Significance
The genetic disease congenital erythropoietic porphyria (CEP) results from the accumulation of toxic porphyrins owing to an enzymatic uroporphyrinogen III synthase (UROS) defect. We have already shown that the UROSC73R and UROSP248Q mutants, usually associated with a severe phenotype, conserve partial intrinsic enzymatic activity but suffer from kinetic instability and premature degradation by the proteasome pathway. We now demonstrate that treatment with a clinically approved proteasome inhibitor could rescue UROS mutant expression in human erythroid cells and reverse the skin photosensitivity in homozygous UrosP248Q CEP mice. Based on our understanding of the biochemical mechanism of UROS enzymatic deficiency, these innovative results open the way for a pharmacologic treatment of CEP disease as an alternative to bone marrow transplantation.
Keywords: protein misfolding, Günther's disease, enzyme instability, heme biosynthetic pathway, pharmacological therapy
Abstract
Congenital erythropoietic porphyria (CEP) is a rare autosomal recessive disorder characterized by uroporphyrinogen III synthase (UROS) deficiency resulting in massive porphyrin accumulation in blood cells, which is responsible for hemolytic anemia and skin photosensitivity. Among the missense mutations actually described up to now in CEP patients, the C73R and the P248Q mutations lead to a profound UROS deficiency and are usually associated with a severe clinical phenotype. We previously demonstrated that the UROSC73R mutant protein conserves intrinsic enzymatic activity but triggers premature degradation in cellular systems that could be prevented by proteasome inhibitors. We show evidence that the reduced kinetic stability of the UROSP248Q mutant is also responsible for increased protein turnover in human erythroid cells. Through the analysis of EGFP-tagged versions of UROS enzyme, we demonstrate that both UROSC73R and UROSP248Q are equally destabilized in mammalian cells and targeted to the proteasomal pathway for degradation. We show that a treatment with proteasomal inhibitors, but not with lysosomal inhibitors, could rescue the expression of both EGFP-UROS mutants. Finally, in CEP mice (UrosP248Q/P248Q) treated with bortezomib (Velcade), a clinically approved proteasome inhibitor, we observed reduced porphyrin accumulation in circulating RBCs and urine, as well as reversion of skin photosensitivity on bortezomib treatment. These results of medical importance pave the way for pharmacologic treatment of CEP disease by preventing certain enzymatically active UROS mutants from early degradation by using proteasome inhibitors or chemical chaperones.
Congenital erythropoietic porphyria (CEP; MIM 263700), or Günther’s disease, is a rare autosomal recessive disease resulting from markedly deficient uroporphyrinogen III synthase (UROS; EC 4.2.1.75) enzymatic activity. UROS catalyzes the cyclization of the linear tetrapyrrole hydroxymethylbilane into uroporphyrinogen III. UROS deficiency results in a specific and massive overproduction of nonphysiologic and pathogenic type I isomers of uroporphyrin (URO I) and coproporphyrin (COPRO I) in bone marrow and erythroid cells. URO I is a highly photocatalytic molecule that cannot be converted to heme and accumulates in erythrocytes, inducing hemolysis (1). The released type I porphyrins disseminate throughout the body, especially in the skin, where they are responsible for skin lesions after sunlight exposure. The clinical severity of the anemia and cutaneous lesions varies widely, and prognosis is poor for some patients, with death occurring in early life or the neonatal period (2). Classical treatments are only symptomatic and unsatisfactory (1, 2).
Because the predominant site of metabolic expression of the disease is the erythropoietic system, bone marrow transplantation (BMT) offers a curative treatment for patients with severe phenotypes, as long as an HLA-compatible donor is available (3). As an alternative strategy, we have successfully used autologous hematopoietic stem cell (HSC) gene therapy with integrative lentivectors in a murine model of CEP (4). Serious oncogenic and abnormal proliferation events have been observed in several gene therapy clinical trials, however (5). Investigations of pharmacologic therapeutic approaches are underway for many genetic diseases, based on an understanding of the protein dysfunction mechanisms.
Molecular study of the UROS gene in CEP patients has highlighted a variety of mutations spreading all along the 10 coding exons, including missense or nonsense mutations, splicing defects, and large deletions/insertions, as well as mutations in the erythroid-specific promoter region (6, 7). In disease related to protein dysfunction, it is diagnostically and therapeutically essential to understand the multiple mechanisms that explain the pathogenicity of specific mutants. The degree of UROS activity impairment and the concomitant accumulation of photoreactive URO I and derived porphyrins may be correlated with the severity of the CEP disease (8, 9).
Given the abnormal UROS enzymatic activity levels in CEP patients, we considered mechanisms of protein alterations owing to inherited mutations. At the protein level, some of these defects may alter the catalytic machinery of the enzyme, whereas other mutations can undermine the stability of the folded conformation. In a recent study, prokaryotic expression showed that most of the missense UROS mutants described in CEP patients presented reduced protein yield recovery compared with WT, likely because of altered protein stability (10). Functional analyses indicated that the altered kinetic parameters were related mainly to a drop in expression and purification yield rather than to alterations of the intrinsic specific enzymatic activity of the mutants.
Whenever gene mutations are responsible for misfolding or unfolding proteins, the reduced thermodynamic stability and accumulation lead to a reticulum endoplasmic (RE) stress. The unfolded protein response (UPR) attenuates translation and enhances proteasomal degradation of abnormal proteins (11). These mechanisms suggest that recovery of protein stability in vivo should be considered as possible therapy owing to a beneficial increase in intracellular enzymatic activity. The two missense C73R and P248Q mutations are retrieved with a higher frequency in CEP patients (6). The C73R mutation is found in ∼30% of the disease alleles retrieved in CEP patients of Caucasian origin. Structural studies and modeled complexes show that Cys-73 is far from the enzyme–substrate interaction site, suggesting that it does not play a critical role in catalysis (12).
Consistent with this idea, we recently demonstrated that purified recombinant human UROSC73R retains partial catalytic activity (30% of that of WT), but suffers from rapid and irreversible unfolding aggregation (10, 13, 14). In a cellular reporter assay, the misfolded UROSC73R is targeted to the proteasomal degradation pathway and becomes undetectable in human cells (13). Interestingly, treatment with a proteasome inhibitor (MG132) restored UROSC73R protein expression.
In the present work, we have extended these promising results to UROSP248Q in vitro and in vivo using a knock-in CEP mouse model (UrosP248Q/P248Q mice; hereinafter CEP mice). Using EGFP-tagged versions of UROS, we showed that both UROSC73R and UROSP248Q are unstable proteins that become rapidly degraded in erythroid K562 cells. Interestingly, UROSC73R and UROSP248Q expression were rescued after treatment with proteasome inhibitors, but not with lysosome inhibitors, in transfected human erythroleukemic cells. Furthermore, CEP mice were treated with the clinically qualified drug bortezomib (PS-341; Velcade) to evaluate the therapeutic potential of proteasomal inhibition in vivo. After a 9-wk period of repeated bortezomib injection, we observed a significant decrease in uroporphyrin accumulation in circulating RBCs and urine, accompanied by the disappearance of skin photosensitivity in CEP mice. We demonstrate that a pharmacologic approach could be successfully used to modulate the expression of the CEP disease phenotype in vivo. These results pave the way for the development of pharmacologic agents (e.g., chemical chaperones) that would stabilize certain UROS mutants to improve the course of the disease, and especially the devastating skin lesions affecting patients with CEP.
Results
P248Q Mutation Induces UROS Destabilization in Vitro.
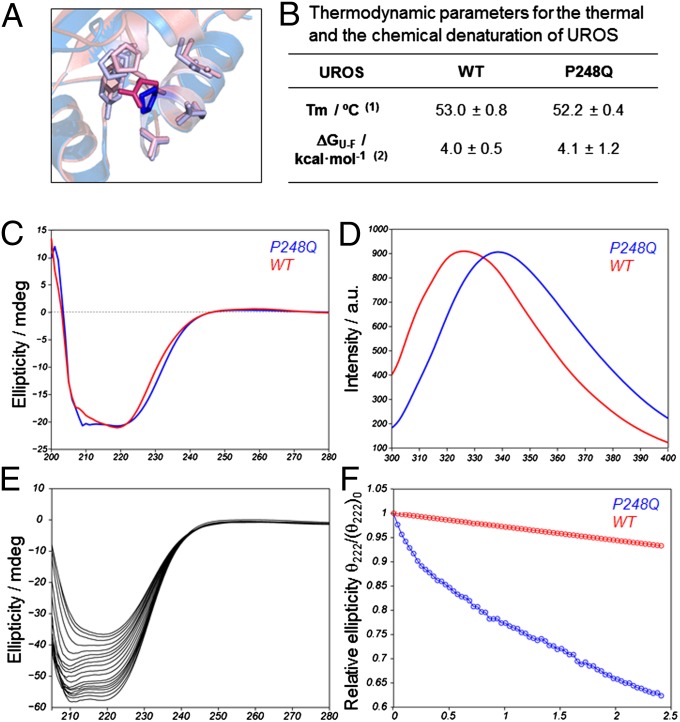
We performed in vitro characterization of the structure and relative stability of UROSP248Q. We used homology modeling to simulate the structure of the folded conformation of UROSP248Q, which disclosed a slightly different orientation of the side chain within the hydrophobic pocket at the mutation site (Fig. 1A). The circular dichroicism spectrum of UROSP248Q was very similar to that of WT UROS, indicating that the secondary structure content is largely preserved on mutation (Fig. 1C); however, small changes in the tertiary structure may occur on mutation, as indicated by a shift in the tryptophan fluorescence spectrum toward a more pronounced solvent-exposed environment in UROSP248Q (Fig. 1D), a trait also observed in deleterious mutants, such as UROSC73R (13).
Fig. 1.
Reduced kinetic stability of UROSP248Q in vitro. (A) Overlay of the WT UROS X-ray structure (1jr2, blue) and the modeled structure for UROSP248Q (pink). The highlighted side chain corresponds to the mutation site. This side chain forms contacts with other nearby residues, also highlighted: P18, Y19, E22, P246, T247, Q249, L251, and A252. (B) Thermodynamic parameters for the thermal and the chemical denaturation of UROSP248Q. (1) Melting temperature (Tm) monitoring the secondary structure change by circular dichroism at 1 °C/min. (2) Unfolding free energy in the absence of denaturant. (C and D) Far utraviolet-circular dichroicism spectrum (C) and natural abundance tryptophan emission spectrum (D) for the two proteins under consideration. (E) Evolution of the folded conformation over time at physiological temperature. The spectral change reflects the loss of α-helical (signal at 222 nm) toward β-sheet (signal at 210 nm) conformation over time for UROSP248Q. (F) Signal decay over time (at 222 nm) for UROSWT (red) and UROSP248Q (blue).
Thermodynamic parameters obtained from thermal unfolding or the use of chemical denaturants (Fig. 1B) were comparable to WT values, reinforcing the idea that both the WT and mutant enzymes share a similar fold. Analysis of the time dependence of the folded conformation revealed that the lifetime of the bioactive conformation is restricted by kinetic stability (Fig. 1E), with UROSP248Q unfolding approximately threefold faster than WT UROS (Fig. 1F). This decreased kinetic stability observed in vitro could be related to a loss of protein homeostasis in vivo.
C73R and P248Q UROS Mutations Lead to Enhanced Protein Degradation That Can Be Rescued by Proteasome Inhibitors, but Not by Lysosome Inhibitors, in Human Erythroid Cells.
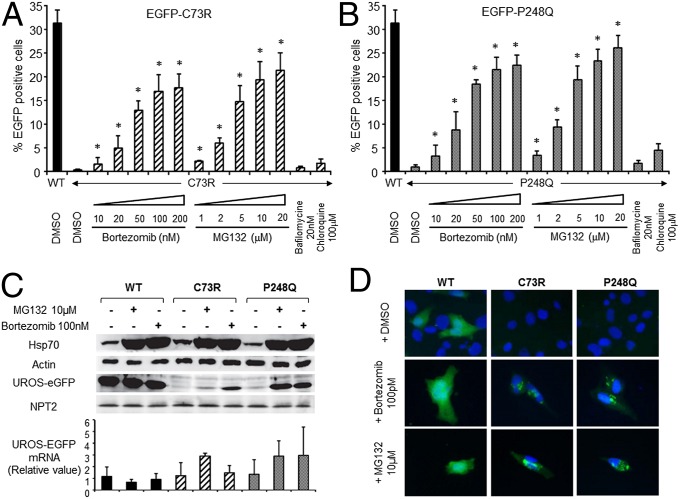
In a previous study, we demonstrated that the C73R mutation is associated with enhanced UROS degradation in mammalian cells, accounting for the almost undetectable UROS activity observed in CEP patients harboring the UROSC73R/C73R genotype (13). To compare the consequences of C73R and P248Q mutations on intracellular UROS stability, we stably introduced C-terminal EGFP-tagged versions of WT, C73R, and P248Q mutants (hereinafter EGFP-WT, EGFP-C73R, and EGFP-P248Q, respectively) into human erythroleukemic K562 cells. The expression of EGFP-tagged UROS versions was analyzed by flow cytometry and Western blot analysis after neomycin selection (Fig. 2 A and B). Neomycin phosphotransferase 2 (NPT2) expression was equally detected in stably transfected cell lines by Western blot analysis, indicating a similar integrated plasmid copy number in all conditions (Fig. 2C). A significantly higher EGFP expression level was detected by flow cytometry analysis in EGFP-WT compared with EGFP-C73R and EGFP-P248 (31 ± 3%, 0.4 ± 0.2%, and 1 ± 0.5% EGFP+ cells, respectively) (Fig. 2 A and B). EGFP-UROS mRNA levels were not statistically different among these conditions, excluding a transcriptional activation process (Fig. 2C, Lower). In agreement with the results obtained from thermodynamic and structural analyses, these data suggest that UROSP248Q and UROSC73R are unstable proteins with shortened intracellular half-lives (13) (Fig. 1).
Fig. 2.
UROSP248Q and UROSC73R trigger premature degradation by the proteasome in human erythroid K562 cells. (A and B) Human erythroleukemic K562 cells were stably transfected with plasmids expressing EGFP fused to the C terminus of WT, C73R (A), or P248Q (B) UROS cDNA. Stably transfected cells were treated with DMSO or the indicated concentration of lysosome (bafilomycine or chloroquine) or proteasome (MG132 or bortezomib) inhibitors for 16 h. EGFP expression was monitored by flow cytometry analysis. Results are expressed as the mean of three independent experiments; error bars represent SD. *Significant difference (P < 0.001) vs. EGFP-UROS mutant treated with DMSO. (C) Western blot (Upper) and RT-PCR (Lower) analyses of stably transfected K562 clones expressing UROS-EGFP. UROS-EGFP, NPT2, HSP70, and actin protein levels from stably transfected K562 cell lysates were analyzed by Western blot analysis using specific antibodies. UROS-EGFP mRNAs were analyzed by quantitative real-time RT-PCR. Results are expressed as mean of three independent experiments; error bars represent SD. (D) Subcellular localization of EGFP-UROS proteins was analyzed by epifluorescence microscopy in transiently transfected human fibroblasts exposed to proteasome inhibition (100 nM bortezomib or 10 μM MG132 for 16 h). DMSO 0.1% served as a control. Merged EGFP fluorescence (green) and DAPI nuclear staining (blue) of representative cells is shown. (Original magnification, 40×.)
We next investigated the contributions of several protein degradation pathways to intracellular UROS mutant behavior. K562 cells stably expressing EGFP-UROS were incubated with proteasome (i.e., MG132 and bortezomib) or lysosome (i.e., bafilomycin and chloroquine) inhibitors (Fig. 2 A and B). Whereas inhibitors of the endosomal-lysosomal pathways did not significantly increase EGFP-UROS expression, proteasome inhibitors dramatically increased EGFP-C73R and EGFP-P248Q expression in a dose-dependent manner (up to a 53-fold increase for EGFP-C73R and a 30-fold increase for EGFP-P248Q compared with DMSO) (Fig. 2 A and B). The increased HSP70 expression confirms the efficient proteasome inhibition on treatment with MG132 and bortezomib (Fig. 2C). In EGFP-WT, a minor (1.5-fold) increase was observed after proteasome inhibition. Western blot and quantitative RT-PCR analyses of EGFP-UROS expression in stably transfected K562 clones confirmed UROS protein stabilization as the main mechanism (Fig. 2C). Using N-terminal EGFP-tagged versions of WT, C73R, and P248Q-UROS produced similar results (Fig. S1).
Because proteasome inhibitors can lead to abnormal protein aggregation in cells, we analyzed the intracellular distribution of EGFP-tagged UROS in human fibroblasts by epifluorescence microscopy (Fig. 2D). As expected, EGFP-UROS mutant expression was rescued after treatment with 100 nM bortezomib or 10 μM MG132. Although a homogenous cytosolic staining was observed for EGFP-WT protein, EGFP-C73R and EGFP-P248Q appeared to be aggregated in fibroblasts (Fig. 2D). Taken together, these results suggest that proteasome inhibitors can partially restore UROS expression by protecting UROSP248Q and UROSC73R from premature degradation.
Bortezomib Significantly Reduces Porphyrin Accumulation in Vivo in UROS-Deficient (UrosP248Q/P248Q) Mice.
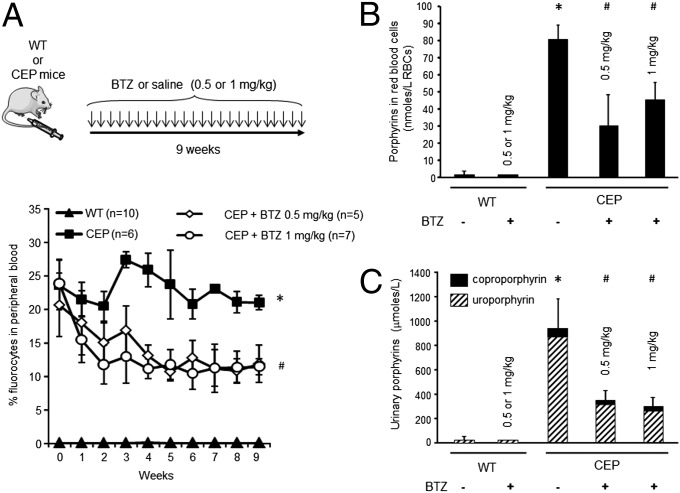
We next investigated whether bortezomib delivery can affect porphyrin accumulation and rescue the skin phenotype in CEP mice in vivo (15). We began by performing a dose–response analysis of proteasome inhibition after bortezomib injection into WT congenic BalbC/J mice. Previous studies from oncology-related research work have highlighted a narrow therapeutic window in vivo (0.7–1.3 mg/kg) owing to bortezomib-induced neuropathy in mouse models (16). After a single bortezomib injection (0.2–2 mg/kg i.p.), we observed a dose-dependent proteasome inhibition in RBCs, reaching a maximum of 70% (Fig. S2) as measured by the Suc-Leu-Leu-Val-Tyr-amido-4-methylcoumarin (Suc-LLVY-AMC) cleavage assay. A kinetic analysis of proteasome inhibition revealed that the 26S proteasome activity recovered rapidly after administration and returned to normal values in 48 h (Fig. S2). Based on these results, and considering the potential neurotoxicity of chronic bortezomib administration, CEP and normal mice were given 0.5 or 1 mg/kg every 48 h for 9 consecutive wk (Fig. 3A). Body weight was measured periodically to detect any eventual side effects. We found no differences between the two groups, suggesting an absence of acute toxic adverse effects of long-term bortezomib administration (Fig. S3).
Fig. 3.
Reduced porphyrin accumulation in CEP mice treated with bortezomib. (A) WT and CEP mice were treated by i.p. injection of bortezomib (BTZ) (0.5 or 1 mg/kg) or saline over a 9-wk period. Porphyrin-accumulating RBCs (i.e., fluorocytes) were analyzed by flow cytometry in WT and CEP mice over time. Results are expressed as mean of individual mice; error bars represent SDs. *Significant difference (P < 0.01) vs. control CEP mice. (B and C) Porphyrin accumulation was quantified in peripheral blood (B) and urine (C) of WT and CEP mice treated with 0.5 or 1 mg/kg bortezomib. Results are expressed as mean of individual mice; error bars represent SDs. *Significant difference (P < 0.0001) vs. WT mice; #Significant difference (P < 0.001) vs. control CEP mice.
Uroporphyrin I accumulation is a hallmark of the disease and leads to the spontaneous fluorescence of RBCs (i.e., fluorocytes), which can be monitored by flow cytometry analysis (15). We observed a progressive decrease in porphyrin accumulation in peripheral RBCs after serial bortezomib injections, as demonstrated by the decreased fluorocyte count over time (Fig. 3A). After 9 wk of treatment, porphyrin accumulation in RBCs was reduced by 63% in CEP mice treated with 0.5 mg/kg and by 43% in those treated with 1 mg/kg compared with untreated mice (Fig. 3B).
Uroporphyrin I is an hydrophobic compound that is massively eliminated in urine. Urinary porphyrins were reduced by 64% and 70% in CEP mice treated with 0.5 and 1 mg/kg bortezomib, respectively, compared with controls (Fig. 3C). Interestingly, bortezomib administration did not disturb the heme biosynthesis pathway, as indicated by the normal porphyrin values in treated WT mice (Fig. 3C). These results demonstrate that proteasome inhibition in vivo, albeit partial, is able to significantly decrease porphyrin accumulation and excretion in CEP mice.
Bortezomib Does Not Cure Hemolytic Anemia, but Does Prevent Skin Photosensitivity in UROS-Deficient Mice.
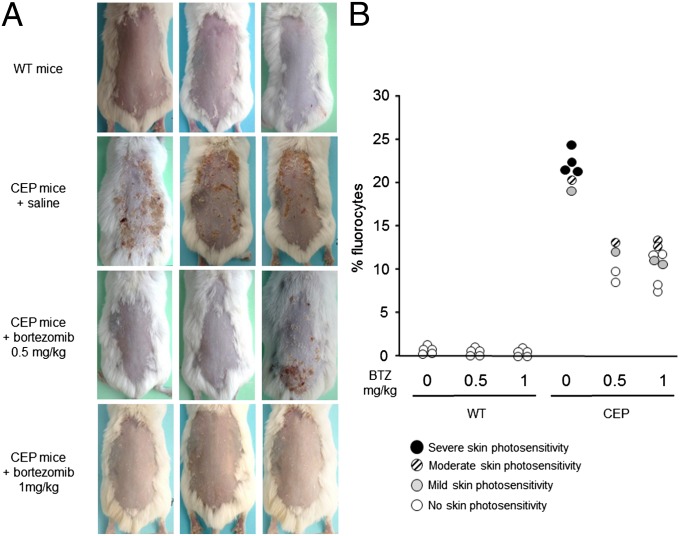
Based on our finding of massive uroporphyrin I accumulation leading to hemolytic anemia and skin photosensitivity, we investigated the consequences of bortezomib administration on these parameters in CEP mice. A skin photosensitivity assay based on UVA light exposure (8 J/cm2) revealed macroscopic skin lesions in the CEP mice, but no changes in the WT and most of the CEP mice treated with bortezomib (Fig. 4A). These results are correlated with porphyrin accumulation in RBCs; massive accumulation was present in untreated mice, which was reduced by fourfold, accompanied by major improvements in skin photosensitivity, after bortezomib administration. Interestingly, some CEP mice treated with proteasome inhibitors were completely free of macroscopic skin lesions (Fig. 4B).
Fig. 4.
Skin photosensitivity reversion in CEP mice treated with bortezomib. (A) Mice were depilated and exposed to UVA irradiation (8 J/cm2). Representative macroscopic pictures of dorsal skin at 5 d after UVA irradiation are shown. (B) Porphyrin-accumulating RBCs (fluorocytes) were analyzed by flow cytometry in WT and CEP mice treated with 0.5 or 1 mg/kg bortezomib (BTZ). Each individual mouse is represented by a circle according to the macroscopic severity of skin photosensitivity: clear-open to dark-filled circles indicate the intensity of cutaneous lesions.
We also investigated the effects of long-term administration of proteasome inhibitors on hematologic parameters. CEP mice demonstrated a typical hemolytic anemia characterized by decreased hemoglobin, hematocrit, and RBC count and significant spleen enlargement (Table S1) (15). The administration of bortezomib during a 9-wk period did not induce hematotoxicity, as indicated by blood cell counts. Unfortunately, however, bortezomib injection failed to improve anemia; RBC, hemoglobin, and hematocrit values remained unchanged in treated CEP mice. However, these mice demonstrated a moderate decrease in spleen size, which is a good indicator of improved stress erythropoiesis related to porphyrin accumulation. These results could indicate poor targeting of erythroid cells by proteasome inhibitors, resulting in moderate metabolic correction, with sustained erythropoiesis triggered by the hemolytic process.
Discussion
CEP is a genetic disease characterized by UROS deficiency leading to impaired heme biosynthesis and accumulation of toxic porphyrins in the erythroid lineage. To date, more than 40 disease-causing mutants have been reported in CEP, most of which are missense variants (6). Missense substitutions result in proteins that are unable to fold efficiently into their native conformation, that is, the most energetically favorable state. This may lead to either accumulation of toxic aggregates or increased degradation and loss of metabolic or cellular functions (17, 18).
UROS is a thermolabile enzyme that suffers from irreversible denaturation over time (14). Biophysical analyses of UROS mutants have clearly shown that thermodynamic destabilization is the main mechanism by which the enzyme losses activity on amino acid substitutions (10). In the present study, we have shown that UROSP248Q and UROSC73R are similarly destabilized and trigger enhanced proteasome degradation that may be rescued with proteasome inhibitors in human erythoid cells. Owing to the aggregation pattern of UROS mutants in cells, it was not possible to precisely quantify the extent of UROSP248Q enzymatic recovery after treatment with proteasome inhibitors; however, in prokaryotic expression experiments followed by enzyme purification, we found a 29% residual specific enzymatic activity for the UROSP248Q in vitro (10). Furthermore, UROSP248Q and UROSC73R expression could not be rescued after exposure to lysosome inhibitors; these results may explain why such molecules failed to improve CEP patients (19).
We previously generated a knock-in CEP mouse model by introducing the P248Q mutation to embryonic stem cells using homologous recombination, and used this animal model to demonstrate the feasibility of gene therapy in CEP (4, 15). As a proof of concept, we then evaluated the therapeutic potential of proteasomal inhibition in CEP mice chronically treated with bortezomib (Velcade; PS-341). Bortezomib belongs to the family of reversible peptide boronates that inhibit proteasome chymotrypsin-like activity. Velcade is already approved by the Food and Drug Administration and the European Medicines Agency for the treatment of multiple myeloma and mantle cell lymphoma. The side effects have been fully explored and managed in clinical trials. Preliminary experiments were performed in WT mice to determine the optimal dose and timing of bortezomib injection to reach optimal proteasome inhibition in vivo in the erythroid lineage. A transient proteasome inhibition in peripheral blood cells was obtained after a single administration in vivo. Inhibition was maximal during the first 2 h after injection, before proteasome activity returned to basal levels within 48 h.
Because the bone marrow erythroid lineage and mature RBCs express high levels of cytoplasmic UROS protein, preventing long-term UROS degradation in vivo using proteasome inhibitors posed a real challenge. To mimic the regimen used in clinical trials, we used chronic bortezomib administeration in the CEP mice. Considering the relatively long life of RBCs in vivo (60 d), bortezomib was administered over a 9-wk period. To our knowledge, this is the longest period of uninterrupted administration for a therapeutic purpose in a genetic disease in mice yet reported.
Proteasome inhibition has been used successfully to correct protein misfolding in several genetic diseases (20–25). Several structural abnormalities of the sarcolemma responsible for muscle dystrophy could be salvaged in primary cultures or in mouse models by treatment with proteasomal inhibitors (26, 27). Gazzero et al. injected bortezomib every 72 h for 2 consecutive wk in Duchene Dystrophy model mice, and was able to restore the membrane expression of dystrophin as well as dystrophin glycoprotein complex members, and to improve the phenotype (26). In a spinal muscular atrophy mouse model, a daily injection of bortezomib 0.15 mg/kg from day 5 to day 13 of life increased the level of survival of motoneuron 1 protein and improved motor function (27). We observed a progressive decrease in porphyrin accumulation in peripheral RBCs of CEP mice treated with bortezomib, but increased dosing had no further effect. This observation may be related to the transient effect of proteasome inhibition worsened by sustained production of erythroid cells owing to chronic hemolysis. However, a partial metabolic correction was sufficient to significantly improve the skin photosensitivity in CEP mice. These results are in accordance with our previous results using hematopoietic stem cell gene therapy, which showed that an ∼50% reduction in fluorocytes could convert a severe skin photosensitivity phenotype into a mild/moderate phenotype (4). However, the regimen used failed to improve the features of hemolytic anemia, which likely were related to the moderate reduction in porphyrin accumulation in peripheral RBCs. We previously demonstrated that at least a 75% decrease in RBC porphyrin content is required to moderately improve the pathological process of hemolytic anemia (4).
Based on previous unfolding kinetics and thermodynamic studies, we can hypothesized that additional UROS missense mutants (A66V, H173Y, I219S, and L237P) with preserved intrinsic enzymatic activity (>85% vs. WT) may be rescued by proteasome inhibitors (10). As proof of principle, we have demonstrated here that a treatment aimed at reversing the stability defect of certain UROS mutants could constitute a promising therapeutic option for CEP (Fig. 5). However, we also have shown that efficient and long-term proteasome inhibition is difficult to obtain in vivo, especially in the erythroid lineage, even with clinical-grade drugs. Furthermore, such long-term unspecific inhibition of abnormal protein degradation could lead to serious adverse toxic effects, especially in the central nervous system, and would not constitute a safe therapeutic choice for CEP (16, 28). Recently, mutations in genes involved in the ubiquitin-proteasome system have been associated with the pathogenesis of neurodegenerative and other brain diseases (29).
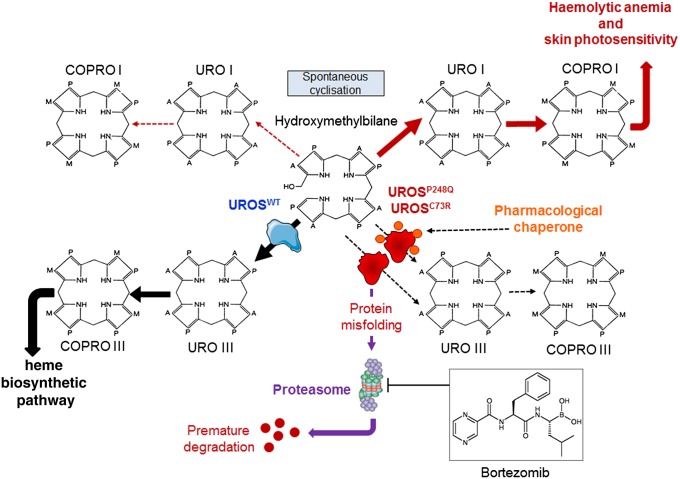
Fig. 5.
Putative mechanism of action of proteasome inhibitors and pharmacologic chaperones in congenital erythropoietic porphyria. UROSC73R and UROSP248Q proteins are misfolded and subjected to premature degradation by the proteasome pathway, leading to a profound UROS enzymatic activity deficiency and the accumulation of URO I and COPRO I. Proteasome inhibitors could partially rescue UROSC73R and UROSP248Q expression and restore the metabolic heme biosynthetic pathway by preventing proteolysis. PCT ideally would stabilize UROSP248Q and UROSC73R proteins and prevent premature degradation by the proteasome pathway.
Therapeutic strategies aimed at modulating protein folding using pharmacologic chaperones have emerged for various genetic diseases. Pharmacologic chaperone therapy (PCT) is based on the concept that ligands may assist the folding of mutated enzymes that retain some catalytic activity and prevent their recognition by the quality control system. PCT approaches have been proposed for loss-of-function genetic disorders including cystic fibrosis (30), phenylketonuria (31), hyperoxaluria type I (32), and lysosomal storage disorders (33).
Following some important therapeutic discoveries, efforts are underway to identify drugs that can repair the underlying molecular defects in cystic fibrosis transmembrane conductance regulator (CFTR) caused by different mutations in cystic fibrosis. Recently, a CFTR potentiator agent, ivacaftor (VX-770), was shown to improve chloride transport by potentiating the open probability of the G551D-CFTR channel, and was proposed in clinical trials for cystic fibrosis (34, 35). The discovery of pharmacologic molecules able to stabilize UROS mutants in vivo would represent a promising therapeutic approach for CEP in patients not eligible for bone marrow transplantation, especially those with prominent skin involvement.
Materials and Methods
Computational Modeling.
The UROSP248Q mutant was modeled computationally using the SWISS-MODEL server (2). PyMOL software was used to optimize side chain orientation, inter-side chain distance measurements, and data representation.
Circular Dichroism and Fluorescence Spectroscopy.
Circular dichroism experiments were performed on a JASCO J-810 spectropolarimeter in quartz cuvettes (0.2-cm path length, 4-nm bandwidth), with collection of data every 0.2 degrees. The interval scanning measurements were run at 37 °C for up to 68 h, with freshly purified samples (5 μM). The guanidinium chloride denaturation experiments used an initial volume of 1.7 mL containing the protein (0.6–1.3 μM). Protein denaturation was achieved by the addition of aliquots (from an automated titrator) of a solution with identical protein concentration and buffer conditions, including a denaturing agent. Data analysis was performed using in-house–built scripts in Matlab.
Cell Culture and Reagents.
Normal fibroblasts were derived from skin biopsy specimens collected during reconstruction surgery. As indicated, cells were cultured for 16 h in the presence of an appropriate concentration of MG132 (Sigma-Aldrich), bortezomib (Velcade), bafilomycin (Sigma-Aldrich), chloroquine (Sigma-Aldrich), or DMSO (0.1%) as a control.
DNA Constructs.
Mammalian expression plasmids encoding human UROS cDNA (WT, C73R, and P248Q mutants) fused to the C terminus or N terminus end of EGFP cDNA in pEGFP-C3 vector or pEGFP-N1, respectively (Promega), were generated as described previously (13).
Transfection and Generation of Stably Transfected Mammalian Cell Lines.
For stable transfection, 1.106 K562 cells were electroporated (Amaxa Nucleofector Technology; Lonza) with 5 μg of C-terminal or N-terminal EGFP-tagged UROS (WT, C73R, or P248 mutant) encoding plasmids and selected with G418 (0.5 g/L) over 3 wk. Normal fibroblasts were transiently transfected by electroporation with EGFP-tagged UROS (WT, C73R, or P248Q)-encoding plasmids and were seeded on a Lab-Tek II chamber slide system (Thermo Fischer Scientific). At 36 h after transfection, the medium was changed, and fibroblasts were treated with 100 nM bortezomib, 10 μM MG132, or DMSO for another 16 h. Cells were then fixed with paraformaldehyde 4% and mounted in DAPI medium for epifluorescence microscopy.
Western Blot Analysis and Antibodies.
For Western blot analysis, equal amounts of protein were separated by SDS/PAGE electrophoresis, and PVDF membranes were probed with rabbit polyclonal anti-EGFP (Santa Cruz Biotechnology; clone 9996), mouse monoclonal anti-NPT2 (Abcam; clone 4B4D1), anti-Hsp70 (Stressgen; clone C92F3A5), and anti–β-actin antibodies (Sigma-Aldrich; clone AC15). The signal was revealed with HRP-linked secondary antibody and visualized using the Enhanced ChemiLuminescence Kit (Amersham) according to the manufacturer’s instructions.
RT-PCR Analysis of EGFP-UROS Expression in K562 Cells.
A quantitative real-time PCR assay was used to quantify EGFP-UROS mRNA in stably transfected K562 cells. The relative expression was normalized to endogenous GAPDH mRNA. The following primers were used: for EGFP-UROS: forward, 5′-GGCATCAGGAAGGCTCTC-3′; reverse, 5′-CCGTAGGTCAGGGTGGTC-3′; for human GAPDH: forward, 5′-CCATCTTCCAGGAGCGAG-3′; reverse, 5′-GAGATGATGACCCTTTTGGC-3′.
Mice and Bortezomib Regimen.
CEP knock-in mice (UrosP248Q/P248Q), described previously (15), were maintained in the animal facility of Bordeaux Segalen University. All animal procedures were approved by the Institutional Animal Care and Use Committee of Bordeaux Segalen University. Mice (age 6–10 wk) were treated by i.p. injection of 0.5 or 1 mg/kg bortezomib (Velcade) or saline diluted in phosphate-buffered saline. Bortezomib was administered every 48 h for 9 consecutive wk, and mice were weighted before each injection.
Flow Cytometry Analysis.
FACS analyses were performed on a FACScanto II analyzer (BD Biosciences). Peripheral blood porphyrin-accumulating RBCs (fluorocytes) and EGFP expression were analyzed as described previously (4).
Hematologic Analysis.
Blood was collected by puncture of the orbital sinus vein. Total hemoglobin, RBC count, and hematocrit were measured with a scil Vet abc animal blood counter (15).
Biochemical Measurements.
Urinary porphyrins were quantified by reverse-phase HPLC with fluorimetric detection (4).
Skin Photosensitivity Assay.
Reversal of skin photosensitivity was assayed as described previously (4).
Proteasome Activity Measurement in Blood Lysates.
The chymotryptic activity of proteasomes was assessed using a fluorogenic assay with the chymotrypsin substrate Suc-LLVY-AMC (Sigma-Aldrich). AMC release from the synthetic peptide substrate LLVY-AMC was analyzed by fluorescence spectroscopy, with excitation at 360 nm and emission at 465 nm.
Statistical Analyses.
Nonparametric Mann–Whitney tests were used to fit to the small sample sizes. Statistical significance was set at P < 0.05.
Supplementary Material
Acknowledgments
We thank the staff of the Unit for Reconstitution of Chemotherapy Agents at the University Hospital Bordeaux for providing bortezomib (Velcade). The Institut National de la Santé et de la Recherche Médicale U1035 laboratory is supported by the Association Française contre les Myopathies and the Agence Nationale de la Recherche.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314177110/-/DCSupplemental.
References
- 1.de Verneuil H, Ged C, Moreau-Gaudry F. Congenital erythropoietic porphyria. In: Kadish KM, Smith KM, Guilard R, editors. The Porphyrin Handbook. Vol 14. New York: Academic Press; 2003. pp. 43–63. [Google Scholar]
- 2.Katugampola RP, et al. A management algorithm for congenital erythropoietic porphyria derived from a study of 29 cases. Br J Dermatol. 2012;167(4):888–900. doi: 10.1111/j.1365-2133.2012.11154.x. [DOI] [PubMed] [Google Scholar]
- 3.Richard E, Robert-Richard E, Ged C, Moreau-Gaudry F, de Verneuil H. Erythropoietic porphyrias: Animal models and update in gene-based therapies. Curr Gene Ther. 2008;8(3):176–186. doi: 10.2174/156652308784746477. [DOI] [PubMed] [Google Scholar]
- 4.Robert-Richard E, et al. Effective gene therapy of mice with congenital erythropoietic porphyria is facilitated by a survival advantage of corrected erythroid cells. Am J Hum Genet. 2008;82(1):113–124. doi: 10.1016/j.ajhg.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C, Dunbar CE. Stem cell gene therapy: The risks of insertional mutagenesis and approaches to minimize genotoxicity. Fr Medecine. 2011;5(4):356–371. doi: 10.1007/s11684-011-0159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ged C, Moreau-Gaudry F, Richard E, Robert-Richard E, de Verneuil H. Congenital erythropoietic porphyria: Mutation update and correlations between genotype and phenotype. Cell Mol Biol (Noisy-le-grand) 2009;55(1):53–60. [PubMed] [Google Scholar]
- 7.Solis C, Aizencang GI, Astrin KH, Bishop DF, Desnick RJ. Uroporphyrinogen III synthase erythroid promoter mutations in adjacent GATA1 and CP2 elements cause congenital erythropoietic porphyria. J Clin Invest. 2001;107(6):753–762. doi: 10.1172/JCI10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.To-Figueras J, et al. ALAS2 acts as a modifier gene in patients with congenital erythropoietic porphyria. Blood. 2011;118(6):1443–1451. doi: 10.1182/blood-2011-03-342873. [DOI] [PubMed] [Google Scholar]
- 9.Ged C, et al. Congenital erythropoietic porphyria: report of a novel mutation with absence of clinical manifestations in a homozygous mutant sibling. J Invest Dermatol. 2004;123(3):589–591. doi: 10.1111/j.0022-202X.2004.23401.x. [DOI] [PubMed] [Google Scholar]
- 10.Fortian A, et al. Uroporphyrinogen III synthase mutations related to congenital erythropoietic porphyria identify a key helix for protein stability. Biochemistry. 2009;48(2):454–461. doi: 10.1021/bi801731q. [DOI] [PubMed] [Google Scholar]
- 11.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4(3):181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 12.Schubert HL, Phillips JD, Heroux A, Hill CP. Structure and mechanistic implications of a uroporphyrinogen III synthase-product complex. Biochemistry. 2008;47(33):8648–8655. doi: 10.1021/bi800635y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortian A, González E, Castaño D, Falcon-Perez JM, Millet O. Intracellular rescue of the uroporphyrinogen III synthase activity in enzymes carrying the hotspot mutation C73R. J Biol Chem. 2011;286(15):13127–13133. doi: 10.1074/jbc.M110.205849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortian A, et al. Structural, thermodynamic, and mechanistical studies in uroporphyrinogen III synthase: molecular basis of congenital erythropoietic porphyria. Adv Protein Chem Struct Biol. 2011;83:43–74. doi: 10.1016/B978-0-12-381262-9.00002-1. [DOI] [PubMed] [Google Scholar]
- 15.Ged C, et al. A knock-in mouse model of congenital erythropoietic porphyria. Genomics. 2006;87(1):84–92. doi: 10.1016/j.ygeno.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Bruna J, et al. Neurophysiological, histological and immunohistochemical characterization of bortezomib-induced neuropathy in mice. Exp Neurol. 2010;223(2):599–608. doi: 10.1016/j.expneurol.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Chaudhuri TK, Paul S. Protein-misfolding diseases and chaperone-based therapeutic approaches. FEBS J. 2006;273(7):1331–1349. doi: 10.1111/j.1742-4658.2006.05181.x. [DOI] [PubMed] [Google Scholar]
- 18.Arakawa T, Ejima D, Kita Y, Tsumoto K. Small molecule pharmacological chaperones: From thermodynamic stabilization to pharmaceutical drugs. Biochim Biophys Acta. 2006;1764(11):1677–1687. doi: 10.1016/j.bbapap.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Moore MR, et al. The biosynthesis of haem in congenital (erythropoietic) porphyria. Int J Biochem. 1978;9(12):933–938. doi: 10.1016/0020-711x(78)90073-3. [DOI] [PubMed] [Google Scholar]
- 20.Azakir BA, Di Fulvio S, Kinter J, Sinnreich M. Proteasomal inhibition restores biological function of mis-sense mutated dysferlin in patient-derived muscle cells. J Biol Chem. 2012;287(13):10344–10354. doi: 10.1074/jbc.M111.329078. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Deuquet J, et al. Hyaline fibromatosis syndrome inducing mutations in the ectodomain of anthrax toxin receptor 2 can be rescued by proteasome inhibitors. EMBO Mol Med. 2011;3(4):208–221. doi: 10.1002/emmm.201100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gioia R, et al. Impaired osteoblastogenesis in a murine model of dominant osteogenesis imperfecta: A new target for osteogenesis imperfecta pharmacological therapy. Stem Cells. 2012;30(7):1465–1476. doi: 10.1002/stem.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, et al. Missense mutations in the NF2 gene result in the quantitative loss of merlin protein and minimally affect protein intrinsic function. Proc Natl Acad Sci USA. 2011;108(12):4980–4985. doi: 10.1073/pnas.1102198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pipalia NH, et al. Histone deacetylase inhibitor treatment dramatically reduces cholesterol accumulation in Niemann-Pick type C1 mutant human fibroblasts. Proc Natl Acad Sci USA. 2011;108(14):5620–5625. doi: 10.1073/pnas.1014890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady RO, Yang C, Zhuang Z. An innovative approach to the treatment of Gaucher disease and possibly other metabolic disorders of the brain. J Inherit Metab Dis. 2013;36(3):451–454. doi: 10.1007/s10545-012-9515-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazzerro E, et al. Therapeutic potential of proteasome inhibition in Duchenne and Becker muscular dystrophies. Am J Pathol. 2010;176(4):1863–1877. doi: 10.2353/ajpath.2010.090468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon DY, Motley WW, Fischbeck KH, Burnett BG. Increasing expression and decreasing degradation of SMN ameliorate the spinal muscular atrophy phenotype in mice. Hum Mol Genet. 2011;20(18):3667–3677. doi: 10.1093/hmg/ddr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mujtaba T, Dou QP. Advances in the understanding of mechanisms and therapeutic use of bortezomib. Discov Med. 2011;12(67):471–480. [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Q, Figueiredo-Pereira ME. Ubiquitin/proteasome pathway impairment in neurodegeneration: Therapeutic implications. Apoptosis. 2010;15(11):1292–1311. doi: 10.1007/s10495-010-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Koulov AV, Kellner WA, Riordan JR, Balch WE. Chemical and biological folding contribute to temperature-sensitive DeltaF508 CFTR trafficking. Traffic. 2008;9(11):1878–1893. doi: 10.1111/j.1600-0854.2008.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pey AL, et al. Identification of pharmacological chaperones as potential therapeutic agents to treat phenylketonuria. J Clin Invest. 2008;118(8):2858–2867. doi: 10.1172/JCI34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopper ED, Pittman AM, Fitzgerald MC, Tucker CL. In vivo and in vitro examination of stability of primary hyperoxaluria-associated human alanine:glyoxylate aminotransferase. J Biol Chem. 2008;283(45):30493–30502. doi: 10.1074/jbc.M803525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parenti G. Treating lysosomal storage diseases with pharmacological chaperones: From concept to clinics. EMBO Mol Med. 2009;1(5):268–279. doi: 10.1002/emmm.200900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Goor F, et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc Natl Acad Sci USA. 2009;106(44):18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsey BW, et al. VX08-770-102 Study Group A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N Engl J Med. 2011;365(18):1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.