Abstract
The S85 type strain of Fibrobacter succinogenes, a major ruminal fibrolytic species, was isolated 49 years ago from a bovine rumen and has been used since then as a model for extensive studies. To assess the validity of this model, we compared the cellulase- and xylanase-degrading activities of several other F. succinogenes strains originating from different ruminants, including recently isolated strains, and looked for the presence of 10 glycoside hydrolase genes previously identified in S85. The NR9 F. intestinalis type strain, representative of the second species of the genus, was also included in this study. DNA-DNA hybridization and 16S rRNA gene sequencing first classified the strains and provided the phylogenetic positions of isolates of both species. Cellulase and xylanase activity analyses revealed similar activity profiles for all F. succinogenes strains. However, the FE strain, phylogenetically close to S85, presented a poor xylanolytic system and weak specific activities. Furthermore, the HM2 strain, genetically distant from the other F. succinogenes isolates, displayed a larger cellulolytic profile on zymograms and higher cellulolytic specific activity. F. intestinalis NR9 presented a higher cellulolytic specific activity and a stronger extracellular xylanolytic activity. Almost all glycoside hydrolase genes studied were found in the F. succinogenes isolates by PCR, except in the HM2 strain, and few of them were detected in F. intestinalis NR9. As expected, the fibrolytic genes of strains of the genus Fibrobacter as well as the cellulase and xylanase activities are better conserved in closely related phylogenetic isolates.
Fibrobacter succinogenes is a major fibrolytic bacterium from the rumen of cattle and sheep. This bacterium is found in large numbers by cultural methods when ruminants are given a poor diet (6), and it is also found to be predominant by the use of molecular tools (37). Pure cultures of F. succinogenes strains S85 and A3C digest more cellulose from intact forage than several strains of Ruminococcus albus and R. flavefaciens, the two other predominant cellulolytic bacterial species in the rumen (9). F. succinogenes also shows a great ability to solubilize other cell wall components such as xylans (27).
The enzymatic system of F. succinogenes type strain S85 has been studied extensively. This bacterium is known to produce at least seven different glucanases, a cellodextrinase, a cellobiosidase, and both a cellobiase and a cellobiose-phosphorylase (3, 13, 14, 22). F. succinogenes S85 produces also at least six xylanases (13), a lichenase (38), an α-glucuronidase (34), and ferrulic acid and acetylxylane esterases and an arabinofuranosidase (26). Some of the genes coding for these activities have been isolated (most of them from strain S85) and sequenced (14).
The S85 strain was originally isolated from the bovine rumen (7) and has been maintained as a pure culture in laboratories ever since. Cultivation of the bacterium in laboratory media for nearly 50 years may have led to genotypic changes in the organism. Indeed, it was suggested that when wild strains of F. succinogenes are grown under laboratory conditions, physiological changes occur, and variants or mutants are selected (36, 40). Furthermore, recent isolates from cellulolytic bacterial species are sometimes more efficient than the reference strains (19). In addition, the genus Fibrobacter is shown to include strains with a high level of genetic variation, as has also been shown for many other groups of rumen bacteria (18, 32). Two species in the Fibrobacter genus are presently known, F. succinogenes and F. intestinalis, and they show less than 20% total DNA homology, indicating a loose relationship between the species (1). The species F. intestinalis comprises strains isolated from the rat and other monogastric animals, but also bovine strains (1). It is therefore probable that several different Fibrobacter strains from both species are present in the rumen and that this mixture represents the dominant Fibrobacter population. It therefore seems important to check that the S85 F. succinogenes strain is truly representative of all rumen Fibrobacter strains, particularly when considering its properties related to plant cell wall degradation.
In this work, we compared the phylogenetic relationships and the cellulolytic or xylanolytic activity of several strains of F. succinogenes and one strain of F. intestinalis. The F. succinogenes strains were isolated from different animals (cows and sheep), and from different parts of the world, and the F. intestinalis strain was isolated from a rat (1). Several of the F. succinogenes strains have been subcultured only a few times since their isolation. The enzyme activities were compared both qualitatively and quantitatively. In parallel, the glycoside hydrolase genes already identified in some strains of F. succinogenes were sought by PCR in all the strains studied here.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The nine Fibrobacter strains used were type strain Fibrobacter succinogenes S85 (ATCC 19169) and strain BL2 (obtained from C. Stewart, Rowett Research Institute, Aberdeen, Scotland), both isolated from the bovine rumen, and strain HM2 (ATCC 43856), isolated from a sheep rumen and belonging to a group different from that of S85 and BL2 (1, 21). F. succinogenes strains H, U, and R, isolated from the bovine rumen, were obtained from K.-J. Cheng, Lethbridge, Alberta, Canada. Strain FE was isolated in our laboratory from a sheep rumen by the roll tube method of Hungate (17). F. intestinalis NR9 (ATCC 43854), the type strain of the second known species of the genus Fibrobacter, was isolated from the rat cecum (29). The Fibrobacter strains were grown on medium containing 30% rumen fluid (4) and 0.5% filter paper cellulose (Whatman 1MM) for enzyme assays or 0.3% glucose or cellobiose for DNA extraction.
The other rumen strains used were Ruminococcus albus 7 (ATCC 27210), Ruminococcus flavefaciens 007, and Prevotella ruminicola 23 (previously obtained from M. P. Bryant, University of Illinois, Urbana-Champaign). These strains and Escherichia coli DH5α were grown as previously described (10).
Fermentation end product assays.
Succinate, acetate, and formate were assayed as described by Matheron et al. (25).
DNA isolation.
DNA extraction from E. coli was performed with the phenol-chloroform procedure (33). DNA extraction from P. ruminicola and from the different strains of Fibrobacter was carried out as described previously for F. succinogenes (12). DNA from R. flavefaciens and R. albus was extracted with the Easy DNA kit (Invitrogen, Groningen, The Netherlands), according to the manufacturer's recommendations.
Quantitative DNA-DNA hybridization.
The relative DNA concentration for each strain was quantified by spectrophotometry and checked by hybridization to a universal oligonucleotide probe, 5′-ACGGGCGGTGTGT(A/G)C-3′ (experimental melting temperature, 46°C), which is complementary to all genes of 16S-like rRNAs (30). The oligonucleotide probe was synthesized by Eurogentec (Seraing, Belgium) and labeled with [γ-32P]ATP and T4 polynucleotide kinase. The hybridization and washing conditions were as described previously (1). DNA (2 μg) from each isolate was denatured, divided into three aliquots of 0.7 μg, and spotted onto nylon membranes (Hybond N+) with a dot-blot device (Bio-Rad). The membranes were then hybridized with [α-32P]dCTP-radiolabeled total DNA (Ready-to-Go kit from Roche) from each strain and washed under conditions similar to those used previously (1). After autoradiography, either the radioactivity in the membranes was counted with an AMBIS counter, or the spots were cut from the membranes and the radioactivity was counted by scintillation spectrometry.
PCR primers and procedure.
The oligonucleotide primers were partly previously designed (4) and purchased from Eurogentec (Seraing, Belgium). The sequence, position, and melting temperature of the primers used to amplify the glycoside hydrolase genes are given in Table 1, as well as the sizes of the amplified PCR fragments. The GH9 degenerate primers were designed to hybridize to family 9 glycoside hydrolase genes. rRNA gene fragments of approximately 1,500 bp were amplified with the 16S rRNA gene bacterial forward primers F8 [5′-AGAGTTTGATC(A/C)TGGCTC-3′] and F500 (5′-CTAACTACGTGCCAGCAGC-3′), associated with the universal reverse R1492 primer (5′-GNTACCTTGTTACGACTT-3′).
TABLE 1.
Primer sequences
| Protein | Glycoside hydrolase family | Gene | Primera | Sequence (5′-3′)b | Position (nucleotide) | Position (codon) | Tm (°C) | Size (bp) |
|---|---|---|---|---|---|---|---|---|
| EGB | 9 | endB | endBF* | CCT-TGG-TGG-ACT-TTG-ATG-CG | 716 | 80 | 62 | |
| endBR* | CGA-TTC-CGT-AGG-CGT-CAC-AA | 1,196 | 240 | 62 | 500 | |||
| GH9R | RTC-NCC-NGC-RTC-RWR-VYA-NCC | 937 | 154 | 68 | 242 | |||
| CedA | 5 | cedA | cedAF* | CCA-CCT-CGC-CAG-CAA-TCA-CG | 1,292 | 108 | 66 | |
| cedAR* | GTT-CGG-GGT-TGT-TGT-ATT-GC | 1,985 | 339 | 60 | 713 | |||
| Cel3 | 5 | cel3 | cel3F* | AGC-GAT-GGT-AAG-GTC-ACT-GC | 1,530 | 452 | 62 | |
| cel3R* | AAG-GAA-TCT-GGA-GGG-TAT-CG | 2,039 | 622 | 60 | 529 | |||
| cel3′F | GAA-AAY-GCM-GAT-GGC-AAG | 1,137 | 321 | 54 | ||||
| cel3′R | TCA-TRT-CCG-GTT-CGT-TCA | 1,513 | 447 | 52 | 394 | |||
| cel3"F | GAA-GAA-GCG-GAC-GGC-AAG | 54 | ||||||
| EGD | 9 | celD | celDF* | ACA-ATC-CGA-ACG-ACG-AAA-AG | 2,668 | 331 | 58 | |
| celDR* | GGT-AAT-CGG-CAC-ACT-TCC-AT | 3,212 | 512 | 60 | 564 | |||
| GH9F | GGN-TRB-YWY-GAY-GCN-GGN-GAY | 2,187 | 171 | 68 | 1,045 | |||
| EGE | 9 | celE | celEF* | CAC-GCT-CCT-TTC-GCT-TTA-CG | 160 | 37 | 62 | |
| celER* | GGC-ATA-AAC-CGC-AAG-ACC-AT | 702 | 218 | 60 | 562 | |||
| celE′F | GGC-TTC-GTT-TGA-CTT-TGC-TG | 400 | 118 | 60 | ||||
| celE′R | CCT-TCT-TGG-CTG-CTT-CGT-AA | 931 | 295 | 60 | 551 | |||
| CelG | 5 | celG | celGF* | CGT-TTA-TTG-ATG-CTG-TCC-GC | 691 | 188 | 60 | |
| celGR* | GTG-CTG-CTT-TCA-TTG-CCC-AT | 1,233 | 369 | 60 | 562 | |||
| celG′F | TTG-AAC-ATC-CAC-CAC-GAA-GG | 468 | 114 | 60 | ||||
| celG′R | TTA-GCA-GCC-GAA-ACG-ACA-TC | 1,182 | 352 | 60 | 734 | |||
| EGF | celF | celFF* | TCG-TGA-AGG-GCA-ACA-AGA-AG | 1,004 | 208 | 60 | ||
| celFR* | GCC-AAC-AGA-GAA-GCG-GAT-TT | 1,562 | 394 | 60 | 578 | |||
| EndA | 9 | endAFS | endAFSF* | GAA-CTT-GAA-CTT-TAC-CGC-AC | 1,178 | 274 | 58 | |
| endAFSR* | TAG-TAA-AGT-CGC-CAG-CAA-TC | 1,721 | 447 | 58 | 563 | |||
| XynC | 11 | xynC | xynCF* | CTC-TAA-GGG-TAA-CGG-CAA-CG | 1,696 | 196 | 62 | |
| xynCR* | ATG-GAG-GGC-TGC-TGG-ACA-CG | 2,180 | 285 | 66 | 504 | |||
| xynC′F | TAA-CAC-CCT-CGC-CAA-TGA-CA | 1,285 | 148 | 60 | ||||
| xynC′R | TAC-CCT-GCT-TGC-TCC-ACT-TG | 1,993 | 384 | 62 | 728 | |||
| LichA | 16 | mlg | mlgF* | TGA-AGC-CCG-TAT-GAA-GAT-GG | 282 | 46 | 60 | |
| mlgR* | CGT-CAA-ATG-TCC-AGT-CAC-CC | 813 | 223 | 62 | 551 |
Sequences were from F. succinogenes S85 genes except for endAFS, which was from strain AR1; cel3′ primers, which were from an S85/A3c consensus sequence; and the cel3" primer, which was designed from the F. intestinalis DR7 sequence. *, described previously (4).
R, A or G; N, A or C or G or T; W, A or T; V, A or C or G; Y, C or T; M, A or C; B, C or G or T.
PCRs were carried out with genomic DNA (0.5 μg) and 1 U of AmpliTaq DNA polymerase (Qbiogene) for glycoside hydrolase genes and the Expand High Fidelity PCR system (Roche) for 16S rRNA gene fragments. Each reaction was subjected to 30 cycles of amplification consisting of 30 s denaturation at 94°C, 2 min of annealing of primers at a temperature 5°C below the melting temperature (Table 1), and 45 s or 2 min of elongation at 72°C for glycoside hydrolase genes and 16S rRNA genes, respectively. A 2-min DNA denaturation at 94°C was performed before the 30 cycles. The final cycle included elongation for 7 min at 72°C. A negative control (without DNA) was included in every PCR procedure.
PCR-generated DNAs were sequenced full length in both directions with an ABI Prism BigDye terminator cycle sequencing kit (Perkin Elmer Corporation, Norwalk, Conn.) with appropriate primers in an ABI Prism 310 genetic analyzer (Perkin Elmer Corporation).
Nucleotide and protein sequence comparison and phylogenetic analyses.
Pairwise nucleic acid and protein sequence similarities were estimated with the global alignment programs ALIGNn and ALIGNp, respectively (http://www.infobiogen.fr).
The phylogenetic tree was constructed by first performing a multiple alignment with the ClustalW 1.8 program (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) with all available Fibrobacter 16S rRNA gene sequences, with the Bacteroides fragilis 16S rRNA gene sequence (EMBL AB050106) used as the outgroup. Optimization of the alignment was performed manually with sequences of equal lengths (nucleotides 9 to 1315 of the S85 16S rRNA gene sequence) before running the Phylip package 3.5c programs (11). A distance matrix was generated with the Kimura model with the DNAdist program following a bootstrap analysis based on 1,000 replicates with the Seqboot program. Final trees were obtained by neighbor-joining analyses with the Consense (11) and Treeview (31) programs.
Harvesting of cultures and preparation of cells.
All Fibrobacter strains were subcultured three times on filter paper rumen fluid medium before harvesting. For each strain, three tubes of 10-ml cultures grown for 48 h at 39°C were first centrifuged for 5 min at 500 × g to eliminate filter paper cellulose. Culture media containing free cells were pooled and centrifuged at 12,000 × g for 20 min. The supernatant was concentrated about sixfold with a Macrosep 10K Omega centrifugal device (PALL Life Sciences). The cell pellet was suspended in 1/5 volume of 50 mM sodium phosphate buffer (pH 7.0). The triplicate cultures were repeated at least twice. Supernatants and cells were kept frozen at −20°C.
Enzyme and protein assay.
The enzyme preparations were incubated at 39°C for 30 min in 50 mM sodium phosphate buffer, pH 7.0, with 0.5% carboxymethyl cellulose (CMC sodium salt, medium viscosity), 1% Sigmacell 101 cellulose, and 1% oat spelt xylans (all from Sigma Chemical Co., St. Louis, Mo.) or with 1% Avicel PH101 microcrystalline cellulose (Fluka). The amount of enzyme extract and the incubation time were chosen to give conditions in which the measured activity was proportional to these two parameters. Reducing sugars released from polysaccharide hydrolysis were assayed as described previously (12) with glucose and xylose as the standards. Results are given in nanokatals, where 1 kat corresponds to 1 mol of glucose or xylose equivalent produced per s. For each culture and for each substrate, assays were performed at least twice.
Protein was determined by the method of Bradford (5), with bovine serum albumin as the standard.
PAGE and zymogram.
Polyacrylamide gel electrophoresis (PAGE) and zymograms for carboxymethyl cellulase (CMCase) and xylanase activity were performed as described previously (10, 12).
Statistical analyses.
All data were analyzed by one-way analysis of variance, and the differences between the means of a row were compared with the Student-Newman-Keuls multiple-comparisons test.
Nucleotide sequence accession numbers.
Nearly complete 16S rRNA gene sequences were obtained for Fibrobacter strains S85 (AJ496032), BL2 (AJ505937), HM2 (AJ496186), H (EMBL AJ496447), R (EMBL AJ505938), U (EMBL AJ496448), FE (AJ496566), and NR9 (AJ496284) and deposited in the EMBL data library under the indicated accession numbers. PCR-DNA fragments of glycoside hydrolases were also deposited in the EMBL database under the accession numbers AJ548833 (F. succinogenes U partial xynC gene), AJ548832 (F. intestinalis NR9 partial xynC gene), and AJ548831 (F. intestinalis NR9 partial celF gene).
RESULTS
Substrates and end products.
All the Fibrobacter strains, like the S85 type strain, grew on media containing either glucose, cellobiose or filter paper cellulose as the carbon source (not shown); this is consistent with previous work (24, 25). None of them was able to grow on xylans, possibly because of an inability to utilize xylose, as shown in S85 (23). The fermentation end product patterns of all isolates were similar to that of S85 (succinate, acetate, and formate).
Grouping of strains.
The relationships between the eight isolates of Fibrobacter were analyzed by DNA-DNA hybridization. The results are presented in Table 2. The estimated genomic DNA similarity determined here between strains S85, HM2, and NR9 were consistent with the values previously obtained with the same technique (1), except for the S85-HM2 DNA similarity, which we found to be higher. The S85 strain presented about 30% DNA similarity with strains BL2 and HM2, whereas 40 to 50% DNA similarity was observed between strains S85 and H, U, R, and FE. This suggested that the four newly isolated bovine or sheep strains morphologically identified as F. succinogenes strains actually belong to this species and are closest to the S85 type strain. Thus, the H, U, and R bovine strains presented more than 70% similarity, suggesting that the animal origin could be related to their phylogenetic proximity. In the same way, the FE and HM2 strains showed about 70% DNA similarity; both of these strains were isolated from the rumen of sheep. Amann et al. (1) have already described subspecies of F. succinogenes constituted exclusively of members from one habitat. F. intestinalis NR9 was distant from all the other strains (2 to 11% DNA similarity) due to the phylogenetic distance between the two species of the genus Fibrobacter.
TABLE 2.
16S rRNA gene identity and DNA/DNA hybridization value for the Fibrobacter strainsa
| Strain | % Identity
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S85 | BL2 | H | U | R | FE | HM2 | NR9 | |
| S85 | 100 | 99.6 | 99.6 | 99.4 | 99.9 | 99.3 | 96.1 | 89.9 |
| BL2 | —/32 | 100 | 99.3 | 99.1 | 99.6 | 99.0 | 96.2 | 89.7 |
| H | 47/48 | 38/— | 100 | 99.7 | 99.7 | 99.4 | 95.6 | 89.5 |
| U | 42/— | 36/37 | 65/77 | 100 | 99.6 | 99.4 | 95.7 | 89.6 |
| R | 49/61 | 38/— | —/70 | 70/64 | 100 | 99.3 | 95.9 | 89.8 |
| FE | 36/40 | 39/— | 39/29 | 50/50 | 58/39 | 100 | 95.6 | 89.4 |
| HM2 | 37/34 | 41/— | 29/36 | 31/27 | 45/27 | 75/78 | 100 | 89.3 |
| NR9 | 3.5/4 | 11/— | 5/4 | 9/4 | 10/3 | 0/2 | 5/3 | 100 |
Values in italics are percent identity between 16S rRNA gene sequences from two strains. Alignments of equal length were made (nucleotides 9 to 1407 from the S85 type strain). The other values are reciprocal DNA/DNA hybridization data with the left value corresponding to use of the DNA from the organism in the first row as the probe. —, not determined.
This grouping was extended by sequencing and comparing the 16S rRNA gene sequence of the strains studied (Table 2). As for Amann et al. (1), the 16S rRNA gene similarity of F. succinogenes S85 was greater with strain BL2 (99.6%) than with strain HM2 (96.1%); the lowest value was obtained with F. intestinalis NR9 (89.9%). The rRNA gene sequence comparison of the new bovine isolates, like the DNA-DNA similarities, suggested that these strains are closer to the S85 type strain (99.3 to 99.9%) than to strain BL2 (99.0 to 99.6%) or HM2 (95.6 to 95.9%). Surprisingly, the rRNA gene sequence from the FE strain was closer to that of the recent bovine isolates (99.0 to 99.4%) than to that of the ovine ruminal strain HM2 (95.6%). According to the DNA-DNA reassociation, FE probably belongs to the same group as HM2, whereas the strain is classified within the bovine ruminal strains with rRNA gene comparison.
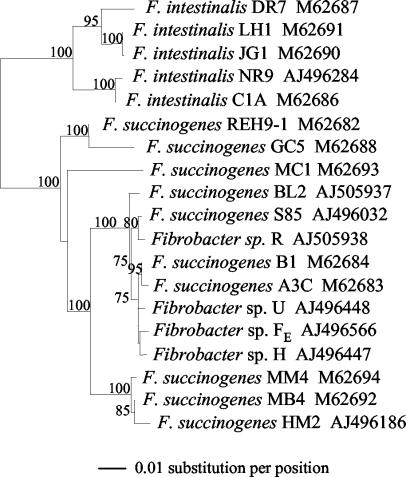
A phylogenetic tree was constructed for the rRNA gene sequences from all the Fibrobacter strains studied here and available in the nucleotide databases, with Bacteroides fragilis (DSM 2151T) as the outgroup sequence (Fig. 1). The 19 Fibrobacter sequences included F. succinogenes strains S85, A3c, B1, BL2, MC1, HM2, MB4, MM4, GC5, and REH9-1, F. intestinalis strains NR9, C1A, DR7, JG1, and LH1, and the Fibrobacter sp. strains from this study (H, U, R, and FE). The tree is consistent with that of Lin and Stahl (20) except for two nodes. The first one, supported by high bootstrap values (85 to 100%), differs in the phylogenetic position of strain HM2, which was closest to strain MB4 in this study. We used a new HM2 16S rRNA gene sequence which was longer and more complete than the previous one (20). The second different node, also supported by a strong bootstrap value (100%), consists of the phylogenetic position of the GC5/REH9-1 branch, which was found to be distant from the other F. succinogenes strains. Additional new 16S rRNA gene sequences in the phylogenetic analysis could have resulted in reorganization of this genetic branch.
FIG. 1.
Phylogenetic tree for 16S rRNA gene sequences from Fibrobacter strains. Phylogenetic relationships were based on ClustalW alignments, optimized manually, and analyzed with the Phylip programs. A total of 1,000 bootstrap replicates were performed, and bootstrap values greater than 75 are indicated above each branch. rRNA gene sequences are shown with their species and EMBL or GenBank accession numbers.
Zymograms.
Six Fibrobacter strains representative of different phylogenetic groups (Fig. 1) were compared by zymograms for CMCase and xylanase activities (Fig. 2). Extracellular culture fluid (ECCF) and cells were analyzed separately.
FIG. 2.
CMCase (A) and xylanase (B) zymogram analyses of bacterial (P) and extracellular (S) proteins from several strains of the genus Fibrobacter. Lanes M, broad-range molecular mass standards (in kilodaltons) (Bio-Rad). Protein migration was performed with a 10% acrylamide gel.
The CMCase activity pattern observed for cells (Fig. 2A, P) was different from that of the ECCF (Fig. 2A, S). The activity profiles obtained for strains U and R were very similar. The strains studied all presented a majority of bacterial CMCases of between 60 and 120 kDa (Fig. 2A, P). Several lower bands of activity could also be detected for strains FE and HM2, at approximately 35 and 45 kDa, but were not detected or detected only weakly for the other strains. The NR9 profile did not differ from those of the F. succinogenes strains.
Comparison of the extracellular CMCases from the six strains revealed three types of profiles (Fig. 2A, S). Strains S85, U, and R presented extracellular CMCases of between 35 and 150 kDa, with two major bands at about 50 and 95 kDa, whereas the FE and HM2 strain profiles included an additional band at 60 kDa. The CMCase profile of F. intestinalis NR9 did not correspond to any of those of the F. succinogenes strains.
The xylanase activity of cells and ECCF of all strains was also examined by zymograms (Fig. 2B). The xylanolytic profile of each strain revealed fewer bands than for the CMCases. Again, the strain U and R profiles looked similar. The F. succinogenes strains revealed several xylanases of between 35 and 130 kDa, while F. intestinalis NR9 showed two large xylanases at about 65 and 95 kDa. Several lower-molecular-mass xylanases were weakly detected.
The cells (Fig. 2B, P) could be separated into three different profiles: the first of these included strains S85, U and R, the second one included strains FE and HM2, and the last profile was specific for F. intestinalis NR9. Except for strains U and R, the extracellular xylanase profiles (Fig. 2B, S) were different for each strain studied. Type strain S85 presented a major xylanase band at about 65 kDa which could correspond to the known F. succinogenes xylanase XynB (about 65 kDa) or XynC (63 kDa) (13).
Quantification of cellulase and xylanase activities of cells and ECCF.
The CMCase, cellulase, and xylanase activities were quantified with CMC, Sigmacell, Avicel, and oat spelt xylans as the substrate. The relative CMCase and xylanase activities of cells and ECCF in an equal volume of 48-h cultures of the Fibrobacter strains are reported in Fig. 3. The activity distribution between bacteria and ECCF was generally similar for all the strains. The relative CMCase activity of each strain was mostly extracellular (60 to 75%) (Fig. 3A). The Sigmacellase relative activity of each strain showed the same profile (not shown). The xylanase activity (Fig. 3B) was also mostly extracellular (55 to 70%) for each strain except for strain U. This activity was almost totally extracellular (90%) for NR9.
FIG. 3.
CMCase (A) and xylanase (B) activity distribution between cells (□) and ECCF (▪) for several strains of Fibrobacter grown on filter paper cellulose. The CMCase activities varied between 3.6 and 10.7 nkat in cells in the initial culture and from 7.0 to 20.0 nkat in ECCF. The xylanase activities varied between 8.3 and 42.7 nkat of total culture volume in cells and from 17.3 to 175.0 nkat in ECCF.
The CMCase, Sigmacellase, Avicelase, and xylanase specific activities of the cells and ECCF were calculated. Bacterial activities are reported in Table 3. Strains HM2 and NR9 showed significantly higher (double) CMCase and Sigmacellase specific activity than the other strains. A weak Avicelase activity could be detected for all the strains. The xylanolytic activity of all the strains was much higher than the cellulolytic activity except for FE, which showed the lowest xylanase specific activity.
TABLE 3.
Enzyme specific activities of bacteria grown on cellulosea
| Strain | Mean sp act (nkat eq/mg of protein) ± SD
|
|||
|---|---|---|---|---|
| CMC | Xylans | Sigmacell | Avicel | |
| S85 | 2.5 ± 0.5a | 15.5 ± 2.9a | 2.6 ± 0.4d,e,f | 0.10 ± 0.04a |
| U | 2.1 ± 0.6a | 12.6 ± 4.6a | 1.8 ± 0.6a,d | 0.07 ± 0.01a |
| R | 1.9 ± 0.7a | 12.0 ± 4.4a | 1.4 ± 0.4a,e | 0.06 ± 0.01a |
| HM2 | 3.8 ± 0.4b | 15.3 ± 1.0a | 3.6 ± 1.1b,c,f | 0.12 ± 0.02a |
| FE | 1.3 ± 0.7a | 4.0 ± 1.5b | 1.1 ± 0.8a | 0.08 ± 0.01a |
| NR9 | 4.1 ± 0.6b | 16.9 ± 1.8a | 4.3 ± 0.6b | 0.13 ± 0.07a |
Specific activities are given as nanokatals of glucose (incubation with CMC or cellulose) or xylose (incubation with xylans) equivalent produced per milligram of protein. Each value is the mean of assays for at least two different cultures. Values bearing different letters within a row differ significantly (P < 0.05).
The ECCF specific activities were almost similar to those of bacteria measured with the same substrates (Table 4). As for the bacteria, Avicelase activity was weakly detectable, suggesting that neither ECCF proteins nor harvested cells are able to degrade crystalline cellulose efficiently. The highest activity (45.1 nkat of xylose equivalent/mg of protein) was obtained for F. intestinalis NR9 ECCF; this could signify a particular ability of this strain to degrade hemicellulosic substrates compared to the F. succinogenes ruminal strains.
TABLE 4.
Enzyme specific activities of ECCF of strains grown on cellulosea
| Strains | Mean sp act (nkat eq/mg of protein) ± SD
|
|||
|---|---|---|---|---|
| CMC | Xylans | Sigmacell | Avicel | |
| S85 | 2.4 ± 0.4a | 18.2 ± 2.3b | 3.3 ± 0.4c,d | 0.17 ± 0.04a |
| U | 2.9 ± 0.8a | 13.0 ± 1.4a | 2.6 ± 0.5a,d | 0.12 ± 0.04a |
| R | 3.3 ± 0.5a | 11.1 ± 1.6a | 2.4 ± 0.7a,d | 0.06 ± 0.06a |
| HM2 | 4.3 ± 0.5b | 16.9 ± 1.5b | 2.7 ± 1.4a,d | 0.18 ± 0.04a |
| FE | 2.3 ± 0.2a | 7.1 ± 1.4c | 1.4 ± 0.5a | 0.05 ± 0.02a |
| NR9 | 5.0 ± 0.2b | 45.1 ± 0.9d | 5.6 ± 1.1b | 0.17 ± 0.06a |
See Table 3, footnote a.
In conclusion, the fibrolytic activities of the recently isolated F. succinogenes strains (U, R, and FE) were similar, according to the nature of the substrate, to those of the S85 type strain except for FE, which showed the lowest levels of activity. The cellulase activities of HM2 and NR9 were usually higher than those of the other Fibrobacter strains that were more distant phylogenetically.
Detection of glycoside hydrolase genes.
We searched for copies of the 10 glycoside hydrolase genes from F. succinogenes S85 in the genomes of the eight other strains by PCR. These genes, which were studied previously (4), include eight cellulase genes (endB, cedA, cel3, celD, celE, celF, celG, and AFS), the xynC gene, encoding a xylanase; and the lichenase gene mlg. To detect homologous genes in all the strains, several pairs of primers had to be used (Table 1). The primers were chosen to amplify DNA fragments of approximately 500 bp, or 700 bp in the case of cedA, celG, and xynC (Table 1). These fragments were located in regions of the gene coding for the catalytic domain of the enzymes except for the celF gene.
Table 5 reports the detection of the genes in the Fibrobacter strains. With the exception of celG, which was not specifically detected in strain FE, all the glycoside hydrolase genes studied were detected in the strains phylogenetically closest to S85 (BL2, H, U, R, and FE) (Table 5, Fig. 1). Only two hydrolase genes were specifically detected in F. succinogenes HM2, the celF cellulase gene and the lichenase gene mlg. In addition, only two cellulase genes, celF and cedA, and the xynC xylanase gene were detected by PCR in F. intestinalis NR9.
TABLE 5.
PCR detection of glycoside hydrolase genes in the Fibrobacter strains
| Gene | Amplificationa in strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S85 | BL2 | H | U | R | FE | HM2 | NR9 | |
| endB | + | + | + | + | + | + | − | − |
| cel3 | + | + | + | + | + | + | NS | NS |
| celD | + | + | + | + | + | + | − | − |
| celE | + | + | + | + | + | + | NS | − |
| celF | + | + | + | + | + | + | + | + |
| celG | + | + | + | + | + | NS | − | NS |
| endAFS | + | + | + | + | + | + | − | NS |
| cedA | + | + | + | + | + | + | − | + |
| mlg | + | + | + | + | + | + | + | − |
| xynC | + | + | + | + | + | + | NS | + |
+, specific amplification; −, no PCR product; NS, nonspecific amplification.
celF is the only glycoside hydrolase gene to be specifically detected in all strains of the two Fibrobacter species. The PCR-DNA fragment of celF amplified from F. intestinalis NR9 was sequenced and showed 80.9% identity with the corresponding sequence (about 580 nucleotides) of celF from F. succinogenes S85 (Table 6). The 192-amino-acid translated protein sequence EGF showed 83.2% identity between the two strains. As the xynC xylanase gene was also detected in F. intestinalis NR9, the sequence of the PCR fragment (725 nucleotides) was determined and compared with that of F. succinogenes S85 and U (Table 6). The partial xynC gene sequence from F. succinogenes S85 showed 98.1% DNA identity (99.2% over 242 amino acids) with strain U and 92.9% (93.8% amino acid identity) with F. intestinalis NR9; this is in agreement with the phylogenetic relationships of these strains (Fig. 1). The PCR-DNA fragment of xynC from F. succinogenes U showed 94% DNA identity (94.6% amino acid identity) with F. intestinalis NR9.
TABLE 6.
Partial glycoside hydrolase gene and deduced amino acid sequence comparisons for several Fibrobacter isolatesa
| Gene | Fibrobacter strains compared | DNA PCR fragment identity (%) | Protein identity (%) |
|---|---|---|---|
| celF | S85-NR9 | 80.9 | 83.2 |
| xynC | S85-U | 98.1 | 99.2 |
| S85-NR9 | 92.9 | 93.8 | |
| U-NR9 | 94.0 | 94.6 | |
| cel3 | S85-A3c | 93.6 | 97.2 |
| S85-REH9-1 | 84.5 | 89.8 | |
| S85-DR7 | 72.6 | 72.3 |
The DNA identities of partial sequences were calculated based on alignments of equal length with F. succinogenes S85 bases 1004 to 1581 for celF, 1285 to 2012 for xynC, and 1530 to 2058 for cel3 and the corresponding sequences (amplified by PCR or available in the GenBank database) in the other strains.
The cel3 endoglucanase gene of F. succinogenes S85 was previously sequenced from several F. succinogenes ruminal isolates, S85, A3c, and REH9-1, and from porcine cecum strain F. intestinalis DR7 (20). Based on the comparison of a 530-bp sequence, surrounded by the positions of the cel3R and cel3F primers (Table 1) and encoding a part of the endoglucanase catalytic domain, cel3 presented 93.6% identity (97.2% protein identity) with the corresponding sequence from the genetically close F. succinogenes strain A3c but only 84.5% (89.8% protein identity) with REH9-1 and 72.6% (72.3% protein identity) with F. intestinalis DR7 (Table 6). This finding is consistent with the phylogenetic distances between these strains (Fig. 1). The DNA and amino acid sequences of cel3/EG3 appeared more divergent between the two Fibrobacter species (about 72% identity) compared to celF and xynC glycoside hydrolase sequences (81 to 93%). This result could explain the fact that even the degenerate primer cel3′, designed with oligonucleotide sequences conserved between F. succinogenes S85, A3c, and REH9 and F. intestinalis DR7, did not allow the gene to be detected in F. succinogenes HM2 or F. intestinalis NR9, which are genetically related to strains REH9-1 and DR7, respectively.
The degenerate family 9-specific glucoside hydrolase primers that we designed allowed the endB and celD genes to be detected in six strains (S85, BL2, H, U, R, and FE) of F. succinogenes (Table 5), but did not enable these genes to be detected specifically in the genome of F. succinogenes HM2 or F. intestinalis NR9. In fact, modifications of the hybridization temperature during the PCR procedures to detect several glycoside hydrolase genes of F. succinogenes FE and HM2 and F. intestinalis NR9 produced nonspecific PCR fragments after amplification (Table 5).
The primers for four F. succinogenes glycoside hydrolase genes (endB, cel3, celF, and xynC) did not allow these genes to be amplified in other fibrolytic ruminal species such as R. albus, R. flavefaciens, and Prevotella ruminicola (not shown). This confirms their specificity for the Fibrobacter fibrolytic system. PCR was negative for all glycoside hydrolase gene-specific primers with E. coli DH5α DNA as a negative control.
DISCUSSION
Previous DNA hybridization and 16S rRNA gene similarity analyses indicate broad genetic diversity in the genus Fibrobacter (1, 21). This work was carried out, first, to extend such studies by characterizing new ruminal Fibrobacter isolates and, second, to determine whether strain S85 is a good model for ruminal F. succinogenes and whether it has been subjected to phenotypic alteration over time.
Both DNA-DNA hybridization and 16S rRNA gene sequence comparisons were used to determine phylogenetic relationships between the Fibrobacter strains studied, as it is known that 16S rRNA gene sequence analysis is insufficient to discriminate among closely related organisms (35, 39). Thus, as four Fibrobacter isolates (H, U, R, and FE) had been proposed by morphological criteria to belong to the same species (F. succinogenes), the phylogenetic positions of seven F. succinogenes strains had to be distinguished in the present study. Equal-length, nearly complete (1,400 bp) 16S rRNA gene sequences were compared and associated with DNA-DNA hybridization to evaluate the similarity of the strains. A good correlation was observed except for strain FE (Fig. 1), which appeared to be close to F. succinogenes HM2 by DNA-DNA reassociation but more distant from the same strain by rRNA gene sequence comparison. Noncongruence between prokaryotic whole-genome and 16S rRNA gene sequences or between genotyping methods has already been documented (15, 19). Nevertheless, our results suggest that all of the newly studied strains belong to F. succinogenes and are distinct from F. intestinalis. Furthermore, the phylogenetic tree confirms the genetic diversity of Fibrobacter isolates.
The fibrolytic systems of the eight Fibrobacter strains studied, including F. intestinalis NR9, revealed common characteristics, such as qualitative cellulolytic and xylanolytic system (visualized by zymograms), homogenous specific activities towards four different polysaccharide substrates, and common hydrolase genes. However, some strains, such as HM2 and NR9, which are more distantly related to S85 than the others presented specific phenotypic characteristics.
The qualitative and quantitative analyses of the fibrolytic activities could be correlated for several strains with respect to the substrate, especially for phylogenetically related F. succinogenes strains S85, U, and R. CMCase zymograms of cells and ECCF revealed common activity patterns for these strains, which are consistent with similar CMCase specific activities. The cell xylanase specific activities of these strains were also concordant with the qualitative profiles observed in zymograms. However, an extracellular xylanase of F. succinogenes S85, appearing as a strong band on the xylan zymogram with an apparent molecular mass of 65 kDa, could explain why the specific activity of the S85 ECCF was higher than that of strains H and U.
The lowest specific activity measured on xylans was that of strain FE and was well correlated with the xylan zymogram, as very few and weakly detectable bands of activity were observed. On the contrary, the highest xylanase activity was obtained from F. intestinalis NR9, which was concordant with the strong bands of activity on the zymogram.
All of the Fibrobacter cells and ECCF studied presented a very low common activity on crystalline cellulose. Forsberg et al. have shown previously (13) that disrupted cell extracts or ECCF were not able per se to degrade crystalline cellulose efficiently, whereas bacteria can grow well on this substrate. This property thus seems to be associated with living bacteria.
The CMCase, sigmacellase, and xylanase activity distribution showed that major enzyme activities of all strains were extracellular for all the substrates studied. This is in agreement with a previous study that showed major endoglucanase and xylanase activities of strain S85 associated with membrane vesicles in the ECCF of cellulose-grown cultures (16).
Molecular study of endoglucanase, cellodextrinase, lichenase, and xylanase genes in eight Fibrobacter isolates provided evidence for the presence of homologous genes in all F. succinogenes strains studied (with one exception, celG). The sequence of the partial xynC gene amplified from strain U confirmed its homology with the F. succinogenes S85 type strain and suggested that the glycoside hydrolase genes of the type strain could be widespread in other close phylogenetic isolates. Moreover, conserved DNA fragments from celF and xynC were detected in the most distantly related strain, F. intestinalis NR9. These genes were conserved between the two strains, whose DNA-DNA similarity is less than 5%. The EGF endoglucanase from F. succinogenes S85 is a complex modular enzyme displaying CMCase activity and comprising a cellulose binding module preceding a catalytic domain homologous to that of the family 51 glycoside hydrolase (28). The ubiquitous detection of the partial celF gene in all Fibrobacter strains studied was probably due to particularly strict conservation of the celF primer sequences, chosen in the N-terminal coding region of EGF, whose function is unknown and which has no homology with any sequence present at this time in the databases. Similar nucleotide conservation was observed with the xynC primers except that no amplification was obtained with F. succinogenes HM2 DNA, suggesting nucleotide substitutions in the HM2 primer targeted sequences or absence of the gene in this strain which may possess homologous xylanases.
The choice of pairs of primers is of particular importance when assessing the distribution of glycoside hydrolase genes in the Fibrobacter genus, and this is illustrated by the fact that, despite the use of several pairs of primers, we did not detect the celE and celG genes in several strains, probably because of mismatches in the sequences targeted by the chosen primers. Similarly, the cel3 gene was not detected in strains HM2 and NR9, whereas the gene was present in other closely related strains (20). This gene may be widespread in Fibrobacter strains, but this was not demonstrated here. Several sets of PCR primers were also necessary to detect homologous xylanase genes in several strains of another ruminal bacterium, Butyrivibrio fibrisolvens (8).
The experimental findings presented here suggest that F. succinogenes strains could be considered a homogeneous group in terms of phylogeny but also because of similar patterns of activities and glycoside hydrolase components. Strains HM2 and FE presented some peculiarities, with more complex zymogram profiles for HM2 and lower glycanase specific activities for FE. A previous study, which compared the polypeptide patterns of 15 strains of F. succinogenes by sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrated homogeneous patterns between the strains (2). However, many of these strains were isolated from the same cow. A previous work demonstrated that the carbon metabolism of strains S85, H, and HM2 of F. succinogenes was very similar (24, 25), including some unusual features, also suggesting marked homogeneity in the carbon metabolism of F. succinogenes (23, 25).
The S85 type strain of F. succinogenes is thus a good model for studying the fibrolytic properties of the species, as the strain has conserved its enzymatic characteristics compared to several phylogenetically close isolates and is representative of recent isolates. Furthermore, our study provide evidence for different enzymatic activities of the F. intestinalis isolate with significantly higher xylanolytic specific activity and extracellular location. This may be related to the origin of the strain, and its function could have been adapted during evolution to the nature of the substrates available in the host digestive tract (monogastric versus polygastric animals).
Acknowledgments
We thank G. Andant for excellent technical assistance in performing the zymograms and quantification of enzyme activities. We thank K.-J. Cheng for the gift of strains H, U, and R and C. Stewart for the gift of strain BL2. We are also grateful to G. Gaudet for helpful discussions.
REFERENCES
- 1.Amann, R. I., C. Lin, R. Key, L. Montgomery, and D. A. Stahl. 1992. Diversity among Fibrobacter isolates: towards a phylogenetic classification. Syst. Appl. Microbiol. 15:23-31. [Google Scholar]
- 2.Begbie, R., and C. S. Stewart. 1984. Polyacrylamide gel electrophoresis of Bacteroides succinogenes. Can. J. Microbiol. 30:863-866. [DOI] [PubMed] [Google Scholar]
- 3.Béra-Maillet, C., V. Broussolle, P. Pristas, J.-P. Girardeau, G. Gaudet, and E. Forano. 2000. Characterisation of endoglucanases EGB and EGC from Fibrobacter succinogenes. Biochim. Biophys. Acta 1476:191-202. [DOI] [PubMed] [Google Scholar]
- 4.Béra-Maillet, C., G. Gaudet, and E. Forano. 2000. Endoglucanase activity and relative expression of glycosyl-hydrolase genes of Fibrobacter succinogenes S85 grown on different substrates. Biochim. Biophys. Acta 1543:77-85. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid sensitive method for the quantification of microgram quantities of protein, utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, M. P., and L. A. Burkey. 1953. Cultural methods and some characteristics of some of the more numerous groups of bacteria in the bovine rumen. J. Dairy Sci. 36:205-217. [Google Scholar]
- 7.Bryant, M. P., and R. N. Doetsch. 1954. A study of actively cellulolytic rod-shaped bacteria of the bovine rumen. J. Dairy Sci. 37:1176-1183. [Google Scholar]
- 8.Dalrymple, B. P., Y. Swadling, I. Layton, K. S. Gobius, and G. P. Xue. 1999. Distribution and evolution of the xylanase genes xynA and xynB and their homologues in strains of Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 65:3660-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehority, B. A. 1993. Microbial ecology of cell wall fermentation, p. 425-453. In H. G. Jung et al. (ed.), Forage cell wall structure and digestibility. American Society for Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, Wis.
- 10.Devillard, E., C. J. Newbold, K. P. Scott, E. Forano, R. J. Wallace, J.-P. Jouany, and H. J. Flint. 1999. A xylanase produced by the rumen anaerobic protozoan Polyplastron multivesiculatum shows close sequence similarity with family 11 xylanases from Gram-positive bacteria. FEMS Microbiol. Lett. 181:145-152. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP — phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Forano, E., V. Broussolle, G. Gaudet, and J. A. Bryant. 1994. Molecular cloning, expression and characterization of a new endoglucanase gene from Fibrobacter succinogenes S85. Curr. Microbiol. 28:7-14. [Google Scholar]
- 13.Forsberg, C. W., J., Malburg, Jr. L. M. Zhu, M. Iyo, A. Cheng, K. J. Krell, and J. P. Philipps. 1994. Cellulases and hemicellulases of Fibrobacter succinogenes and their roles in fibre digestion, p. 125-136. In K. Shimada et al. (ed.), Genetics, biochemistry and ecology of lignocellulose degradation. Unipublishers Co. Proceedings of the MIE Bioforum, Japan.
- 14.Forsberg, C. W., Forano E., and A. Chesson. 2000. Microbial adherence to the plant cell wall and enzymatic hydrolysis. p. 79-97. In N. Casey et al. (ed.), Ruminant physiology: proceedings of the IX International Symposium on Ruminant Physiology. CABI Publishing, Boston, Mass.
- 15.Gogarten, J. P., W. F. Doolittle, and J. G. Lawrence. 2002. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19:2226-2238. [DOI] [PubMed] [Google Scholar]
- 16.Gong, J., and C. W. Forsberg. 1993. Separation of outer and cytoplasmic membranes of Fibrobacter succinogenes and membrane and glycogen granule locations of glycanase and cellobiase. J. Bacteriol. 175:6810-6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hungate, R. E. 1969. A roll-tube method for cultivation of strict anaerobes. Methods Microbiol. 3B:117-132.
- 18.Kopecny, J., M. Zorec, J. Mrazek, Y. Kobayashi, and R. Marinsek-Logar. 2003. Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov., butyrate-producing bacteria from the rumen. Int. J. Syst. Evol. Microbiol. 53:201-209. [DOI] [PubMed] [Google Scholar]
- 19.Krause, D. O., R. J. Bunch, W. J. M Smith, and C. S. McSweeney. 1999. Diversity of Ruminococcus strains: a survey of genetic polymorphisms and plant digestibility. J. Appl. Microbiol. 86:487-495. [Google Scholar]
- 20.Lin, C., and D. A. Stahl. 1995. Comparative analyses reveal a highly conserved endoglucanase in the cellulolytic genus Fibrobacter. J. Bacteriol. 177:2543-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, C., and D. A. Stahl. 1995. Taxon-specific probes for the cellulolytic genus Fibrobacter reveal abundant and novel equine-associated populations. Appl. Environ. Microbiol. 61:1348-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matheron, C., A.-M. Delort, G. Gaudet, and E. Forano. 1996. Simultaneous but differential metabolism of glucose and cellobiose in Fibrobacter succinogenes cells, studied by in vivo 13C-NMR. Can. J. Microbiol. 42:1091-1099. [DOI] [PubMed] [Google Scholar]
- 23.Matheron, C., A.-M. Delort, G. Gaudet, and E. Forano. 1997. Re-investigation of glucose metabolism in Fibrobacter succinogenes, using NMR spectroscopy and enzymatic assays. Evidence for pentose phosphates phosphoketolase and pyruvate formate lyase activities. Biochim. Biophys. Acta 1355:50-60. [DOI] [PubMed] [Google Scholar]
- 24.Matheron, C., A.-M. Delort, G. Gaudet, and E. Forano. 1998. In vivo 13C NMR study of glucose and cellobiose metabolism by four strains of the genus Fibrobacter. Biodegradation 9:451-461. [DOI] [PubMed] [Google Scholar]
- 25.Matheron, C., A-M. Delort, G. Gaudet, E. Forano, and T. Liptaj. 1998. 13C and 1H nuclear magnetic resonance study of glycogen futile cycling in strains of the genus Fibrobacter. Appl. Environ. Microbiol. 64:74-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDermid, K. P., C. R. McKenzie, and C. W. Forsberg. 1990. Esterase activities of Fibrobacter succinogenes subsp. succinogenes S85. Appl. Environ. Microbiol. 56:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miron, J., and D. Ben-Ghedalia. 1993. Digestion of cell-wall monosaccharides of ryegrass and alfalfa hays by the ruminal bacteria Fibrobacter succinogenes and Butyrivibrio fibrisolvens. Can. J. Microbiol. 39:780-786. [DOI] [PubMed] [Google Scholar]
- 28.Mitsumori, M., and H. Minato. 2000. Identification of the cellulose-binding domain of Fibrobacter succinogenes endoglucanase F. FEMS Microbiol. Lett. 183:99-103. [DOI] [PubMed] [Google Scholar]
- 29.Montgomery, L., and J. M. Macy. 1982. Characterization of rat cecum cellulolytic bacteria. Appl. Environ. Microbiol. 44:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pace, N. R., D. A. Stahl, D. J. Lane, and C. J. Olsen. 1986. The use of rRNA sequences to characterize natural microbial populations. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 31.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biol. Sci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 32.Ramsak, A., M. Peterka, K. Tajima, J. C. Martin, J. Wood, M. E. A. Johnston, R. I. Aminov, H. J. Flint, and G. Avgustin. 2000. Unravelling the genetic diversity of ruminal bacteria belonging to the CFB phylum. FEMS Microbiol. Ecol. 33:69-79. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J. S., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Smith, D. C., and C. W. Forsberg. 1991. α-Glucuronidase and other hemicellulase activities in Fibrobacter succinogenes S85 grown on crystalline cellulose or ball-milled barley straw. Appl. Environ. Microbiol. 57:3552-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 36.Stewart, C. S., C. Paniagua, D. Dinsdale, K.-J. Cheng, and S. Garrow. 1981. Selective isolation and characteristics of Bacteroides succinogenes from the rumen of a cow. Appl. Environ. Microbiol. 41:504-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teather, R. M., and J. D. Erfle. 1990. DNA sequence of Fibrobacter succinogenes mixed-linkage β-glucanase (1,3-1,4-β-d-glucan 4-glucanohydrolase) gene. J. Bacteriol. 172:3837-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalainen, K. Lindstrom, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of rhizobia. J. Bacteriol. 185:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Gylswyk, N. O., and J. J. T. K. van der Toorn. 1986. Enumeration of Bacteroides succinogenes in the rumen of sheep fed maize-straw diets. FEMS Microbiol. Ecol. 38:205-209. [Google Scholar]