR and BOXA1R primers (9), and identical clones from the same animal were eliminated from analyses. Unique isolates were grown overnight in 150 μl of Luria-Bertani liquid medium in microtiter plates and were spot inoculated, with a multiple inoculator, onto tryptic soy agar (Difco Laboratories, Detroit, Mich.) supplemented with 0, 5, 10, 20, 40, 70, 93, 117, 175, and 233 μg of tetracycline per ml (Sigma Chemicals, St. Louis, Mo.). The plates were incubated overnight at 37°C and visually examined for growth. MICs were determined from growth patterns, and average values are shown in Fig. 1. If the tetracycline MIC for an isolate was <5 μg /ml, the isolate was considered sensitive to the antibiotic; if it was 10 to 70 or >90 μg/ml, the isolate was considered resistant or highly resistant, respectively. For statistical analysis, a MIC of >233 μg/ml was considered to be 233 μg/ml.
TABLE 1.
E. coli isolates used in this study and their animal sources
| Animal source of E. coli | No. of isolates used for MIC analysis | No. of isolates used for multiplex PCR |
|---|---|---|
| Cat | 46 | 9 |
| Cow | 158 | 24 |
| Deer | 74 | 1 |
| Turkey | 82 | 30 |
| Duck | 70 | 1 |
| Human | 176 | 30 |
| Sheep | 48 | 15 |
| Goose | 122 | 3 |
| Dog | 47 | 9 |
| Pig | 182 | 131 |
| Horse | 66 | 3 |
| Chicken | 151 | 66 |
| Goat | 41 | 3 |
| Total | 1,263 | 325 |
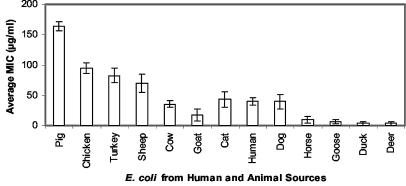
FIG. 1.
Average MICs of tetracycline for E. coli isolates obtained from pigs, chickens, turkeys, sheep, cows, goats, cats, humans, dogs, horses, geese, ducks, and deer, as determined by the plate dilution method.
Of the 1,263 E. coli isolates examined, 31% were resistant to tetracycline (MICs, >10 μg/ml). Forty-two, 21, 17, and 4% of the isolates from livestock, humans, companion animals (cats, dogs, and horses), and wild animals, respectively, were resistant to tetracycline. More than 78, 47, and 41% of the E. coli isolates from pigs, chickens, and turkeys were resistant or highly resistant to tetracycline, respectively. Together these resistant isolates represent about 20% of the 1,263 isolates examined. In contrast, about 22, 30, 3, 3, 21, 33, 7, 23, 6, and 12.2% of the E. coli isolates from cats, cows, deer, duck, humans, sheep, geese, dogs, horses, and goats were resistant or highly resistant to tetracycline, respectively. Moreover, the tetracycline MICs for 61, 29, and 29% of E. coli isolates from pigs, chickens, and turkeys, respectively, were ≥233 μg/ml. In contrast, the lowest numbers of E. coli strains showing resistance or a high level of resistance to tetracycline were those from goats, horses, ducks, geese, and deer. Our results may be explained by the potential exposure of livestock, humans, and companion and wild animals to tetracyclines. Tetracycline is often continuously fed to livestock at subtherapeutic levels for the purpose of growth promotion. For example, up to 70% of U.S. cattle and pig operations use feeds supplemented with antibiotics for growth promotion, and the majority are tetracyclines (2). In contrast, humans and companion animals are most often treated therapeutically, for a limited time, for bacterial infections, perhaps reflecting the intermediate level of resistance to tetracycline (average MICs, 10 to 70 μg/ml) of the isolates from these organisms. This resistance level may be changing, however, as other uses of antibiotics become more common, such as the treatment of parasitic and noninfectious diseases (21). The low level of occurrence of tetracycline resistance among isolates from wild animals is presumably due to their low exposure to these antibiotics. Most of these isolates either had a high level of resistance or none at all, suggesting that the acquisition of a mobile genetic element accounts for resistance.
Epidemiology of tet genes.
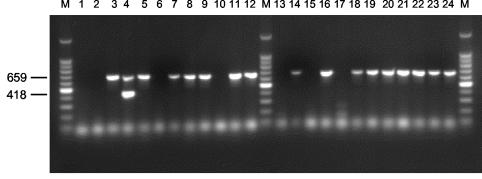
All isolates for which the tetracycline MIC was ≥93 μg/ml (which we considered to indicate a high level of resistance) (n = 325) were examined further by use of a multiplex PCR for the presence of the tetA, tetB, tetC, tetD, tetE, tetG, tetK, tetL, tetM, tetO, tetS, tetA(P), tetQ, and tetX genes (18). Single-colony isolates were streaked onto plate count agar (Difco), picked using disposable 10-μl sterile loops, and suspended in 50 μl of sterile H2O. One microliter of the standardized cell suspension served as a template DNA for colony-based multiplex PCR. The primers used for PCR amplification of the 14 tetracycline resistance genes were as described by Ng et al. (18). The primers were aliquoted into four groups: group I contained primers for tetB, tetC, and tetD; group II contained primers for tetA, tetE, and tetG; group III contained primers for tetK, tetL, tetM, tetO, and tetS; and group IV contained primers for tetA(P), tetQ, and tetX. PCR was performed with 96-well plates and an MJ Research (Waltham, Mass.) model PTC100 thermocycler, by using the following conditions as described previously (18): 5 min of initial denaturation at 94°C, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min. The PCR products were separated by gel electrophoresis in 1% (wt/vol) agarose gels in 1× Tris-acetate-EDTA buffer, stained with ethidium bromide, and visualized under UV illumination. The validity of multiplex PCRs and product sizes was ascertained by using the following positive control plasmids: pSL18, pRT11, pBR322, pSL106, pSL1504, pJA8122, pAT102, pVB.A15, pJ13, pUOA1, pAT451, pJIR39, pNFD13-2, and pBS5, for the genes tetA, tetB, tetC, tetD, tetE, tetG, tetK, tetL, tetM, tetO, tetS, tetA(P), tetQ, and tetX, respectively (18). The sizes of the PCR products were determined by comparison to the migration of a 100-bp ladder (Gibco BRL). The identity of all tet genes in a representative sample of nonclinical isolates was ascertained by DNA sequencing of the PCR products, following extraction from agarose gels. A representative agrose gel of PCR products obtained using primer group I, amplifying tetB, tetC, and tetD, is shown in Fig. 2.
FIG. 2.
Representative agarose gel of PCR products from nonclinical E. coli isolates, using primer group I, containing primers for tetB, tetC, and tetD. Lanes: 1, no template control; 2, E. coli H25; 3, E. coli H45; 4, E. coli H77; 5, E. coli P282; 6, E. coli P284; 7, E. coli P285; 8, E. coli P286; 9, E. coli P289; 10, E. coli P290; 11, E. coli P291; 12, E. coli P293; 13, E. coli P294; 14, E. coli P295; 15, E. coli P296; 16, E. coli P297; 17, E. coli P298; 18, E. coli P300; 19, E. coli P304; 20, E. coli P307; 21, E. coli P308; 22, E. coli P309; 23, E. coli P310; and 24, E. coli P312. E. coli isolate numbers beginning with P and H were isolated from pigs and horses, respectively. Lane M, molecular weight markers (100 bp ladder). The sizes of the amplicons in base pairs are indicated on the left.
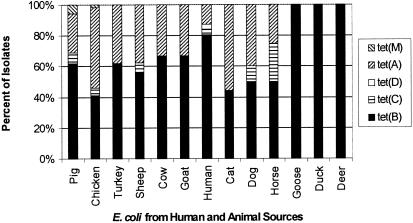
Of the 325 strains analyzed by PCR, 97% contained at least 1 of 14 [tetA, tetB, tetC, tetD, tetE, tetG, tetK, tetL, tetM, tetO, tetS, tetA(P), tetQ, and tetX] tetracycline resistance determinants. The most common determinants were Tet B (63% of isolates) and Tet A (35% of isolates) (Fig. 3). However, Tet C, Tet D, and Tet M were also found with various frequencies. The frequencies of tetA, tetB, tetC, and tetD in the tested isolates (Fig. 3) were consistent with those previously reported for lactose-fermenting coliforms based on colony hybridization (11). In contrast, Sengeløv and coworkers (22) reported that 71 and 25% of 100 isolates from the diseased and healthy pigs, cattle, and chickens that they tested for five tetracycline resistance determinants contained tetA and tetB, respectively. None of the tested strains contained tetE, tetG, tetK, tetL, tetO, tetS, tetA(P), tetQ, or tetX. Since our studies analyzed only highly resistant isolates by PCR, it is possible that additional resistance genes were present in the E. coli populations but were nonfunctional or provided only intermediate or low-level resistance.
FIG. 3.
Frequency of tetM, tetA, tetD, tetC, and tetB in E. coli isolates obtained from pigs, chickens, turkeys, sheep, cows, goats, humans, cats,dogs, horses, geese, ducks, and deer, as determined by colony multiplex PCR. The tetracycline genes tetE, tetG, tetK, tetL, tetO, tetS, tetA(P), tetQ, and tetX were not found among any of the 325 E. coli isolates tested.
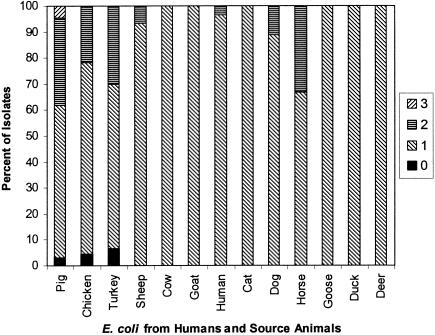
Isolates from pigs and chickens were the only strains to contain tetM and commonly had more than one tetracycline resistance determinant per strain (Fig. 4). The greatest number of strains for which the MICs were high were E. coli isolates from these animals. Over 30% of E. coli isolates from turkeys, pigs, and horses contained two tetracycline resistance determinants, and 4.5% of the pig isolates contained three tet genes. However, the presence of more than one resistance determinant did not lead to noticeably higher MICs. It is possible that strong selection pressures provided by environments containing elevated levels of tetracycline lead to the acquisition of more than one tetracycline gene in a given strain due to their prevalence in the environment, rather than to a selective advantage. The results of our studies also showed that 22.2 and 1.9% of the isolates contained two and three tet genes, respectively. This is in contrast to results from previous studies, in which only 3.5% (16) and 5.4% (22) of isolates had two genes, perhaps due to our use of a larger number and variety of isolates and to the greater number of genes examined.
FIG. 4.
Percentages of E. coli isolates obtained from pigs, chickens, turkeys, sheep, cows, goats, humans, cats, dogs, horses, geese, ducks, and deer, containing multiple tetracycline resistance genes as determined by multiplex PCR using primers for tetA, tetB, tetC, tetD, tetE, tetG, tetK, tetL, tetM, tetO, tetS, tetA(P), tetQ, and tetX.
To our knowledge, this is the first report documenting the presence of the tetM gene in E. coli (7). Due to the uniqueness of these results, the presence of tetM in one of our E. coli isolates from pigs was verified by sequencing the PCR product produced using tetM-specific primers. BLAST analysis (3) indicated that of the 386 bp of high-quality and continuous sequence examined, there was 98% nucleotide sequence identity to the tetM gene from Enterococcus faecalis (GenBank accession number M85225). The tetM gene, which imparts resistance to tetracyclines by encoding a ribosomal protection mechanism, commonly occurs in transposons Tn916 and Tn1545. The tetM gene is widely dispersed among various gram-positive organisms, but it has only rarely been documented in gram-negative bacteria (19, 21). The presence of tetM in E. coli is most likely due to genetic transfer from Enterococcus, a common carrier of tetM (8). Evidence for this possibility is provided by the studies of Poyart et al. (19), who demonstrated the in vitro transfer of Tn916 from E. faecalis to E. coli (16).
In summary, by examining the frequency and distribution of tetracycline resistance genes among diverse natural E. coli populations present in different animal species, a picture of the selection pressures in the various host animals can be inferred. Not only did those animal hosts that presumably had continuous exposure to tetracycline have a higher percentage of tetracycline-resistant E. coli isolates, but also those isolates carried a greater diversity of resistance genes. Moreover, these isolates often had more than one tetracycline resistance determinant and contained a tet gene previously thought not to be present in E. coli. This suggests that human activity provides environments that select for resistant strains and encourages the transfer of genetic information from unrelated bacterial species. Although this study examined only nonclinical E. coli isolates, the prevalence of tetracycline resistance genes among these unrelated bacteria, and circumstantial and direct evidence of horizontal gene transfer, suggests that these same resistance determinants may also be present in animal and human pathogens.
Acknowledgments
We thank LeeAnn Johnson, John Ferguson, Mary Brown, and Priscilla Dombek for help with the E. coli strains. We also thank Lai-King Ng for the generous gift of positive control strains and cloned tet resistance genes.
This study was supported in part by grants from the University of Minnesota Agricultural Experiment Station and from the Legislative Commission on Minnesota Resources, through the Environment and Natural Resources Trust Fund and the Minnesota Future Resource Fund (to M.J.S.), and by funding from the Life Sciences Summer Undergraduate Research Program (to A.B.).
REFERENCES
- 1.Aarestrup, F. M., A. M. Seyfarth, H. D. Emborg, K. Pedersen, R. S. Hendriksen, and F. Bager. 2001. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 45:2054-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akkina, J. E., and R. Johnson. 1999. Antibiotic use in U.S. livestock production, p. 17-29. In Antimicrobial resistance issues in animal agriculture. USDA/APHIS/VS/CEAHI/CEI, Washington, D.C.
- 3.Altschul, S. F., T. L. Madden, A Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arzese, A. R., L. Tomasetig, and G. A. Botta. 2000. Detection of tetQ and ermF antibiotic resistance genes in Prevotella and Porphyromonas isolates from clinical specimens and resident microbiota of humans. J. Antimicrob. Chemother. 45:577-582. [DOI] [PubMed] [Google Scholar]
- 5.Blake, D. P. R. W. Humphry, K. P. Scott, K. Hillman, D. R. Fenlon, and J. C. Low. 2003. Influence of tetracycline exposure on tetracycline resistance and the carriage of tetracycline resistance genes within commensal Escherichia coli populations. J. Appl. Microbiol. 94:1087-1097. [DOI] [PubMed] [Google Scholar]
- 6.Boyce, J. M. 2001. Consequences of inaction: importance of infection control practices. Clin. Infect. Dis. 33(Suppl.):S133-S137. [DOI] [PubMed]
- 7.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFlaun, M. F., and S. B. Levy. 1989. Genes and their varied hosts, p. 1-32. In S. B. Levy and R. V. Miller (ed.), Gene transfer in the environment. McGraw-Hill, New York, N.Y.
- 9.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuPont, H. L., and J. H. Steele. 1987. The human health implication of the use of antimicrobial agents in animal feeds. Vet. Q. 9:309-320. [DOI] [PubMed] [Google Scholar]
- 11.Guillaume, G., D. Verbrugge, M.-L. Chasseur-Libotte, W. Moens, and J.-M. Collard. 2003. PCR typing of tetracycline resistance determinants (Tet A-E) in Salmonella enterica serotype Hadar and in the microbial community of activated sludges from hospital and urban wastewater treatment facilities in Belgium. FEMS Microbiol. Ecol. 32:77-85. [DOI] [PubMed] [Google Scholar]
- 12.Jones, M. E., J. A. Karlowsky, D. C. Draghi, C. Thornsberry, D. F. Sahm, and D. Nathwani. 2003. Epidemiology and antibiotic susceptibility of bacteria causing skin and soft tissue infections in the USA and Europe: a guide to appropriate antimicrobial therapy. Int. J. Antimicrob. Agents 22:406-419. [DOI] [PubMed] [Google Scholar]
- 13.Lenart, J., A. A. Andersen, and D. D. Rockey. 2002. Growth and development of tetracycline-resistant Chlamydia suis. Antimicrob. Agents Chemother. 45:2198-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy, S. G. 2001. Antibiotic resistance: consequences of inaction. Clin. Infect. Dis. 33(Suppl.):S124-S129. [DOI] [PubMed] [Google Scholar]
- 15.Levy, S. B., G. B. Fitzgerald, and A. B. Macone. 1976. Changes in intestinal flora of farm personnel after introduction of tetracycline-supplemented feed on a farm. N. Engl. J. Med. 295:583-588. [DOI] [PubMed] [Google Scholar]
- 16.Marshall, B., C. Tachibana, and S. B. Levy. 1983. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob. Agents Chemother. 24:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathew, A. G., D. B. Arnett, P. Cullen, and P. D. Ebner. 2003. Characterization of resistance patterns and detection apramycin resistance genes in Escherichia coli isolated from swine exposed to various environmental conditions. Int. J. Food Microbiol. 89:11-20. [DOI] [PubMed] [Google Scholar]
- 18.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 19.Poyart, C., J. Celli, and P. Trieu-Cuot. 1995. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob. Agents Chemother. 39:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts, M. C. 1996. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 19:1-24. [DOI] [PubMed] [Google Scholar]
- 21.Roberts, M. C. 2003. Tetracycline therapy: update. Clin. Infect. Dis. 36:462-467. [DOI] [PubMed] [Google Scholar]
- 22.Sengeløv, G., B. Halling-Sørensen, and F. M. Aarestrup. 2003. Susceptibility of Escherichia coli and Enterococcus faecium isolated from pigs and broiler chickens to tetracycline degradation products and distribution of tetracycline resistance determinants in E. coli from food animals. Vet. Microbiol. 95:91-101. [DOI] [PubMed] [Google Scholar]
- 23.Smilack, J. D. 1999. The tetracyclines. Mayo Clin. Proc. 74:727-729. [DOI] [PubMed] [Google Scholar]
- 24.U.S. Congress Office of Technology Assessment. 1995. Impacts of antibiotic-resistant bacteria. OTA-H-629, stock no. GPO 052-003-01446-7. Government Printing Office, Washington, D.C.
- 25.Wegener, H. G., F. M. Aarestrup, P. Gerner-Smidt, and F. Bager. 1999. Transfer of antibiotic resistant bacteria from animals to man. Acta Vet. Scand. Suppl. 92:51-57. [PubMed] [Google Scholar]