Significance
Epithelial–mesenchymal transitions play crucial roles in embryonic development, wound healing, and cancer metastasis. It is clearly of interest to quantitatively understand the core circuitry that takes input from the cell’s environment to perform this cell-fate decision. We devised a unique model of the microRNA (miR)-based coupled chimeric modules underlying this core circuit; this effort utilizes a unique theoretical framework for treating miR dynamics. We show that the miR-200/ZEB module functions as a ternary switch, allowing not only for the epithelial and mesenchymal phenotypes but also for a hybrid phenotype with mixed characteristics of collective cell migration, as seen in branching morphogenesis and wound closure. Our results provide valuable clues on how to control and reverse epithelial–hybrid–mesenchymal transitions.
Keywords: multistable decision circuit, partial EMT, cancer systems biology, microRNA modeling, metastable intermediate phenotypes
Abstract
Forward and backward transitions between epithelial and mesenchymal phenotypes play crucial roles in embryonic development and tissue repair. Aberrantly regulated transitions are also a hallmark of cancer metastasis. The genetic network that regulates these transitions appears to allow for the existence of a hybrid phenotype (epithelial/mesenchymal). Hybrid cells are endowed with mixed epithelial and mesenchymal characteristics, enabling specialized capabilities such as collective cell migration. Cell-fate determination between the three phenotypes is in fact regulated by a circuit composed of two highly interconnected chimeric modules—the miR-34/SNAIL and the miR-200/ZEB mutual-inhibition feedback circuits. Here, we used detailed modeling of microRNA-based regulation to study this core unit. More specifically, we investigated the functions of the two isolated modules and subsequently of the combined unit when the two modules are integrated into the full regulatory circuit. We found that miR-200/ZEB forms a tristable circuit that acts as a ternary switch, driven by miR-34/SNAIL, that is a monostable module that acts as a noise-buffering integrator of internal and external signals. We propose to associate the three stable states—(1,0), (high miR-200)/(low ZEB); (0,1), (low miR-200)/(high ZEB); and (1/2,1/2), (medium miR-200)/(medium ZEB)—with the epithelial, mesenchymal, and hybrid phenotypes, respectively. Our (1/2,1/2) state hypothesis is consistent with recent experimental studies (e.g., ZEB expression measurements in collectively migrating cells) and explains the lack of observed mesenchymal-to-hybrid transitions in metastatic cells and in induced pluripotent stem cells. Testable predictions of dynamic gene expression during complete and partial transitions are presented.
Understanding cell-fate decisions during embryonic development and tumorigenesis remains a major research challenge in modern developmental and cancer biology (1). Over recent years, we have witnessed rapid progress in mapping the gene regulatory networks associated with cellular phenotype with applications to transitions from epithelial to mesenchymal modalities, to the differentiation of pluripotent stem cells into progenitor cells, to the existence and role of cancer stem-like cells (CSC), and to the production of induced pluripotent stem cells (iPSCs). Cell-fate determinations in all of these examples involve changes in expression of various transcription factors (TFs) and microRNAs (miRNAs) that regulate cascades of regulatory networks, ultimately generating genome-wide gene-expression patterns and concomitant protein levels corresponding to a particular cell lineage (fate).
The E/M Hybrid Phenotype.
An archetypal example of cell-fate decisions concerns the forward and backward transitions between the epithelial (E) and mesenchymal (M) phenotypes (EMT and MET), transitions that play a critical role in embryonic development, tissue repair, and cancer metastasis (2). The genetic network that regulates epithelial–mesenchymal transitions also allows for partial transitions into a hybrid phenotype, which is endowed with combined epithelial (e.g., cell–cell adhesion) and mesenchymal (e.g., motility) capabilities. These mixed traits of the E/M hybrid phenotype enable cells to undergo directed collective cell migration, instead of the solitary migration of purely mesenchymal cells (3). The E/M hybrid state, the result of what is commonly referred to as partial EMT (pEMT), can revert to enable reepithelialization when required, for example, during the completion stages of wound healing and branching morphogenesis (4–6).
Relevance to Tumorigenesis.
Aberrantly regulated complete and partial E–M transitions play a crucial role in cancer metastasis (5). These processes begin when primary tumor cells lose their epithelial apico-basal polarity and tight cell–cell adhesion to become migratory and invasive. Upon reaching a proper colonization niche in a distant organ, some of the seeding mesenchymal cells undergo the backward transition, regain their epithelial characteristics, and form micrometastases, some of which later evolve into full-grown macrometastases (2). Traversing the extracellular matrix (ECM) toward blood vessels is a complex task requiring specialized cellular motility. It has been shown that cancer cells can migrate as dispersed individuals of the mesenchymal phenotype, or in coordinated collective motion of the hybrid phenotype (5–7). Collective migration obviates the need for all cells to be able to detect extrinsic signals for migration (8), enabling the cohort of cells to adeptly adapt to different microenvironments. Thus, E/M hybrid cells have an important functional role for successful migration through the ECM. Notably, whereas E/M hybrid cells have been observed during forward E-to-M transitions, they have not been observed to occur during the reverse M-to-E transitions (9, 10). These experiments imply an innate asymmetry in the underlying mechanisms regulating the epithelial–mesenchymal cell fate.
The Core Regulatory Unit.
Cell-fate determination between the three phenotypes can be induced by various internal and external signals such as HIF-1α, p53, TGF-β, HGF, FGF, EGF, Notch, Wnt, and Hedgehog (2). It is now understood that these signals converge on a core regulatory unit composed of four components—two families of E-box binding transcription factors (TFs) SNAIL and ZEB and two families of microRNAs (miRs) miR-34 and miR-200. The unit architecture can be thought of as two highly interconnected modules—the miR-34/SNAIL and the miR-200/ZEB (Fig. 1). Each of these modules is a mutually inhibiting microRNA–TF chimeric feedback loop circuit (11–13). Generally speaking, high levels of miR-34 and miR-200 correspond to the epithelial phenotype, and high levels of SNAIL and ZEB correspond to the mesenchymal phenotype and are associated with higher metastatic capacity (2, 14). The two TFs repress epithelial-specific gene expression, including E-cadherin, the hallmark of the epithelial phenotype, and promote the expression of mesenchymal markers such as N-cadherin and vimentin (14). The core regulatory system is also known to play an important role as “a motor of cellular plasticity,” as it is coupled to a variety of other key cellular features (SI Appendix, Fig. S1), including stemness, cell-cycle arrest, apoptosis and senescence, resistance to chemotherapy, and cell–cell communication (15, 16).
Fig. 1.
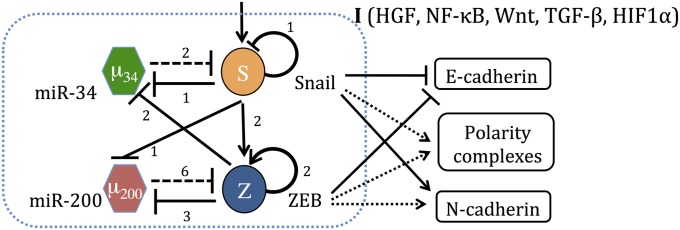
The regulatory network governing epithelial–mesenchymal plasticity. The unit is composed of two highly linked miR-TF chimeric circuits, the miR-34/SNAIL and the miR-200/ZEB mutually inhibiting loops. A solid arrow denotes transcriptional activation, and a solid bar denotes transcriptional inhibition. Dashed lines indicate microRNA-mediated translational regulation, and dotted lines represent indirect regulation. The numbers listed alongside each regulatory line represent the number of binding sites, as deduced from experiments.
Ternary Switches.
The realization that mutually inhibiting TF–TF switches and mutually inhibiting miR–TF chimeric switches play an important role in cell-fate determination has led several groups to conduct theoretical studies of such circuits (17–22). These studies have revealed that inclusion of transcription self-activation can lead to the coexistence of three stable states (tristability) (17–20). More specifically, the classical TF–TF toggle switch has two stable states, (0,1) and (1,0), where state (0) corresponds to relatively low expression (low concentration) and state (1) corresponds to relatively high expression (23). However, when one of the TFs is self-activating, the circuit can in general have three such states—(0,1), (1,0), and (1/2,1/2), and thereby acts as a ternary switch. Recently, Lu et al. (24) devised a theoretical framework to incorporate translation-transcription regulation in the dynamics of microRNA-dependent chimeric circuits. Their dynamical system analysis revealed that inclusion of TF self-activation can also allow the miR–TF chimeric circuit to act as a ternary switch.
Ternary Chimera Switch Driven by Noise-Buffering Integrator.
A generalized version of the framework devised by Lu et al. (24) is used to study the dynamics and task performance of the [miR-34/SNAIL]:[miR-200/ZEB] core regulatory unit. This biologically consistent approach incorporates in detail the dynamics of microRNA silencing, both translational repression and the active mRNA/miR degradation. Consequently, the kinetic equations contain new types of terms playing the roles of the inhibitory and excitatory Hill functions. This approach enables us to investigate the effect of the number of the microRNA binding sites on the mRNA. We found that the [miR-34/SNAIL] module is a monostable circuit that acts as a noise-buffering integrator, reminiscent of the bacterial sporulation sensing system (25–27). The [miR-200/ZEB] module was found to be a tristable circuit that acts as a ternary switch. We show that the different operating characteristics of these two modules are associated with the experimentally known differences in the nonlinearity of their translation-transcription inhibition (high vs. low) and in the TF self-regulation (ZEB self-activation vs. SNAIL self-inhibition) (see SI Appendix, Section 2 for circuit details). The tristable structure of the [miR-200/ZEB] module is retained upon coupling to the other module. We associate the decision module (1/2,1/2) state with the epithelial–mesenchymal hybrid phenotype and discuss the consistency of this picture with current experimental observations as well as make specific testable predictions. In Discussion, we compare our findings with a recently published contrasting view associating the hybrid phenotype with a mixed state of high miR-200 and low miR-34 (28).
Results
Summary View of the Core Regulatory Components.
Before presenting our detailed analysis, we summarize some of our key findings. The core regulatory unit, Fig. 1, is composed of two highly interconnected modules—the miR-34/SNAIL and the miR-200/ZEB microRNA-TF double negative feedback loops. In SI Appendix, Fig. S1, we illustrate how the decision circuit participates in the regulation of other key cellular processes and characteristics including stress response, stemness, genome stability, cell–cell communication, and cellular motility.
We initially considered each loop in isolation. Analyzing the stand-alone dynamics, we found that the miR-200/ZEB generically exhibits tristability (the existence of three distinct stable states) and that the miR-34/SNAIL circuit exhibits monostability (existence of a single stable state). The term “generically” is used to indicate that this is the characteristic behavior for a wide range of realistic parameters inferred from experimental observations (see SI Appendix for details), using as input the known functional character of the translation and transcription inhibition of the two circuits. Consequently we put forward the following interpretation regarding the function of the two modules: (i) the miR-200/ZEB operates as a ternary switch (Fig. 2A); (ii) the miR-34/SNAIL operates as a noise-buffering integrator (Fig. 2B). In this interpretation, the three states of the switch are as follows: (i) the (1,0) state—high miR-200 with low ZEB, which corresponds to the epithelial (E) phenotype; (ii) the (0,1) state—low miR-200 with high ZEB, which corresponds to the mesenchymal (M) phenotype; and (iii) the (1/2,1/2) state—intermediate levels of both miR-200 and ZEB, which corresponds to the epithelial–mesenchymal (E/M) hybrid phenotype.
Fig. 2.
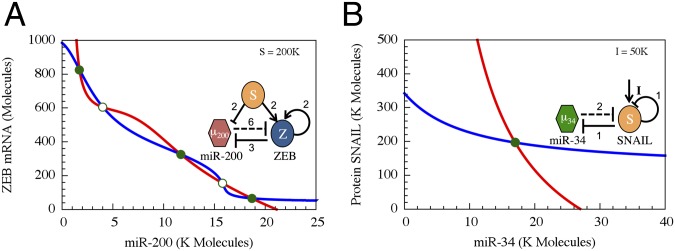
Dynamical system characteristics of the miR-200/ZEB and miR-34/SNAIL modules. The figures show the nullclines and the possible states in the phase-spaces corresponding to the two modules as are determined from Eqs. S4.2 and S4.3 in SI Appendix. (A) The miR-200/ZEB driven by signal SNAIL is tristable. Red nullcline is for dµ200/dt = 0 and dZ/dt = 0, and blue nullcline is for the condition dmZ/dt = 0 and dZ/dt = 0. (B) The miR-34/SNAIL is monostable. Blue nullcline is for the conditions dµ34/dt = 0 and dS/dt = 0, and red for dmS/dt = 0 and dS/dt = 0. The variables µ200, µ34, mZ, mS, Z, and S correspond to the levels of miR-200, miR-24, ZEB mRNA, SNAIL mRNA, ZEB, and SNAIL, respectively, as are defined in Eqs. S4.2 and S4.3. Green solid dots denote stable fixed points, and green unfilled circles denote unstable fixed points. The value of the SNAIL signal for A is taken from the stable fixed point in B. K Molecules, thousand molecules.
Theoretical Framework for Modeling miRNA-based Chimera Circuits.
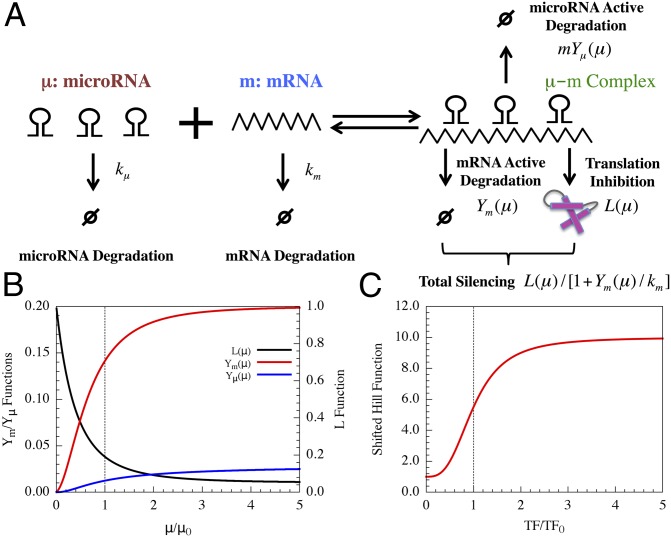
To incorporate the essential feature of miRNA-mediated regulation in the translation–transcription chimeric circuits, we followed and generalized the recent theoretical framework devised by Lu et al. (24). This framework starts with a detailed description of the TF–promoter binding/unbinding dynamics and adds the miRNA–mRNA binding/unbinding dynamics (Fig. 3). The generic deterministic equations for a two-component chimeric circuit with miR (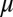 ) targeting the mRNA (m) of a transcribed TF protein B are given by,
) targeting the mRNA (m) of a transcribed TF protein B are given by,
where 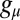 and
and 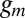 are the synthesis rates (possibly functions of
are the synthesis rates (possibly functions of 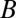 and of external signals) of
and of external signals) of 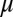 and
and 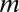 respectively,
respectively, 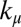 ,
, 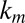 ,
, 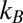 are the innate degradation rates of
are the innate degradation rates of 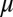 ,
, 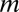 , and
, and 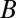 , respectively, and
, respectively, and 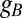 is the translation rate of protein
is the translation rate of protein 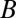 for each mRNA in the absence of miR. The derivation of the functional form of the μ-dependent miRNA–mRNA couplings Yµ, Ym, and L are presented in SI Appendix; in Fig. 3B, we show the typical form of these functions—Yµ is weakly increasing with
for each mRNA in the absence of miR. The derivation of the functional form of the μ-dependent miRNA–mRNA couplings Yµ, Ym, and L are presented in SI Appendix; in Fig. 3B, we show the typical form of these functions—Yµ is weakly increasing with 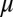 , Ym is strongly increasing, and L is strongly decreasing. The dependence of the functional form on the number of binding sites of
, Ym is strongly increasing, and L is strongly decreasing. The dependence of the functional form on the number of binding sites of 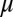 on
on 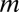 is described in SI Appendix. We note that this system of three-coupled deterministic equations, derived by a more biologically consistent approach, differs from the previously derived models studied in the context of miRNA–TF chimeric circuits (21, 28–31). Because, typically, miRs are more stable than mRNAs (32), in some cases, Eqs. 1 can be reduced to two coupled equations for
is described in SI Appendix. We note that this system of three-coupled deterministic equations, derived by a more biologically consistent approach, differs from the previously derived models studied in the context of miRNA–TF chimeric circuits (21, 28–31). Because, typically, miRs are more stable than mRNAs (32), in some cases, Eqs. 1 can be reduced to two coupled equations for 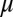 and
and 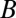 by assuming that
by assuming that 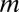 always reaches steady states (SI Appendix, Sections 3–6).
always reaches steady states (SI Appendix, Sections 3–6).
Fig. 3.
The microRNA-based circuit (MBC) modeling approach. (A) Schematic diagram for the MBC model. One or more microRNAs (µ) reversibly bind to mRNA (m) to form a µ–m complex. km and kµ are innate degradation rates of µ and m, respectively. µ can inhibit the translation of m [the translation rate is scaled by L(µ)] and degrade mRNA actively [at rate Ym(µ)]. The combined silencing effects can be characterized by P(µ). Also, µ can itself be degraded actively [at rate mYµ (µ) for µ microRNAs]. (B) The values of the L, Ym, Yµ as the functions of µ (scaled by the threshold µ0). The number of microRNA binding sites on the mRNA is taken to be six. A vertical dotted line is plotted to show the values at µ0. (C) The values of the shifted Hill function versus TF levels (scaled by the threshold TF0). Here, the Hill coefficient is three, and the fold change λ is 10.
Using this theoretical framework, the deterministic equations for the stand-alone driven (by a signal S representing SNAIL) miR-200/ZEB circuit, the stand-alone driven (by a signal I) miR-34/SNAIL circuit and the combined regulatory unit are given by equation sets S4.2, S4.3, and S4.4, respectively (SI Appendix, Section 3). All of the model parameters and the equations for each circuit module can be found in SI Appendix. In our study, we used the continuation method [PITCON7 (33) library] to calculate nullclines and plot bifurcations. See SI Appendix, Sections 4 and 6 for the model parameters.
Stand-Alone Driven Dynamics.
Dynamics of the miR-200/ZEB—a ternary switch.
We performed an analysis of this circuit when it is driven by a fixed input signal S mimicking the effect of SNAIL—the activation of ZEB and inhibition of miR-200. A typical phase-space diagram is shown in Fig. 2A. As can be seen, three stable states coexist—(high miR-200/low ZEB), (low miR-200/high ZEB), and (medium miR-200/medium ZEB), which we denote as (1,0), (0,1), and (1/2,1/2), respectively. As mentioned earlier, the states (1,0) and (0,1) correspond to epithelial (E) and mesenchymal (M) phenotypes, and we propose to associate the (1/2,1/2) state with an epithelial–mesenchymal (E/M) hybrid phenotype.
Dynamics of the miR-34/SNAIL—a noise-buffering integrator.
Next, we performed an analysis of this circuit when SNAIL is driven by an input signal I, mimicking the effect of the variety of input signals known to affect this gene (e.g., HIF-1and TGF-beta). A typical phase-space diagram is shown in Fig. 2B. As can be seen, only a single stable state exists, which corresponds to fixed levels So of SNAIL and μo of miR-34. As the signal I increases, So smoothly increases and μo smoothly decreases. Further investigations (SI Appendix, Section 7) revealed that SNAIL self-inhibition reduces the effect of external noise in incoming signals. This noise-buffering characteristic, combined with the relatively mild input dependence of the operating point (the stable state values of miR-34 and SNAIL), can prevent aberrant activation of EMT due to transient inputs and explain the stability of the epithelial phenotype (14).
The effects of inhibitory nonlinearity and the nature of the TF self-regulation.
The different operating characteristics of the modules are associated with the differences in the nonlinearity of their translation–transcription inhibition (high vs. low) and in their TF self-regulation (activation vs. inhibition). More specifically, the translation inhibition of ZEB by the miR-200 family (which includes miR-200a/b/c, miR-141, and miR-429) is highly nonlinear—at least six binding sites—and the transcription inhibition of miR-200 by ZEB has matched nonlinearity—a Hill function of rank 3. In contrast, the translation inhibition of SNAIL by the miR-34 occurs via only two binding sites, and the transcription inhibition of miR-34 by ZEB is also a weakly nonlinear Hill function of rank 1 (11–13). General study of the requirements for multistability in chimera switches is presented SI Appendix, Section 5. In SI Appendix, Section 8, we present study of the effects of the TF auto-regulations.
The Combined Dynamics of the Core Regulatory Unit.
Bifurcation diagram of the mir-200/ZEB circuit driven by SNAIL.
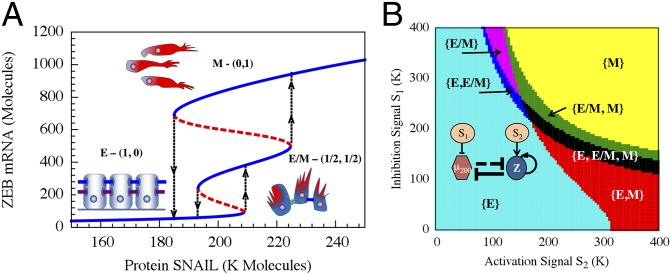
As a first step toward sorting out the combined system, we analyzed the behavior of the decision circuit when it is driven by variable levels of SNAIL. A typical bifurcation diagram illustrating the range of coexistence of and the transition between the three stable states is shown in Fig. 4A (additional bifurcation diagrams are included in SI Appendix, Section 9). Next, we calculated a phase diagram (a 2-dimensional bifurcation diagram) when the action of SNAIL is represented by two independent signals, S1, a transcription inhibitor of miR-200, and S2, a transcription activator of ZEB. We identified, as is shown in Fig. 4B, the existence of seven possible phases (the corresponding nullclines are included in SI Appendix, Section 10): (i) monostable phenotypes—{E}, {M}, and {E/M}; (ii) coexistence of two phenotypes—{E, M}, {E, E/M}, and {E/M, M}; and (iii) coexistence of all three phenotypes—{E, E/M, M}. The richness of these identified phases can help explain experiments regarding the diverse behaviors triggered by SNAIL (34–36) in inducing EMT. According to the rate of SNAIL increase and to the details of how it is increased (e.g., by TFG-β or hypoxia, which also affect the level of ZEB), the cells will follow a different trajectory in the phase diagram and thus will go through different phenotypic transitions. The largest region in the phase diagram corresponds to the epithelial phenotype, consistent with the view that epithelial is the default phenotype (14).
Fig. 4.
Bifurcation and phase-diagram of the driven miR-200/ZEB decision module. (A) Bifurcation of ZEB mRNA levels when driven by a signal S (SI Appendix, Eqs. S4.2) representing SNAIL. The bifurcation illustrates the possible coexistence (for some range of S) of three states: (i) the (1,0) state with high miR-200 and low ZEB, which corresponds to the epithelial (E) phenotype; (ii) the (0,1) state, which corresponds to the mesenchymal phenotype (M); (iii) the (1/2,1/2) state, which corresponds to the hybrid phenotype (E/M). Starting with the (1,0) state (E) and increasing SNAIL, the circuit undergoes a transition to the (1/2,1/2) state (E/M)—the first upward arrows on the right. Further increase in SNAIL leads to a transition from the (1/2,1/2) state to the (0,1) state (M)—the second upward arrows on the right. Starting from the (0,1) state and decreasing SNAIL yields a direct transition to the (1,0) state—the downward arrows further to the left. (B) The phase-diagram of the chimeric circuit when it is driven by two independent signals S1 and S2, as is illustrated in the Inset circuit. Each phase corresponds to a different combination of coexisting states. For example, in phase {E}, only state (1,0) is stable, and, in phase {E,E/M}, the states (1,0) and (1/2,1/2) can coexist.
The effect of feedback from miR-200/ZEB to miR-34/SNAIL.
The complete core regulatory unit includes feedback from the decision circuit to the integrator via inhibition of miR-34 by ZEB. As shown in Fig. 5A, the full unit retains the tristable nature of the decision circuit even when the ZEB–miR-34 feedback is included. In Fig. 5B, we show the bifurcation diagram in response to an external signal I, which acts as an activator of SNAIL. The bifurcation diagram is presented in terms of the level of ZEB mRNA. The corresponding bifurcation diagrams for miR-200, miR-34, SNAIL, and ZEB are shown in SI Appendix, Fig. S9. As is inferred from the bifurcation curve, the functional role of the inhibitory feedback is to break the symmetry between the forward (EMT) and backward (MET) epithelial–mesenchymal transitions: whereas the transition in the forward direction occurs through the hybrid phenotype, the backward transition proceeds directly from the mesenchymal phenotype to the epithelial one. This result can explain the fact that transitions into the hybrid phenotype have not been observed during MET, such as in reprogramming to iPSCs (9, 10).
Fig. 5.
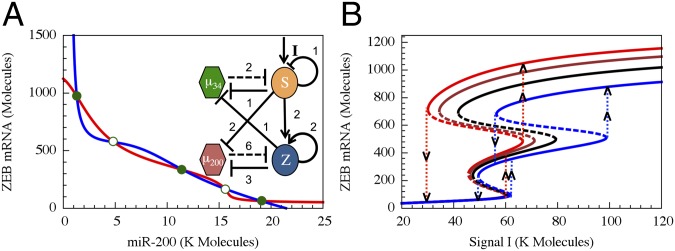
The phase-space and bifurcation diagram of the core regulatory unit. (A) The nullclines and the possible states in the phase-space corresponding to the combined regulatory unit, modeled by Eqs. S4.4 in SI Appendix. The blue nullcline is for the whole circuit when all ODEs are set to 0 except for mZ′ = 0 (red line), and the red nullcline is for the conditions when all ODEs are set to 0 except for µ200′ = 0. Green dots denote stable fixed points, and green unfilled circles denote unstable/saddle fixed points. (B) Bifurcation plots for the ZEB levels with respect to the driving signal I. Different lines show different cases of feedback from ZEB to miR-34 (regulation fold change is denoted by λZEB->miR34). Compared with the case without feedback (black, λZEB->miR34 = 1), strong repression (red, λZEB->miR34 = 0.2) or weaker repression (brown, λZEB->miR34 = 0.5) makes the bifurcation slightly shift to the left whereas activation (blue, λZEB->miR34 = 2.0) makes the bifurcation shift mainly to the right. The vertical dotted lines show the transitions among different states along the bifurcation curves.
Increasing the feedback strength toward stronger repression of miR-34 leads to stronger asymmetry, as the bifurcation curve shifts further to the left, as is illustrated in Fig. 5B. Transitions from the mesenchymal phenotype into the hybrid one can be obtained by replacing the inhibitory ZEB–miR-34 feedback by an activating feedback (Fig. 5B). Finally, in the model presented here, we assumed that ZEB is self-activating. We found that removing this auto-regulation or making it noncooperative (using a Hill function of rank 1) would lead to the absence of any hybrid phenotype, both for the case of the stand-alone miR-200/ZEB circuit and for the combined regulatory unit (when ZEB–miR-34 feedback is included). Thus, cooperative auto-regulation of ZEB is necessary for attaining the hybrid E/M phenotype. See SI Appendix, Section 11 for the discussion of the coupling between modules for different parameters.
Testable Predictions.
Our theoretical analysis of the regulatory circuitry is consistent with several experimental findings, as is discussed in more detail in SI Appendix, Section 2. Below, we discuss possible future experimental verification of the predictions, which emerged from this work.
Asymmetry in forward and backward phenotype transitions.
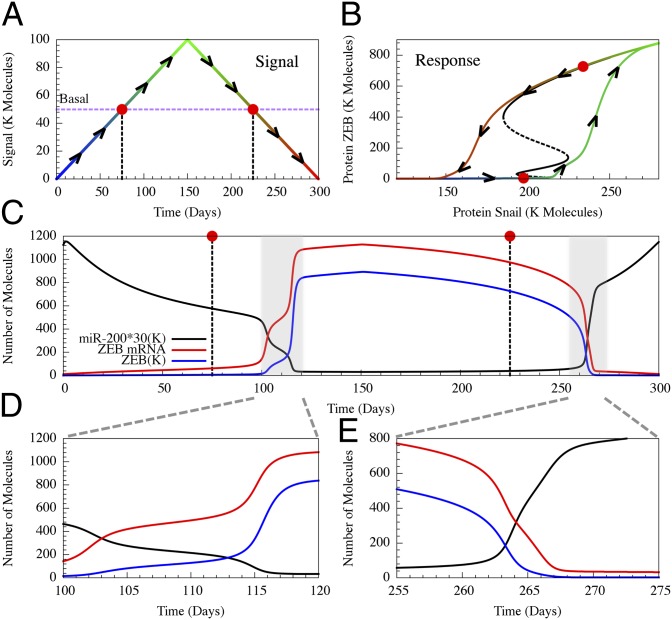
To clarify the predicted asymmetry and to ease comparison with future experiments, we present in Fig. 6 the predicted response of the core regulatory unit to a temporally varying external signal (Fig. 6A). The signal I serves as an activator of SNAIL and can represent the effect of a variety of inputs such as TGF-β and HIF-1. In Fig. 6B, we show the corresponding trajectory in the ZEB-SNAIL phase-space superimposed on the miR-200/ZEB bifurcation diagram (when this decision circuit is driven by SNAIL). We show in Fig. 6 C–E the predicted changes in the levels of miR-200, ZEB mRNA, and ZEB as the external signal is increased and then decreased. It is clear that, upon increasing the signal, the cell makes a transition from the epithelial phenotype to the hybrid phenotype and remains in this state (days 16–18 in Fig. 6D) before making a second transition into the mesenchymal phenotype when the signal is further increased. In contrast, direct transitions from the mesenchymal phenotype to the epithelial phenotype are observed (between days 104–114 in Fig. 6E) when the signal is decreased. A sudden increase of the signal to high level leads to a direct transition from epithelia into mesenchymal phenotype. However, it is possible to induce transitions into the hybrid phenotype by a sudden increase of the signal, but to lower levels. These predictions can be tested by comparing the dynamics of the epithelial–hybrid–mesenchymal transitions by using different temporal applications of the signal, for example, by exposing the cells to different levels of TGF-β and/or different durations of hypoxia. Such tests can reveal some insights into differential regulation of EMT during embryonic development vs. cancer metastasis. We note that high level of noise can also induce mesenchymal-to-hybrid transitions.
Fig. 6.
Temporal dynamics of the complete and partial epithelial–hybrid–mesenchymal transition. (A) We simulated the effect of time-varying external signal. The basal signal level is shown in a purple dotted line. The input signal starts from zero (blue) at day 0 and increases to 100 K molecules (green) at day 25. Then, the input signal linearly decreases until day 50 (red). (B) The dynamics of ZEB in response to the input signal is shown in the SNAIL-ZEB phase-plane. The colors for the signal epoch and the corresponding response are matched. Both A and B also show arrows to clarify the time evolution. A bifurcation plot for ZEB with respect to SNAIL (black) is superimposed. (C) Temporal evolution of miR-200 (navy), ZEB mRNA (red), and ZEB protein (blue, scaled by 0.33 to fit in the plot). In A–C, we labeled the states at the basal signal with red dots. The plot shows that cells at the basal signal level are epithelial during EMT but are still mesenchymal during MET. Thus, the two-way transitions are dynamically asymmetric. The areas marked in gray are expanded in D to show that cells going from epithelial to mesenchymal pass through a partial EMT state with intermediate levels of miR-200 and ZEB during days 16–18, (E) but cells directly return from mesenchymal to epithelial without going through any intermediate state.
There is another manifestation of the asymmetry between the forward and backward transitions. The basal level of the signal (purple dashed line in Fig. 6A and red dots in Fig. 6 A–C) lies below the threshold level of SNAIL transcription activation. Thus, the transition from the epithelial phenotype occurs when the signal rises above the basal level (Fig. 6B). After EMT is complete, ZEB levels remain high (Fig. 6C); the cells maintain the stable mesenchymal state, in agreement with observations (37). Starting from this phenotype, the signal has to be reduced significantly below the basal level to allow for transitions back to the epithelial phenotype. This asymmetry is consistent with experiments showing that lifting the activation of SNAIL (i.e., returning it to basal levels) is insufficient to trigger MET. SNAIL needs to be actively inhibited, e.g., through activation of miR-34, to initiate MET (38).
The existence and character of the hybrid phenotype.
According to the picture presented here, the experimentally observed epithelial–mesenchymal hybrid phenotype is associated with partial expression of both miR-200 and ZEB, which we termed the (1/2,1/2) state. Despite the supportive experimental findings, this core assumption/prediction has not yet been verified by direct expression level measurements. We propose that experiments should be carried out involving collective cell migration similar to those reported in ref. 39, in which the levels of both miR-200 and ZEB would be directly measured (instead of focusing exclusively on downstream effectors such as E-cadherin and vimentin). Furthermore, according to our analysis, the (1/2, 1/2) state, and thus the hybrid phenotype, can exist only if ZEB is self-activating. This prediction can be tested by adding an indirect (e.g., via another gene) yet strong self-inhibition of ZEB. We also predicted that a hypothetical positive feedback from ZEB to miR-34 could allow transitions from the mesenchymal phenotype into the hybrid one. This prediction can be tested by adding an indirect yet strong transcriptional activation of miR-34 when the level of ZEB is elevated. We also found that the existence of the (1/2,1/2) state requires that miR-200 silences ZEB translation by both translational repression and mRNA degradation, with the larger contribution from the repression (see SI Appendix, Fig. S7 for analysis on the role of the two different modes of microRNA silencing). This prediction could be tested by introducing mutations to miR-200 to reduce the degradation part of the silencing.
Discussion
The complete and partial epithelial–hybrid–mesenchymal transitions play crucial roles during embryonic development and during cancer metastasis (4). Considerable research effort has been devoted to untangling the genetic regulation underlying these transitions (40). The miR-200/ZEB double-negative feedback loop with input from the mir-34/SNAIL module is a central regulatory circuit of the transitions that, as already mentioned and detailed in SI Appendix, Section 1, couples to other key cellular properties (e.g., stemness, cell–cell communication, and motility) (15, 16). Although the recognized importance of the miR-200/ZEB decision circuit has led to intense experimental efforts, the circuit has been given limited theoretical attention.
Here, we generalized the recent theoretical framework devised by Lu et al. (24) to study the involvement of microRNAs in decision-making circuits. Compared with previous studies (21, 28–31), the approach taken here incorporates in detail the dynamics of miR-mediated silencing (both translational repression and the active mRNA/miR degradation) and includes the effect of the number of binding sites of miR on mRNA (known for the circuits studied here). This framework was used to unravel the modular design principles of the epithelial–hybrid–mesenchymal core regulatory network. More specifically, we investigated the function of the miR-200/ZEB circuit and the function of its paired miR-34/SNAIL mutual (inhibitory) feedback loop, as well as the function of the combined [miR-34/SNAIL]:[miR-200/ZEB] core regulatory unit. We found that the [miR-34/SNAIL] module is a monostable circuit that acts as a noise-buffering integrator and that the [miR-200/ZEB] module is a tristable circuit that acts as a ternary switch. The three metastable states are (high miR-200/low ZEB), (low miR-200/high ZEB), and (medium miR-200/medium ZEB), which we denote as (1,0), (0,1), and (1/2,1/2), respectively. The states (1,0) and (0,1) correspond to the epithelial (E) and mesenchymal (M) phenotypes. We propose to associate the (1/2,1/2) state with epithelial–mesenchymal (E/M) hybrid phenotype, corresponding to concurrent intermediate levels of expression of both ZEB and miR-200. By intermediate, we mean that the expression level lies between those cells belonging to the epithelial and mesenchymal phenotypes.
Our (1/2,1/2) state hypothesis is consistent with recent studies of gastrulation in Drosophila embryos that show that collectively migrating cells, which correspond to the hybrid state, coexpress ZEB1 and E-cadherin (39). The latter is inhibited by ZEB and SNAIL (Fig. 1) and therefore is expressed in epithelial cells (low levels of ZEB) but not in mesenchymal cells (high levels of ZEB). Its continued expression in hybrid cells is indicative that this phenotype corresponds to an intermediate level of ZEB. The hypothesis is also consistent with the role of miR-200/ZEB in cell–cell communication via the Jag1/Notch/Delta system (Fig. 1 and SI Appendix, Fig. S1). There is evidence of Jagged1-Notch interaction between collectively migrating cells (41). Because miR-200 strongly inhibits Jagged1 (Jag1 mRNA has five binding sites for miR-200) (16), expression of Jagged1 is possible only for either low or intermediate levels of miR-200 and not for the high levels characteristic of epithelial cells.
As mentioned in the Introduction, the (1/2,1/2) state hypothesis presented here differs from the hypothesis suggested by et al. (28) based on studies of the [miR-34/SNAIL]:[miR-200/ZEB] when coupled to TGF-β. In that work, the self-regulations of SNAIL and ZEB were not included, the two double-negative feedback loops were assumed to be identical, and the transcription and translation inhibitions process were not distinguished (they were both modeled by inhibitory Hill functions of rank 2). Under these assumptions, the two modules have similar bistable dynamics and act as binary switches. Analyses of the coupled system revealed the existence of a mixed (high miR-200, low miR-34) state, which the authors associated with the hybrid phenotype. The experimental findings mentioned above and in Results are more consistent with our (medium miR-200, medium ZEB) hypothesis. However, future direct measurements of the expression level of all four components of the regulatory system in the epithelial, mesenchymal, and hybrid phenotypes are needed to fully resolve the issue. It might of course turn out that different cell lines exhibit different hybrid phenotypes.
In SI Appendix, Section 1, we illustrate the participation of the miR-200/ZEB circuit in other key cellular processes. Future theoretical studies of these circuits can provide valuable new insights about how circuits determining other cellular characteristics are coupled with the epithelial–hybrid–mesenchymal transition. In general, we expect that cell–cell interactions between neighboring cells can lead to costabilization of the hybrid phenotype. The hypothesis can be theoretically investigated by dynamical system studies of a coupled circuit model (SI Appendix).
To conclude, better understanding of the transitions into and from the hybrid phenotype holds promise for a better comprehension of cancer progression. It can also help to address long-standing fundamental questions linked to compelling clinical needs—how to distinguish between the aberrant dynamics of epithelial–hybrid–mesenchymal transitions during tumorigenesis vs. the normal programs of embryonic development and tissue regeneration. Of practical importance is the requirement that pregnancy and normal wound healing must continue during antimetastatic therapies, and wound healing augmentation must not promote metastasis if a malignancy is present (42).
Supplementary Material
Acknowledgments
We benefited from useful discussions with I. Tsarfaty, R. Brown, and M. C. Farach-Carson. This work was supported by National Science Foundation (NSF) Center for Theoretical Biological Physics Grant NSF PHY-1308264, by the Cancer Prevention and Research Institute of Texas (CPRIT) Scholar Program of the State of Texas at Rice University, and by the Tauber Family Funds at Tel-Aviv University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318192110/-/DCSupplemental.
References
- 1.Balázsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144(6):910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 4.Nakaya Y, Sheng G. EMT in developmental morphogenesis. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 5.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15(2):117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21(Suppl 7):vii89–vii92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revenu C, Gilmour D. EMT 2.0: Shaping epithelia through collective migration. Curr Opin Genet Dev. 2009;19(4):338–342. doi: 10.1016/j.gde.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Li R, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 10.Samavarchi-Tehrani P, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Bracken CP, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68(19):7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 12.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siemens H, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10(24):4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 14.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 15.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop—a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11(9):670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brabletz S, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30(4):770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrecut M, Halley JD, Winkler DA, Huang S. A general model for binary cell fate decision gene circuits with degeneracy: indeterminacy and switch behavior in the absence of cooperativity. PLoS ONE. 2011;6(5):e19358. doi: 10.1371/journal.pone.0019358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guantes R, Poyatos JF. Multistable decision switches for flexible control of epigenetic differentiation. PLOS Comput Biol. 2008;4(11):e1000235. doi: 10.1371/journal.pcbi.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macía J, Widder S, Solé R. Why are cellular switches Boolean? General conditions for multistable genetic circuits. J Theor Biol. 2009;261(1):126–135. doi: 10.1016/j.jtbi.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Zhang K, Xu L, Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. Proc Natl Acad Sci USA. 2011;108(20):8257–8262. doi: 10.1073/pnas.1017017108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P, Cai S, Liu Z, Wang R. Mechanisms generating bistability and oscillations in microRNA-mediated motifs. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;85(4 Pt 1):041916. doi: 10.1103/PhysRevE.85.041916. [DOI] [PubMed] [Google Scholar]
- 22.Schultz D, Onuchic JN, Wolynes PG. Understanding stochastic simulations of the smallest genetic networks. J Chem Phys. 2007;126(24):245102. doi: 10.1063/1.2741544. [DOI] [PubMed] [Google Scholar]
- 23.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 24.Lu M, et al. Tristability in cancer-associated microRNA-TF chimera toggle switch. J Phys Chem B. 2013 doi: 10.1021/jp403156m. [DOI] [PubMed] [Google Scholar]
- 25.Schultz D, Ben Jacob E, Onuchic JN, Wolynes PG. Molecular level stochastic model for competence cycles in Bacillus subtilis. Proc Natl Acad Sci USA. 2007;104(45):17582–17587. doi: 10.1073/pnas.0707965104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN. Deciding fate in adverse times: Sporulation and competence in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106(50):21027–21034. doi: 10.1073/pnas.0912185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz D, Lu M, Stavropoulos T, Onuchic JN, Ben-Jacob E. Turning oscillations into opportunities: Lessons from a bacterial decision gate. Sci Rep. 2013;3:1668. doi: 10.1038/srep01668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian XJ, Zhang H, Xing J. Coupled reversible and irreversible bistable switches underlying TGFβ-induced epithelial to mesenchymal transition. Biophys J. 2013;105(4):1079–1089. doi: 10.1016/j.bpj.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: Consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci USA. 2008;105(50):19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine E, Ben Jacob E, Levine H. Target-specific and global effectors in gene regulation by MicroRNA. Biophys J. 2007;93(11):L52–L54. doi: 10.1529/biophysj.107.118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai X, Wolkenhauer O, Vera J. Modeling miRNA regulation in cancer signaling systems: miR-34a regulation of the p53/Sirt1 signaling module. Methods Mol Biol. 2012;880:87–108. doi: 10.1007/978-1-61779-833-7_6. [DOI] [PubMed] [Google Scholar]
- 32.Khanin R, Vinciotti V. Computational modeling of post-transcriptional gene regulation by microRNAs. J Comput Biol. 2008;15(3):305–316. doi: 10.1089/cmb.2007.0184. [DOI] [PubMed] [Google Scholar]
- 33.Denheijer C, Rheinboldt WC. On steplength algorithms for a class of continuation methods. SIAM J Numer Anal. 1981;18(5):925–948. [Google Scholar]
- 34.Acloque H, Thiery JP, Nieto MA. The physiology and pathology of the EMT: Meeting on the epithelial-mesenchymal transition. EMBO Rep. 2008;9(4):322–326. doi: 10.1038/embor.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundgren K, Nordenskjöld B, Landberg G. Hypoxia, Snail and incomplete epithelial-mesenchymal transition in breast cancer. Br J Cancer. 2009;101(10):1769–1781. doi: 10.1038/sj.bjc.6605369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanco MJ, et al. Snail1a and Snail1b cooperate in the anterior migration of the axial mesendoderm in the zebrafish embryo. Development. 2007;134(22):4073–4081. doi: 10.1242/dev.006858. [DOI] [PubMed] [Google Scholar]
- 37.Gregory PA, et al. An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Mol Biol Cell. 2011;22(10):1686–1698. doi: 10.1091/mbc.E11-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim NH, et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195(3):417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vannier C, Mock K, Brabletz T, Driever W. Zeb1 regulates E-cadherin and Epcam (epithelial cell adhesion molecule) expression to control cell behavior in early zebrafish development. J Biol Chem. 2013;288(26):18643–18659. doi: 10.1074/jbc.M113.467787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 41.Chigurupati S, et al. Involvement of notch signaling in wound healing. PLoS ONE. 2007;2(11):e1167. doi: 10.1371/journal.pone.0001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leopold PL, Vincent J, Wang H. A comparison of epithelial-to-mesenchymal transition and re-epithelialization. Semin Cancer Biol. 2012;22(5-6):471–483. doi: 10.1016/j.semcancer.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.