Significance
Most people infected with Mycobacterium tuberculosis (Mtb) suppress the pathogen’s replication without eradicating it. It is unknown how Mtb survives for decades in a hostile host environment. Respiration of nitrate to nitrite could help Mtb survive in hypoxic tissues but was not thought to be significant at physiologic oxygen tensions, nor was the resultant nitrite considered consequential to Mtb’s physiology. We found that Mtb infecting human macrophages in vitro produces copious nitrite at physiologic oxygen tensions. This slows Mtb’s growth and consumption of ATP and remodels its transcriptome differently than nitric oxide. Thus, respiration of nitrate and adaptation to nitrite are likely to play a prominent role in Mtb’s pathophysiology, whether or not the Mtb resides in hypoxic sites.
Abstract
In high enough concentrations, such as produced by inducible nitric oxide synthase (iNOS), reactive nitrogen species (RNS) can kill Mycobacterium tuberculosis (Mtb). Lesional macrophages in macaques and humans with tuberculosis express iNOS, and mice need iNOS to avoid succumbing rapidly to tuberculosis. However, Mtb’s own ability to produce RNS is rarely considered, perhaps because nitrate reduction to nitrite is only prominent in axenic Mtb cultures at oxygen tensions ≤1%. Here we found that cultures of Mtb-infected human macrophages cultured at physiologic oxygen tensions produced copious nitrite. Surprisingly, the nitrite arose from the Mtb, not the macrophages. Mtb responded to nitrite by ceasing growth; elevating levels of ATP through reduced consumption; and altering the expression of 120 genes associated with adaptation to acid, hypoxia, nitric oxide, oxidative stress, and iron deprivation. The transcriptomic effect of endogenous nitrite was distinct from that of nitric oxide. Thus, whether or not Mtb is hypoxic, the host expresses iNOS, or hypoxia impairs the action of iNOS, Mtb in vivo is likely to encounter RNS by producing nitrite. Endogenous nitrite may slow Mtb’s growth and prepare it to resist host stresses while the pathogen waits for immunopathology to promote its transmission.
The ability to produce reactive nitrogen species (RNS) is widely distributed among animals, plants, and eubacteria. The high-output pathway of RNS production by host inducible nitric oxide synthase (iNOS) contributes to control of diverse infections (1, 2), including tuberculosis (TB) (3, 4), via impacts on both the pathogen and the host (5). Macrophages in the lungs of humans (6, 7) and macaques (8) with TB express functional iNOS. Correlative evidence suggests that the action of iNOS may contribute to human control of TB (1, 3, 9, 10).
Contributions of bacterial RNS production to host–pathogen interactions have only recently come under study. Bacterial NO synthases or nitrate reductases are sources of RNS that can contribute to antibiotic resistance (11, 12) and increase virulence (13). Some commensal bacteria exploit host iNOS during colonic inflammation by using iNOS-derived nitrate (NO3−) as a terminal electron acceptor (14). Furthermore, host-derived RNS have been proposed to enhance the survival of Mtb within macrophages (15). Nitrate, along with nitrite (NO2−), is an autooxidation product of nitric oxide (·NO), the product of iNOS (16).
In the host, Mtb is thought to survive over decades in hypoxic sites (17, 18). When oxygen is scarce, Mtb reduces nitrate (NO3−) to nitrite (NO2−) as a means to maintain redox homeostasis and energy production (19). The mycobacterial respiratory nitrate reductase, encoded by narGHJI, is constitutively expressed (19) and functions at a low level under aerobic conditions when nitrate is taken up by passive diffusion (20, 21). Hypoxia promotes the induction of narK2, a nitrate transporter and a member of the dormancy survival (DOS) regulon (22–24), such that Mtb cultured at ≤1% oxygen in the presence of nitrate rapidly produces nitrite up to a level of 2.5 mM (20). Further production is restricted in association with nonlethal cessation of bacterial growth (20).
Transcripts from narG, which encodes a subunit of the nitrate reductase, and narX, which lies downstream of narK2 and encodes a nonfunctional nitrate reductase, were identified in granulomas from humans with TB (25, 26). This suggests that populations of mycobacteria residing within the human host are exposed to low oxygen tensions and likely respire nitrate to adapt. Nitrate is a physiologic component of human body fluids, where it arises from the diet and from autooxidation of the NO produced not only by iNOS (NOS2) but also by the constitutively expressed enzymes NOS1 and NOS3. Studies in which nitrate was furnished in vitro revealed that Mtb’s ability to respire nitrate allows it to better withstand acid, nitrosative stress (27), or sudden anaerobiosis (28). The evolutionary success of “modern” Mtb strains, which are more prevalent than “ancestral” strains, has been attributed to their enhanced nitrate reductase activity (29, 30). Although nitrate and nitrite reproduce the bioactivity of nitric oxide in vivo in a wide array of physiologic processes (16, 31, 32), reduction of nitrate to nitrite is rarely considered in studies of the host–pathogen relationship in TB. In fact, nitrate was omitted when the standard growth media were formulated in which almost all in vitro experiments with Mtb are currently conducted.
During the course of modifying standard culture conditions for Mtb-infected human monocyte-derived macrophages (33), we observed abundant accumulation of nitrite in the supernatant. Herein we report the identification of Mtb as both the source of the nitrite and a highly responsive target.
Results
Mtb Respires Nitrate in Human macrophages Cultured at Nonhypoxic Oxygen Tensions.
We differentiated normal donors’ monocytes into macrophages under 10% oxygen for 2 wk, activated them with IFN-γ, and infected them with Mtb, as previously described (33). Cultures of macrophages from multiple donors consistently accumulated nitrite in a time-dependent manner to a median level of 25 μM over 5 d (Fig. 1A). This suggested that the macrophages may have expressed one of the three NOS isoforms. However, we detected no iNOS protein by immunoblot (Fig. S1) and no NOS1, NOS2, or NOS3 transcripts by microarray or RNA Seq. To determine whether Mtb might be the source, we infected macrophages with a strain of Mtb lacking narG. The mutant strain grows normally in aerobic culture and is as virulent as WT Mtb in mice (17, 19) but cannot respire nitrate. Macrophage cultures infected with ΔnarG Mtb accumulated no nitrite, whereas infection with a mutant strain complemented with the WT narG allele led to accumulation of as much nitrite as infection with WT Mtb (Fig. 1B). Accumulation of nitrite was nearly abolished when the cultures were washed gently to replace most of the standard culture medium with medium containing negligible nitrate and was restored when the nitrate-deficient cultures were repleted with 5 mM nitrate (Fig. 1C). Even when we cultured infected macrophages in conventional, hyperoxic levels of oxygen (21%), we observed narG-dependent nitrite accumulation in the supernatant (Fig. 1D). To further confirm that the resultant nitrite was a mycobacterial, rather than a host product, we infected macrophages with Mtb in the presence of the bactericidal antibiotic rifampicin. Addition of rifampicin to macrophages 1 d before infection completely abolished nitrite production after infection. Treatment with rifampicin even 1 or 2 d after infection markedly reduced the production of nitrite (Fig. 1E).
Fig. 1.
Nitrite generation by Mtb within primary human macrophages cultured in nonhypoxic levels of oxygen. (A) Accumulation of nitrite in supernatant of macrophages in monolayer culture in 10% oxygen infected with WT (Wt) Mtb (MOI: 5; 5 × 105 bacteria) for the indicated times. Box and whisker plots depict median and 5th-95th percentiles of results from six donors. (B) As in A, but macrophages were uninfected or infected with narG-deficient Mtb (NarG) or WT or complemented (NarGc) strains. Results for uninfected macrophages and those infected with NarG Mtb overlap near zero. Means ± SD from one experiment representative of >10. (C) Lack of accumulation of nitrite in nitrate-depleted medium. Macrophages were infected in the presence (*) or absence of 5 mM nitrate. Means ± SD for one experiment representative of two. (D) As in B, but with macrophages maintained under either 21% or 10% oxygen. Means ± SD for one experiment representative of two. (E) Impact of rifampicin on nitrite production. Where indicated, macrophages were infected with WT Mtb (MOI: 5) for 3 d in 10% oxygen in the presence or absence of rifampicin (1 μg/mL). Rifampicin was added 1 d before (D-1), 1 d after (D1), or 2 d after (D2) infection. Uninfected macrophages were treated with rifampicin for the duration of the experiment. Nitrite was measured on day 3 after infection. Means ± SD, n = 2 experiments. (F) As in B, but using ∼20-fold higher numbers of macrophages grown on Cytodex surface microcarrier beads in 10% oxygen and then infected with Mtb (MOI: 5; 107 bacteria). Means ± SD, n = 2 experiments.
We wondered what levels of Mtb-derived nitrite might be attained in tissue sites such as granulomas, where the number of macrophages per unit volume vastly exceeds the nonphysiologic ratio attainable in monolayer culture. To better simulate the cell densities of tissues, we cultured macrophages on Cytodex microcarrier beads. The narG-dependent accumulation of nitrite in bead cultures reached 800 μM (Fig. 1F). In both monolayer and bead culture, the cfus of Mtb recovered from the cultures were not significantly different for narG-deficient and WT Mtb (34).
Oxygen Tension Within Mtb-Infected Human Macrophages Functionally Approximates 1%.
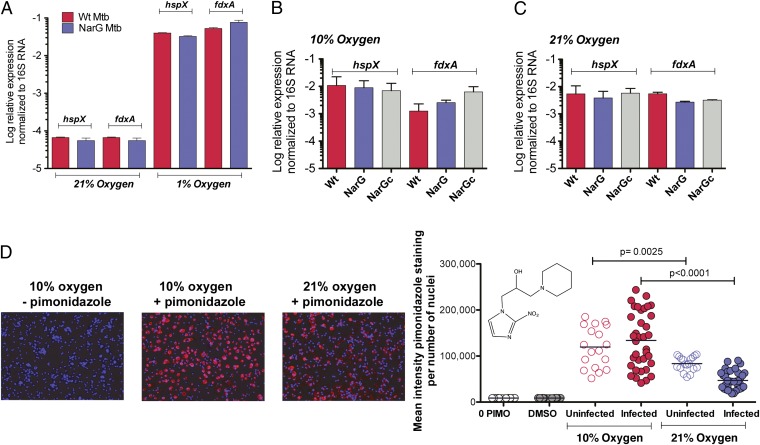
Abundant mycobacterial nitrate reduction within infected macrophages incubated under ambient oxygen tensions of 10% and 21% suggested that Mtb experienced an intracellular oxygen tension that approximated 1%. To test this, we compared the expression of DOS regulon members hspX and fdxA by Mtb cultured axenically or within infected macrophages. As reported previously (23), we found that Mtb cultured axenically in standard medium under 1% oxygen but not 21% oxygen induced both genes by ∼100-fold (Fig. 2A). Mtb residing within macrophages also highly induced the two DOS genes, particularly under 10% oxygen (Fig. 2B) and less so under 21% oxygen (Fig. 2C). To confirm this surprising finding, we treated Mtb-infected macrophages with pimonidazole. At oxygen tensions ≤1%, pimonidazole is reduced to a form that covalently modifies proteins, as detected by immunocytochemistry (35). Macrophages cultured at an oxygen tension of 10% showed intense pimonidazole staining, whether or not they were Mtb-infected (Fig. 2D). Even under hyperoxic conditions (21% oxygen), considerable pimonidazole staining was detected (Fig. 2D). Thus, despite incubation under ambient tensions of 10% or even 21%, intracellular oxygen tensions in the human macrophages functionally approximated 1%.
Fig. 2.
Functional evidence for intracellular hypoxia in primary human macrophages cultured at physiologic tissue level (10%) or hyperoxic (room air) oxygen tensions. (A) Expression of DOS regulon genes hspX and fdxA in axenic cultures of WT (Wt) or narG-deficient (NarG) Mtb in 21% or 1% oxygen in standard medium without nitrate, OD580 0.1 (means ± SD, n = 2 experiments). (B and C) Expression of hspX and fdxA by WT or NarG strains within human macrophages incubated in 21% or 10% oxygen for 10 h (MOI: 40). Means ± SD, n = 2 experiments. (D) Pimonidazole staining of macrophages infected or not with WT Mtb (MOI: 5) and incubated at the oxygen tensions indicated. Photomicrographs (Left): Representative images. Blue, DAPI-stained nuclei; red, pimonidazole adducts. Graph (Right): Quantification of pimonidazole staining intensity. 0 PIMO, uninfected cells treated with neither pimonidazole nor DMSO. DMSO, uninfected cells treated with only the vehicle. Each dot represents the average intensity in ∼500 macrophages. Horizontal lines indicate the means of two independent experiments.
Endogenously Generated Nitrite Represses Mycobacterial Growth and ATP Consumption.
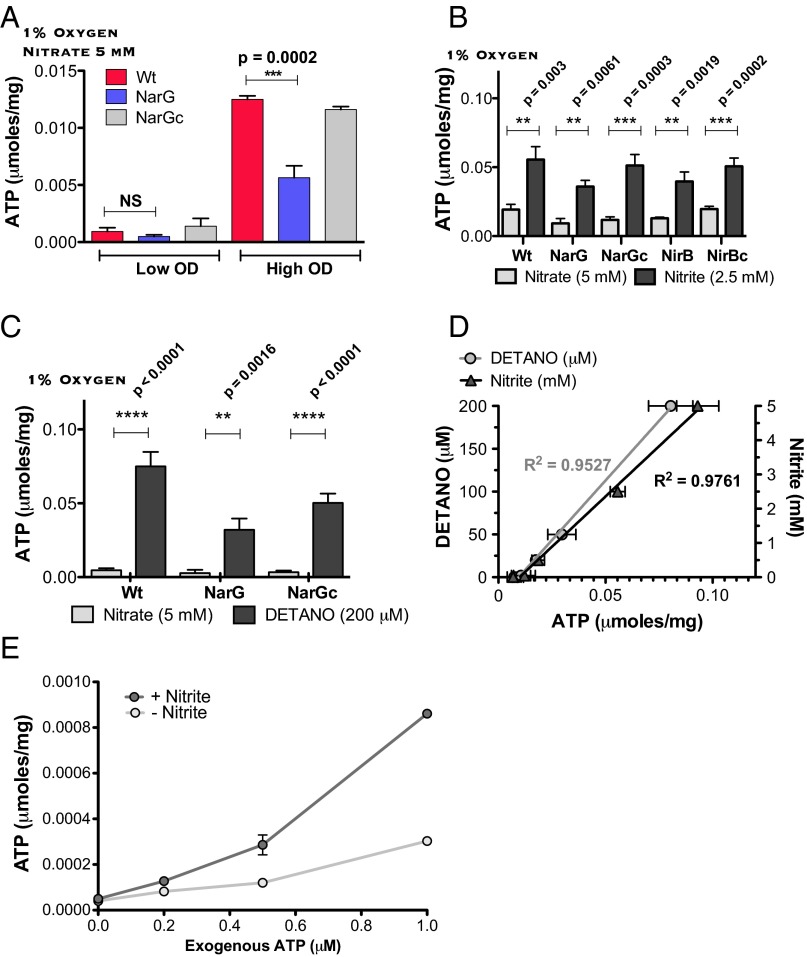
As noted by Wayne and Hayes (20), nitrite, whether endogenously produced or exogenously supplied, suppressed the growth of Mtb under 21% oxygen (Fig. 3A). The concentration-dependence of this effect was similar for WT, narG-deficient, and complemented Mtb (Fig. 3A). In contrast, when we incubated the three strains in 1% oxygen for 3 d with 5 mM nitrate, followed by return to 21% oxygen, only the WT and complemented strains failed to resume growing, reflecting that they generated endogenous nitrite from nitrate, which narG-deficient Mtb could not (Fig. 3B). The failure of cultures of WT Mtb to increase in turbidity after generating nitrite reflected bacteriostasis, not cell death, because the number of cfus recovered was unaffected (Fig. 3C).
Fig. 3.
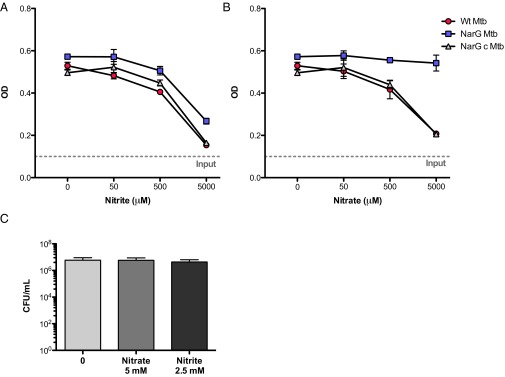
Nitrite represses mycobacterial growth. (A and B) Strains of Mtb designated as in Fig. 1 (OD580 0.1, as marked by horizontal dotted lines) were incubated in the presence of the indicated concentrations of nitrite (A) or nitrate (B) for 3 d in 1% oxygen and then transferred to 21% oxygen for 5 d before growth was assessed by measuring OD580. Means ± SD, n = 2 experiments. (C) As in A and B, except that cfus were determined at the end of the 3-d incubation in 1% oxygen for WT Mtb. Means ± SD, n = 2 experiments.
It is not known how nitrite impairs aerobic mycobacterial growth. We suspected an impact of nitrate respiration on intrabacterial ATP, because nitrite has been shown to inhibit bacterial respiration and prevent ATP synthesis (36). To test this, we cultured WT, narG-deficient, and complemented strains of Mtb axenically for 3 d under 1% oxygen either at high density (OD 0.1, ∼5 × 107 cells/mL) or low density (OD 0.01, ∼5 × 106 cells/mL) in the presence of 5 mM nitrate and measured ATP in their lysates. As expected, in the low-density cultures the concentration of nitrite remained low. Under these conditions, ATP levels in WT and narG-deficient Mtb were not significantly different (Fig. 4A). In contrast, high-density cultures of the WT and complemented strains contained significantly more ATP than the narG-deficient strain, confirming an observation of Tan et al. (27) (Fig. 4A).
Fig. 4.
Endogenously generated or exogenously supplied nitrite elevates ATP in Mtb and suppresses ATP consumption in its lysates. (A) Intrabacterial ATP content measured in WT (Wt), narG-deficient (NarG), or complemented (NarGc) strains of Mtb incubated at high (0.1) or low OD580 (0.01) for 3 d in 1% oxygen in the presence of 5 mM nitrate. Results are expressed per mg protein in the lysate and are means ± SEM from two experiments. (B) Impact of exogenous nitrite or nitrate on ATP content in WT, narG-deficient (NarG), nirB-deficient (NirB), and respective complemented strains (NarGc, NirBc) of Mtb incubated for 3 d in 1% oxygen. (Means ± SEM, n = 3 experiments). (C) As in B, but comparing the impact of single concentrations of nitrate (5 mM) and DETANO (200 μM). Means ± SEM, n = 3 experiments. (D) As in C, but comparing the impact of varied concentrations of nitrite (y axis on the right) and DETANO (y axis on the left). Means ± SEM, n = 2 experiments. (E) Recovery of exogenous ATP added to lysates from WT Mtb that had been incubated in the presence or absence of 2.5 mM nitrite for 3 d at 1% oxygen. ATP was added only after cell lysates had consumed endogenous ATP. Means ± SEM, n = 2 experiments. P values were determined by unpaired t tests.
Tan et al. (27) attributed this result to increased ATP production by Mtb using nitrate as an electron acceptor when oxygen was limiting. However, when we added 2.5 mM exogenous nitrite to low-density cultures of Mtb in 1% oxygen, ATP levels were increased even farther than seen with WT Mtb given twice as much nitrate (Fig. 4B). The ATP-elevating effect of exogenous nitrite was not dependent on the presence of narG or nirBD, which encodes a nitrite reductase (37). Thus, it seemed that nitrite, a product of nitrate respiration, led to accumulation of ATP in Mtb in a manner that did not depend on Mtb’s donation of electrons to nitrate.
To test whether nitrite itself was responsible for this effect, as opposed to nitric oxide, to which nitrite can be converted, we incubated low-density Mtb cultures with 5 mM nitrate as a control or with diethylenetriamine-NO adduct (DETANO), a compound that decomposes to release nitric oxide with a half-life of ∼22 h at pH 7.4. DETANO also elevated intrabacterial ATP levels in WT, narG-deficient and the complemented strains. Both nitrite and DETANO increased intrabacterial ATP in a concentration-dependent manner. DETANO was ∼25-fold more potent than nitrite (Fig. 4 C and D). Thus, either nitrite or nitric oxide potentially arising from it seemed to increase ATP.
We next considered whether nitrite or its possible conversion products might increase ATP production or decrease ATP consumption. Nitrite repressed the growth of Mtb; therefore, we considered it unlikely that nitrite enhanced the synthesis of ATP. Thus, we compared the consumption of exogenous ATP in lysates from nitrite-treated and untreated Mtb, after first demonstrating that neither nitrite nor nitric oxide interfered with the ATP assay (Fig. S2A). WT Mtb was incubated with or without nitrite as described above for 3 d in 1% oxygen. Whole-cell lysates were prepared on ice, moved to room temperature, and allowed to consume endogenous ATP until it was no longer detectable, which required less than 20 min. We then added various amounts of exogenous ATP to the lysates and immediately measured its concentration. We consistently recovered greater amounts of exogenous ATP in lysates from nitrite-treated Mtb than from untreated Mtb (Fig. 4E). Time-course measurements of the disappearance of exogenous ATP (Fig. S2B) supported the interpretation that consumption of ATP was slower in lysates from nitrite-treated Mtb than from untreated Mtb.
Endogenously Generated Nitrite Regulates the Mtb Transcriptome.
Having demonstrated an effect of nitrite on ATP metabolism, we characterized the impact of endogenously generated nitrite more broadly by examining its effect on Mtb gene expression as assessed by RNA Seq. Samples of Mtb were prepared following the same protocol that revealed that mycobacterial nitrate respiration repressed subsequent bacterial growth in air (Fig. 3 A and B). In three independent experiments, we treated axenic cultures of Mtb with or without 5 mM nitrate for 3 d at 1% oxygen to allow for the generation of endogenous nitrite and then transferred the cultures to 21% oxygen overnight. Fig. 5 refers to these samples as “untreated” and “nitrate,” respectively. Two further control cultures were prepared as above, except that one was treated with 2.5 mM nitrite in place of nitrate, and the other was exposed neither to nitrate nor nitrite, but its RNA was collected at the conclusion of the 3-d incubation in 1% oxygen, without an ∼18-h culture in 21% oxygen. Fig. 5 refers to these two samples as “nitrite” and “untreated 1% oxygen,” respectively. Ribosomal RNA was removed from the total RNA by oligonucleotide-based hybridization. Sequences determined in three separate sequencing runs were aligned to the Mtb H37Rv genome (GenBank accession no. NC_000962.3). After culling sequences encoding ribosomal RNA, ∼3 × 106 reads per sample were mapped to coding regions, covering ∼99.9% of the coding genome. Gene expression was reported as reads per kilobase per million mapped reads (RPKM).
Fig. 5.
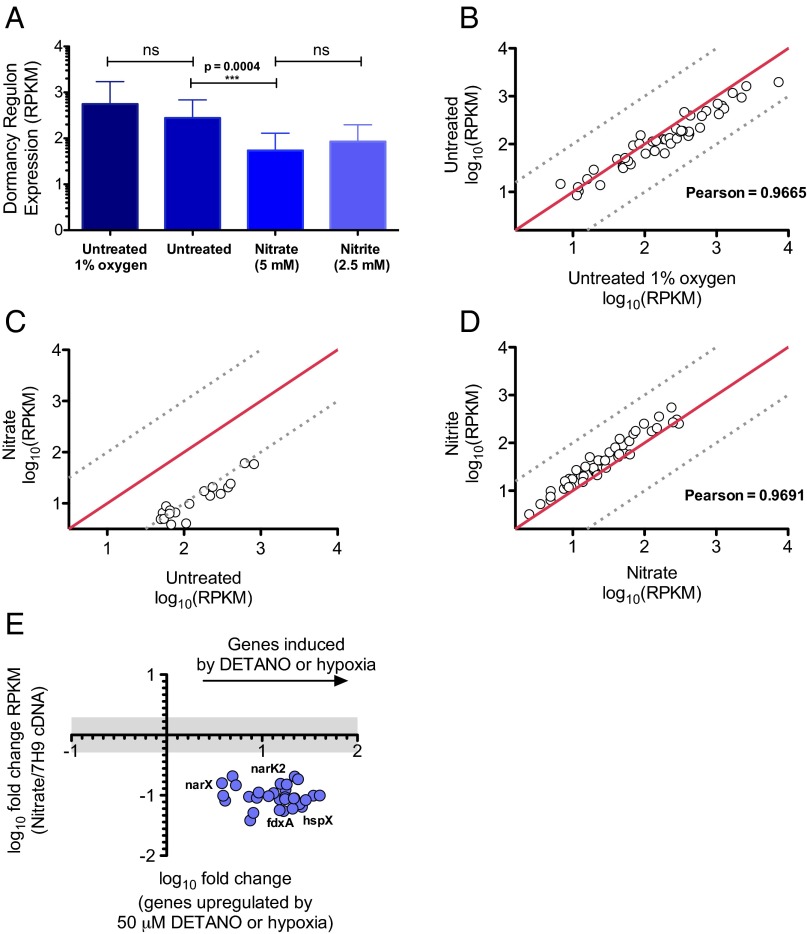
Respiration of nitrate represses expression of the DOS regulon in Mtb. (A–D) RPKM for DOS genes as determined by RNA Seq for WT Mtb incubated in 1% oxygen for 3 d (“untreated 1% oxygen”) or incubated in 1% oxygen for 3 d in the absence (“untreated”) or presence of 5 mM nitrate (“nitrate”) or 2.5 mM nitrite (“nitrite”), as indicated, and transferred to 21% oxygen overnight. (A) The average expression of all 47 DOS genes across all samples (means ± SD) for the “untreated” and “nitrate” samples in 3 independent samples per condition, for the “untreated 1% oxygen ” and “nitrite” samples (n = 1 experiment per condition; Methods). The P value was determined by an unpaired t test. (B) Gene-by-gene comparison of expression in the “untreated 1% oxygen” sample and the “untreated” sample. (C) Expression of the DOS genes that were significantly regulated by nitrate respiration compared with expression of each such gene in the untreated sample. (D) Expression of each DOS gene from WT type Mtb treated with 5 mM nitrate compared with a sample treated with 2.5 mM nitrite. (B–D) Red lines indicate 1:1 correlation; dashed lines indicate a 20% deviation from a 1:1 correlation. (E) Comparison of the expression of individual genes significantly regulated by nitrate in these experiments, with their reported regulation in hypoxia (23) or upon exposure to DETANO as a source of nitric oxide (24). The x axis indicates the regulation of shared genes by the condition of interest, and the y axis indicates their regulation by nitrate respiration. All y values above or below the gray bar were regulated greater than twofold.
We found that 120 genes were significantly regulated by provision of nitrate. Those induced are listed in Table S1, and those repressed are listed in Table S2. Inspection of the list suggested that the most informative comparisons would be with reports of Mtb’s transcriptional adaptation to host-imposed stresses (38), such as hypoxia (23), iron restriction (39), acid (pH 5.5) (40), hydrogen peroxide (10 mM) (41), nitric oxide (50 mm) (24), and residence in mouse macrophages (40, 42). Genes regulated by nitrate respiration but not by any other reported stimulus are listed in Table S3.
We first analyzed expression of the DOS regulon, which is induced by three of these conditions—hypoxia (23), nitric oxide (24), and residence in macrophages (40, 42). The average expression of all 47 DOS regulon genes was nearly the same for the “untreated” and “untreated 1% oxygen” samples (Fig. 5A, first two columns). The close correspondence of DOS induction in these two conditions was also evident for each DOS gene considered individually (Fig. 5B).
Surprisingly, in comparison with the untreated sample, the average expression of the DOS regulon was significantly repressed by provision of nitrate (P < 0.0004) (Fig. 5A). Individually, 27 DOS genes were significantly repressed in nitrate-treated Mtb (Fig. 5C). (To achieve statistical significance for all 47 DOS genes considered individually, we would likely need to sequence more samples.) This effect of nitrate could be attributed to generation of nitrite, because the effect was qualitatively and quantitatively the same when nitrite itself was provided (Fig. 5D). Finally, the effect of nitrite could not be ascribed to conversion to nitric oxide, in that nitric oxide has the opposite effect: it induces the DOS regulon, rather than repressing it. This is shown in Fig. 5E, which plots the present results for 28 DOS genes: 27 that were significantly repressed by nitrite and reported to be induced by hypoxia (23) and 28 that were significantly repressed by nitrite and reported to be induced by 50 mM nitric oxide (24). Hypoxic cultures of Mtb have been reported to accumulate up to 2.5 mM nitrite and no more (20). These results offer an explanation: nitrite negatively regulates its own accumulation by limiting the expression of the DOS regulon and thus narK2 (Fig. S3).
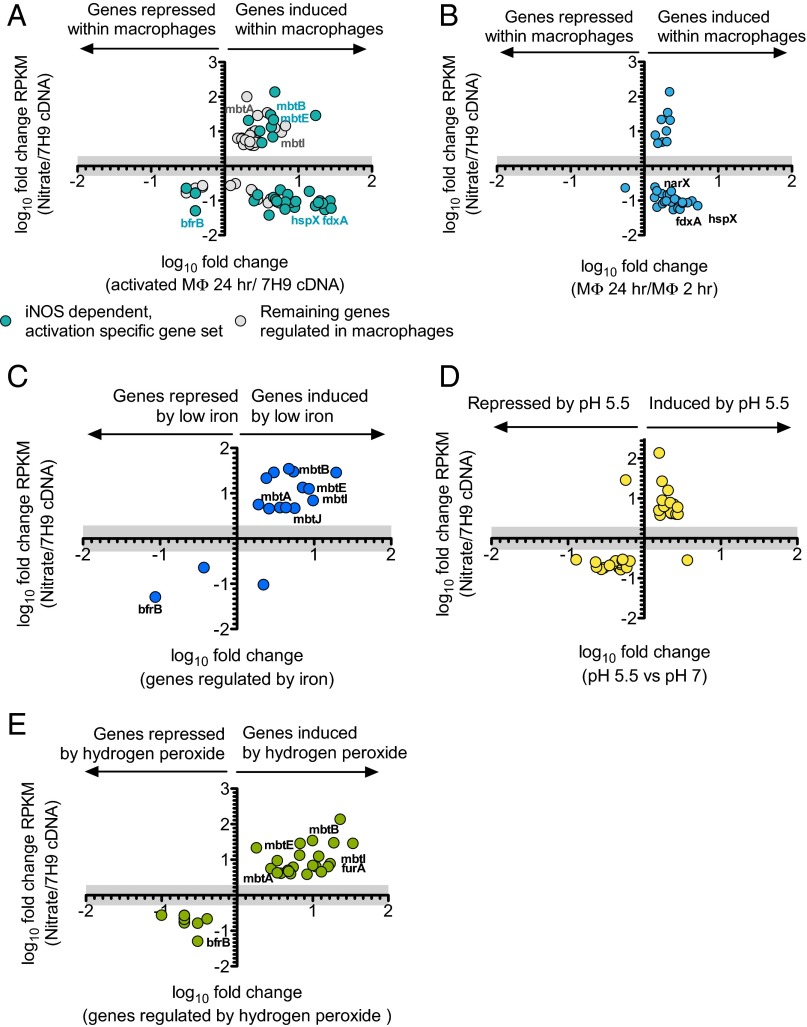
Of the 120 genes that were significantly regulated by nitrate respiration, many overlapped with genes that were regulated by Mtb residing within activated mouse macrophages (40, 42) (Fig. 6 A and B). In addition, Schnappinger et al. (42) reported a subset of genes that were specifically regulated by iNOS. However, there was no correlation between regulation of genes by Mtb’s respiration of nitrate and those regulated specifically by Mtb’s residence in macrophages that expressed iNOS (Fig. 6A). This provided further evidence that the effect of endogenously generated nitrite was not solely mediated by conversion to nitric oxide.
Fig. 6.
Comparison of the impact of nitrate respiration on the Mtb transcriptome with the impact of (A and B) residence within macrophages, (C) iron restriction, (D) exposure to pH 5.5, or (E) treatment with 10 mM hydrogen peroxide. Results of the present experiments and literature reports are graphed as in Fig. 5E. The literature results were from the following studies: (A) (42), (B) (40), (C) (39), (D) (40), and (E) (41).
Many Mtb genes regulated by nitrate respiration were reported to be similarly regulated in Mtb exposed to iron restriction (39) (Fig. 6C), pH 5.5 (40) (Fig. 6D), or treatment with 10 mM hydrogen peroxide (41) (Fig. 6E). Table 1 presents the total number of genes regulated by nitrate respiration and reported to be regulated by these and other physiologic stimuli. After standardization to account for gene lists of differing size, the comparison indicates the number of genes expected to overlap by chance and whether the overlap is statistically significant. This analysis suggested that nitrate regulates the expression of genes that are also regulated when Mtb is exposed to several host-imposed stresses.
Table 1.
Comparison of regulation of expression of Mtb genes by nitrate respiration observed here and by physiologic situations or stresses as reported in the literature
| Condition | Regulation | Genes regulated by the indicated condition | Total no. that overlap (by chance) |
No. induced by nitrate respiration | No. repressed by nitrate respiration | Percent of total gene list that overlaps | Significance of the overlap, P value | |
| Iron depletion (39) (low iron culture: 2 μM FeCl3 vs. high iron culture: 50 μM FeCl3, H37Rv) | Induced | 66 | 14 | (2) | 13 | 1 | 11.67 | 4.17E-08 |
| Repressed | 27 | 2 | (1) | 0 | 2 | 1.67 | 2.05E-01 | |
| Hypoxia (22) (2 h at 0.2% oxygen, H37Rv) | Induced | 47 | 27 | (1) | 0 | 27 | 22.50 | <2.2e-16 |
| Enduring hypoxic response (62) (incubation at 0.2% oxygen over 4 d, H37Rv) | Induced | 230 | 17 | (7) | 12 | 5 | 14.17 | 6.53E-04 |
| Nitric oxide (23) (50 μM DETA/NO for 40 min, H37Rv) | Induced | 48 | 28 | (1) | 0 | 28 | 23.33 | <2.2e-16 |
| Hydrogen peroxide (41) (10 mM, 40 min, clinical isolate 1254) | Induced | 166 | 22 | (5) | 22 | 0 | 18.33 | 1.11E-08 |
| Repressed | 48 | 10 | (1) | 10 | 0 | 8.33 | 4.65E-06 | |
| pH 5.5 (40) (pH 5.5 vs. pH 7, H37Rv) | Induced | 261 | 16 | (8) | 15 | 1 | 13.33 | 5.72E-03 |
| Repressed | 375 | 18 | (11) | 1 | 17 | 15.00 | 3.27E-02 | |
| Intraphagosomal (42) (24 h, H37Rv) | Induced | 454 | 65 | (14) | 35 | 30 | 54.17 | <2.2e-16 |
| Induced, iNOS dependent | 60 | 32 | (2) | 9 | 23 | 26.67 | <2.2e-16 | |
| Repressed | 147 | 11 | (4) | 0 | 11 | 9.17 | 5.63E-03 | |
| Repressed, iNOS dependent | 8 | 3 | (1) | 0 | 3 | 2.50 | 3.31E-03 | |
| Intraphagosomal (40) (24 h, CDC 1551) | Induced | 266 | 32 | (8) | 9 | 23 | 26.67 | 2.39E-11 |
| Repressed | 111 | 1 | (3) | 0 | 1 | 0.83 | 9.65E-01 | |
| DNA damage (63) (induced by mitomycin C, H37Rv) | Induced | 113 | 1 | (3) | 1 | 0 | 0.83 | 9.67E-01 |
| Repressed | 26 | 2 | (1) | 2 | 0 | 1.67 | 1.94E-01 | |
| Starvation (64) (24 h after growth in PBS, H37Rv) | Induced | 252 | 6 | (8) | 6 | 0 | 5.00 | 7.69E-01 |
| Repressed | 301 | 12 | (9) | 6 | 6 | 10.00 | 1.94E-01 | |
| Nitrate respiration (Table S3) (genes regulated by nitrate respiration and no other physiologic stimuli reported above) | — | — | — | — | 6 | 5 | 10.83 | — |
The total number of genes induced or repressed by nitrate respiration is listed along with the total number reported to be regulated by the indicated condition. The number regulated in common is compared with the number that would be expected to show a similar regulation by chance calculated by a Fisher’s exact test. The columns labeled “No. induced/repressed by nitrate respiration” refer only to the number of overlapping genes. “Percent of total gene list that overlaps” indicates the percentage of all genes regulated by nitrate respiration that are also regulated by the condition of interest. The associated significance of this overlap, listed as a P value in the final column, was calculated using a Fisher’s exact test. Of note, genes regulated by mitomycin C-induced DNA damage and starvation did not significantly overlap with genes regulated by nitrate respiration. –, not applicable.
Discussion
Within the host, both host NOSs and mycobacterial nitrate reductase can serve as sources of nitrite. However, whereas mycobacterial nitrate reduction is markedly induced by hypoxia (19), NOSs require oxygen as a substrate (43), and at oxygen tensions of 1%, iNOS loses 80–90% of its activity (44). Therefore, the activities of host iNOS and mycobacterial nitrate reductase could be spatially and temporally segregated, depending on their access to molecular oxygen.
The studies reported here led to four interrelated findings. First, there can be a striking dissociation between the oxygen tension in culture and the functional oxygen concentration in macrophage phagosomes. Although a dissociation was previously noted between oxygen concentration in extracellular fluid and in mouse macrophage phagosomes (45), to our knowledge this is the first report that the level of oxygen in the phagosomes of primary human macrophages cultured at physiologic tissue oxygen levels of 10% or even 21% approximates 1% functionally, as reflected by Mtb’s induction of DOS genes and copious reduction of nitrate. The low functional level of intramacrophage oxygen was not dependent on ingestion of Mtb. In agreement with James et al. (45), we speculate that the steep gradient between atmospheric and intracellular oxygen reflects a rate of mitochondrial oxygen consumption that outpaces diffusion from the gas phase through the extracellular fluid and across the plasma membrane. If Mtb likewise experiences functional hypoxia within human macrophages at physiologic oxygen tensions in vivo, then Mtb’s respiration of nitrate to nitrite is likely to be much more widespread than previously appreciated, and the convention of culturing Mtb in room air levels of oxygen in the absence of nitrate is likely to be misleading with regard to Mtb’s physiology.
Second, nitrite, whether endogenously produced or exogenously supplied, affects Mtb profoundly, although we may have only scratched the surface in characterizing its effects. Besides halting Mtb’s growth, impairing its consumption of ATP, and altering its transcriptome, mycobacterial nitrite also markedly reduces Mtb’s susceptibility to isoniazid (34). We do not know the major ATP-consuming pathway(s) in Mtb that function(s) in whole-cell lysates and that nitrite inhibits. This contrasts with the reported effect of nitrite to reduce the amount of intrabacterial ATP in Pseudomonas aeruginosa (36).
Third, the effects of nitrite and nitric oxide partially overlap but in some respects are diametrically opposed. The effect of nitrite is also distinct from the effect of the respiration of nitrate, from which the nitrite arises. The ability of nitrite to affect biologic systems in its own right, rather than through conversion to more reactive forms, has been proposed (46) and is supported by these results. For example, nitrite can oxidize ferrous iron to the ferric state and displace iron from iron–sulfur clusters (47, 48). The induced expression of mycobactin synthase genes suggests that Mtb-derived nitrite inactivated iron-containing enzymes, rendering the Mtb functionally iron deficient despite the high iron content of the medium. DOS regulon expression is coordinated by the heme-containing kinases DosS and DosT, which function as redox and hypoxia sensors, respectively. Hypoxia, carbon monoxide, and nitric oxide are physiologic ligands that induce DOS regulon expression by modulating the oxidation and ligation states of the iron atoms in DosS and DosT (49). Nitrite may repress DOS regulon expression by oxidizing the iron of these sensor kinases in a manner that prevents their regulation by other physiologic ligands. Oxidation of iron in additional enzymes, such as ribonucleotide reductase, may also contribute to the ability of nitrite to slow or halt Mtb’s growth (50–52).
Last, the shared regulation of genes by nitrate and by the potentially mycobactericidal conditions of iron restriction and exposure to acid and hydrogen peroxide suggests that nitrate respiration may benefit Mtb in vivo by preparing it to withstand the latter stresses. Nitrite is highly diffusible, as evidenced by its accumulation in the plasma of Mtb-infected individuals (53, 54). Endogenously produced nitrite may diffuse within infected tissue and contribute to a preemptive induction of Mtb genes that benefit Mtb’s survival in a hostile host environment.
To date, in vivo experiments to investigate the consequence of mycobacterial nitrate respiration have been limited to the mouse, where “granulomas” are areas of pneumonitic consolidation lacking the layered, fibrous-capped structure found in the granulomas of humans and nonhuman primates with TB. Mtb-infected granulomatous tissue in the mouse had a measured oxygen tension averaging ∼40 mm Hg (or 2–9% oxygen) (17) and did not stain with a hypoxia-sensitive 2-nitroimidazole probe (55). Furthermore, Mtb in mice did not express the only DOS regulon member tested, narK2, until the onset of the adaptive immune response almost 3 wk after infection, such that DOS regulon expression may have depended on the activation of iNOS (56) rather than hypoxia. When mouse macrophages cultured in 21% oxygen were infected with Mtb in vitro, DOS regulon induction depended on expression of iNOS (42). In contrast, in the present studies, exposure of human macrophages to Mtb over an interval long enough to allow for phagocytosis but too brief to allow for nitrite accumulation led to marked induction of the DOS regulon in the absence of any NOS. Therefore, to the extent that DOS regulon induction is an indicator of hypoxia, human macrophages as cultured here seem to achieve a greater degree of functional intracellular hypoxia in vitro than mouse macrophages as usually cultured. The same may be true in vivo, insofar as iNOS expressed in mouse macrophages during Mtb infection seems to access sufficient oxygen to restrain Mtb growth (3, 42). Therefore, experiments with NarG-deficient Mtb in the mouse may not reflect the functional relevance of mycobacterial nitrate respiration in the human host.
RNS can act as double-edged swords, depending on their level (57). The present results bring this truism into sharp relief with respect to Mtb. High levels of RNS can kill Mtb, but low levels may do the pathogen a service. First, nitrite of host or mycobacterial origin may induce genes with whose help Mtb can withstand other host stresses. Second, nitrite slows or stops Mtb’s aerobic replication. Mtb may thus avoid killing a host that has not yet destroyed enough lung tissue to provide Mtb with a path back to the air. Third, nitrite increases intrabacterial ATP. Once Mtb is expelled from the host within aerosolized droplets, this reserve of metabolic energy may facilitate colonization of new hosts. Drugs that inhibit Mtb’s defenses against RNS may therefore be useful even in settings where immunodeficiency impairs the expression of host iNOS or hypoxia limits its action.
Methods
Isolation and Differentiation of Primary Human Monocytes.
Heparinized peripheral blood was collected by venepuncture from healthy human donors who provided informed consent under an institutional review board-approved protocol. Monocytes were isolated and differentiated as previously described (33). Briefly, mononuclear cells isolated by centrifugation over Ficoll-Paque (GE Healthcare) were subject to positive selection using magnetic beads coupled to anti-CD14 antibodies (Milteyi Biotec) and cultured in 60% (vol/vol) RPMI, supplemented with 1% glutamax, 40% (vol/vol) human plasma, and GM-CSF and TNF-α (0.5 ng mL−1 each) at 5 × 105 cells mL−1. Thirty percent of the total culture volume was replaced with fresh medium and cytokines twice per week. For Cytodex1 bead culture, Cytodex 1 beads were prepared according to the instructions provided by the manufacturer (GE Healthcare). The cells were plated at a density of 2 × 106 cells mL−1 per well of a 24-well plate, and 50% of the medium was replaced three times per week. After 2 wk at 10% or 21% O2 and 5% CO2 at 37 °C in a humidified atmosphere, the cells were activated with IFN-γ (5 ng mL−1) and infected with Mtb the following day. No antibiotics were used at any point.
To remove most nitrate from the medium, 1 d before infection the cells were washed three times with PBS, and the medium was replaced with DMEM with 10% human plasma, GM-CSF, 0.5 ng mL−1 TNF-α, and 5 ng mL−1 IFN-γ.
Measurement of Nitrite.
Nitrite levels in the supernatants of macrophage cultures were measured by the Griess assay (58) using nitrite standards prepared in the same medium used to culture the macrophages.
Mtb.
M. tuberculosis H37Rv was grown in Middlebrook 7H9 broth supplemented with 0.2% glycerol, 0.5% BSA, 0.2% dextrose, and 0.085% NaCl (ADNaCl) with 0.05% Tween 80. The narG, nirB, and complemented strains were generously provided by F. Bange, Department of Medical Microbiology and Hospital Epidemiology, Medical School Hannover, Hannover, Germany (59). Single-cell suspensions were collected in the supernatant after centrifugation at 120 × g for 10 min. For macrophage infection, ∼2 × 107 bacteria were pelleted by centrifugation and resuspended in PBS. Cfus were determined by serial dilution in 0.1% Triton X-100 and plating on Middlebrook 7H11 agar with 10% oleic acid-albumin-dextrose-catalase enrichment (Difco, BD) supplemented with 0.5% glycerol for 2–3 wk at 37 °C, 5% CO2.
Pimonidazole Staining.
Uninfected or Mtb-infected [multiplicity of infection (MOI): 5] macrophages were treated with or without 200 mM pimonidazole for 24 h, washed, and stained with anti-pimonidazole antibody coupled to allophycocyanin (APC) (Hypoxyprobe-RedAPC Kit) as recommended by the manufacturer and counterstained with the nuclear stain DAPI. APC and DAPI signals were quantified at the Cell Screening Core Facility, Weill Cornell Medical College and analyzed using MetaXpress High Content Image Acquisition & Analysis Software (Molecular Devices). The APC signal was normalized to the number of nuclei per image.
Intrabacterial ATP.
Single-cell suspensions of Mtb (7 × 108 per condition) were incubated in 7H9/ADNaCl with 0.05% Tween 80 at an OD580 of 0.07 in 1% O2, 5% CO2 at 37 °C in a humidified atmosphere, pelleted by centrifugation, washed once in PBS, pelleted, and processed immediately for ATP measurement or frozen at −80 °C for later use. For a single endpoint measurement, the pellet was resuspended in 220 μL of standard lysis buffer: 100 mM Tris·OH and 1 mM phenylmethylsulfonyl fluoride (pH 7.4), with 1× protease inhibitor (Roche: cOmplete ULTRA Tablets, Mini, EDTA-free, EASYpack catalog no. 05892791001) added immediately before bacterial lysis in a bead-beating homogenizer with silica beads for two cycles with incubation on ice between them. After brief centrifugation to pellet silica beads, 1:1:2 (vol/vol/vol) ratio of cell lysate (50 mL), lysis buffer, and ATP reagent was added in triplicate to a black 96-well opaque plate for the BacTiter-Glo Microbial Cell Viability Assay (Promega). Luminescence was read immediately using a SpectraMax L Luminescence Microplate Reader (Molecular Devices). For rate measurements, the lysate was filter-sterilized (0.22 μm Corning Costar Spin-X Plastic Centrifuge Tube Filters at 10,000 × g for 20–30 min). ATP was measured in an aliquot, and reagent ATP was added to the remaining lysate, from which triplicate 10-uL aliquots were immediately dispensed to the assay plate. At the indicated times, 90 μL of lysis buffer and 100 μL of ATP reagent were added and luminescence measured. ATP standards diluted in lysis buffer were prepared fresh for each experiment. The intrabacterial ATP for each sample was normalized to the protein concentration of each sample (Bio-Rad Protein Assay).
Quantitative RT-PCR.
Macrophages (5 × 106 per T25 flask) were cultured in 10% or 21% oxygen and infected with Mtb (MOI: 40) for 10 h. The monolayer was washed twice with PBS. TRIzol (2 mL) was added to each flask and the cells detached using a rubber scraper. Alternatively, ≥2 × 109 bacteria were incubated per condition in 1% or 21% oxygen for 3 d in standard medium, in the absence of nitrate. An equal volume of buffer containing 5 M guanidinium thiocyanate, 25 mM sodium citrate, 20 mM N-lauryl-sarcosine, and 0.7% (vol/vol) β-mercaptoethanol was added. The bacteria were pelleted by centrifugation, 1 mL of TRIzol added per sample, and the suspension beaten with silica beads. RNA extraction was conducted using an RNeasy kit (Qiagen) in accordance with the manufacturer’s instructions, except that off-column DNase digestion was performed for 2 h at 37 °C. One mg of total RNA was reverse-transcribed using the GeneAmp RNA PCR Kit (Applied Biosystems). Quantitative RT-PCR was performed using gene-specific primers (TaqMan Gene Expression Assay, Life Technologies) and the SuperScript III Platinum Two Step qRT-PCR Kit (Life Technologies) with a 7900HT Fast Real Time PCR System (Applied Biosystems). Each experiment was performed in triplicate. Values within the linear range of the primers were normalized to values for 16S rRNA.
RNA Sequencing.
At least 5 × 109 bacteria per sample were incubated in 1% oxygen and left untreated or treated with 5 mM nitrate or 2.5 mM nitrite. After 3 d RNA was collected from one flask of untreated Mtb. The remaining flasks (untreated or nitrite- treated) were transferred to 21% oxygen for an additional overnight incubation before RNA collection. RNA samples with an RNA Integrity Number [Bioanalyzer (Agilent Technologies 2100)] value >8 were processed further. Ribosomal RNA was removed by hybridization with magnetic bead-coupled oligonucleotides (MicrobExpress Kit, Life Technologies). Libraries were prepared according to the manufacturer’s instructions and resultant RNA sequenced (HiSeq2000/1000, Illumina). Single-ended sequencing reads were aligned to the reference genome of Mtb H37Rv using the Burrows-Wheeler Alignment tool (60) (GenBank accession no. NC_000962.3). Cufflinks (61) was used to measure transcript abundances in RPKM as well as to find differentially expressed genes. Regulation was considered significant for changes with a P value corrected for multiple comparisons (Pcorr) <0.05 (Table S1 and S2). The samples labeled “nitrate” and “untreated” were prepared in three independent experiments. Only one sample was available for the “nitrite” and “untreated 1% oxygen” conditions. These samples were only used to evaluate the expression all 47 genes of the DOS regulon as a unit, with the expression of each gene serving as a control for the expression of the others.
Statistical Analysis.
Statistical analysis was performed as indicated in the figure legends using Prism 5.0f for Macintosh (GraphPad Software).
Supplementary Material
Acknowledgments
We thank the blood donors for their participation; Guillaume Vogt for his initial observation of nitrite accumulation; Franz Bange for generously providing Mtb strains; Jenny Xiang and the Weill Cornell Genomics Resources Core Facility for assistance sequencing mycobacterial RNA; and Kristin Burns, Ben Gold, Ruslana Bryk, Sabine Ehrt, Dirk Schnappinger, Poonam Rath, and Michele Fuortes for advice. This study was supported by the Milstein Program in Chemical Biology of Infectious Disease. A.C.-B. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health under Award T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. The Weill Cornell Medical College Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper (RNA seq data on Mycobacterium tuberculosis) have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE51037).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316894110/-/DCSupplemental.
References
- 1.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97(16):8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 3.MacMicking JD, et al. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci USA. 1997;94(10):5243–5248. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175(4):1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra BB, et al. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1β. Nat Immunol. 2013;14(1):52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicholson S, et al. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183(5):2293–2302. [Google Scholar]
- 7.Choi HS, Rai PR, Chu HW, Cool C, Chan ED. Analysis of nitric oxide synthase and nitrotyrosine expression in human pulmonary tuberculosis. Am J Respir Crit Care Med. 2002;166(2):178–186. doi: 10.1164/rccm.2201023. [DOI] [PubMed] [Google Scholar]
- 8.Mattila JT, et al. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J Immunol. 2013;191(2):773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schön T, et al. Arginine as an adjuvant to chemotherapy improves clinical outcome in active tuberculosis. Eur Respir J. 2003;21(3):483–488. doi: 10.1183/09031936.03.00090702. [DOI] [PubMed] [Google Scholar]
- 10.Schön T, et al. Effects of a food supplement rich in arginine in patients with smear positive pulmonary tuberculosis—a randomised trial. Tuberculosis (Edinb) 2011;91(5):370–377. doi: 10.1016/j.tube.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325(5946):1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCollister BD, Hoffman M, Husain M, Vázquez-Torres A. Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob Agents Chemother. 2011;55(5):2189–2196. doi: 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Sorge NM, et al. Methicillin-resistant Staphylococcus aureus bacterial nitric-oxide synthase affects antibiotic sensitivity and skin abscess development. J Biol Chem. 2013;288(9):6417–6426. doi: 10.1074/jbc.M112.448738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467(7314):426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung JY, et al. The intracellular environment of human macrophages that produce nitric oxide promotes growth of mycobacteria. Infect Immun. 2013;81(9):3198–3209. doi: 10.1128/IAI.00611-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundberg JO, Weitzberg E, Cole JA, Benjamin N. Nitrate, bacteria and human health. Nat Rev Microbiol. 2004;2(7):593–602. doi: 10.1038/nrmicro929. [DOI] [PubMed] [Google Scholar]
- 17.Aly S, et al. Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J Pathol. 2006;210(3):298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 18.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76(6):2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sohaskey CD, Wayne LG. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J Bacteriol. 2003;185(24):7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wayne LG, Hayes LG. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber Lung Dis. 1998;79(2):127–132. doi: 10.1054/tuld.1998.0015. [DOI] [PubMed] [Google Scholar]
- 21.Wayne LG, Doubek JR. Classification and identification of mycobacteria. II. Tests employing nitrate and nitrite as substrate. Am Rev Respir Dis. 1965;91:738–745. doi: 10.1164/arrd.1965.91.5.738. [DOI] [PubMed] [Google Scholar]
- 22.Giffin MM, Raab RW, Morganstern M, Sohaskey CD. Mutational analysis of the respiratory nitrate transporter NarK2 of Mycobacterium tuberculosis. PLoS ONE. 2012;7(9):e45459. doi: 10.1371/journal.pone.0045459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman DR, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci USA. 2001;98(13):7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198(5):705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rachman H, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74(2):1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenhalls G, et al. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect Immun. 2002;70(11):6330–6338. doi: 10.1128/IAI.70.11.6330-6338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan MP, et al. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS ONE. 2010;5(10):e13356. doi: 10.1371/journal.pone.0013356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sohaskey CD. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J Bacteriol. 2008;190(8):2981–2986. doi: 10.1128/JB.01857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goh KS, Rastogi N, Berchel M, Huard RC, Sola C. Molecular evolutionary history of tubercle bacilli assessed by study of the polymorphic nucleotide within the nitrate reductase (narGHJI) operon promoter. J Clin Microbiol. 2005;43(8):4010–4014. doi: 10.1128/JCM.43.8.4010-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Homolka S, Niemann S, Russell DG, Rohde KH. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: Delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog. 2010;6(7):e1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlström M, et al. Dietary inorganic nitrate reverses features of metabolic syndrome in endothelial nitric oxide synthase-deficient mice. Proc Natl Acad Sci USA. 2010;107(41):17716–17720. doi: 10.1073/pnas.1008872107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsen FJ, Ekblom B, Sahlin K, Lundberg JO, Weitzberg E. Effects of dietary nitrate on blood pressure in healthy volunteers. N Engl J Med. 2006;355(26):2792–2793. doi: 10.1056/NEJMc062800. [DOI] [PubMed] [Google Scholar]
- 33.Vogt G, Nathan C. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J Clin Invest. 2011;121(10):3889–3901. doi: 10.1172/JCI57235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham-Bussel A, Bange FC, Nathan CF. Nitrite impacts the survival of Mycobacterium tuberculosis in response to isoniazid and hydrogen peroxide. Microbiologyopen. 2013 doi: 10.1002/mbo3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arteel GE, Thurman RG, Raleigh JA. Reductive metabolism of the hypoxia marker pimonidazole is regulated by oxygen tension independent of the pyridine nucleotide redox state. Eur J Biochem. 1998;253(3):743–750. doi: 10.1046/j.1432-1327.1998.2530743.x. [DOI] [PubMed] [Google Scholar]
- 36.Rowe JJ, Yarbrough JM, Rake JB, Eagon RG. Nitrite inhibition of aerobic bacteria. Curr Microbiol. 1979;2(1):51–54. [Google Scholar]
- 37.Malm S, et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology. 2009;155(Pt 4):1332–1339. doi: 10.1099/mic.0.023275-0. [DOI] [PubMed] [Google Scholar]
- 38.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: Defence against host stresses. Cell Microbiol. 2009;11(8):1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in mycobacterium tuberculosis: Role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70(7):3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: Linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2(5):352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011;2:105. doi: 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnappinger D, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J Exp Med. 2003;198(5):693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: Structure, function and inhibition. Biochem J. 2001;357(Pt 3):593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCormick CC, Li WP, Calero M. Oxygen tension limits nitric oxide synthesis by activated macrophages. Biochem J. 2000;350(Pt 3):709–716. [PMC free article] [PubMed] [Google Scholar]
- 45.James PE, Grinberg OY, Michaels G, Swartz HM. Intraphagosomal oxygen in stimulated macrophages. J Cell Physiol. 1995;163(2):241–247. doi: 10.1002/jcp.1041630204. [DOI] [PubMed] [Google Scholar]
- 46.Bryan NS, et al. Nitrite is a signaling molecule and regulator of gene expression in mammalian tissues. Nat Chem Biol. 2005;1(5):290–297. doi: 10.1038/nchembio734. [DOI] [PubMed] [Google Scholar]
- 47.Wright RO, Lewander WJ, Woolf AD. Methemoglobinemia: Etiology, pharmacology, and clinical management. Ann Emerg Med. 1999;34(5):646–656. doi: 10.1016/s0196-0644(99)70167-8. [DOI] [PubMed] [Google Scholar]
- 48.Reddy D, Lancaster JR, Jr, Cornforth DP. Nitrite inhibition of Clostridium botulinum: Electron spin resonance detection of iron-nitric oxide complexes. Science. 1983;221(4612):769–770. doi: 10.1126/science.6308761. [DOI] [PubMed] [Google Scholar]
- 49.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJC. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci USA. 2007;104(28):11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg ED. Iron and susceptibility to infectious disease. Science. 1974;184(4140):952–956. doi: 10.1126/science.184.4140.952. [DOI] [PubMed] [Google Scholar]
- 51.Harris AB. Inhibition of growth and nucleic acid synthesis in iron-deficient Mycobacterium smegmatis. J Gen Microbiol. 1967;47(1):111–119. doi: 10.1099/00221287-47-1-111. [DOI] [PubMed] [Google Scholar]
- 52.De Voss JJ, Rutter K, Schroeder BG, Barry CE., 3rd Iron acquisition and metabolism by mycobacteria. J Bacteriol. 1999;181(15):4443–4451. doi: 10.1128/jb.181.15.4443-4451.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dlugovitzky D, et al. Influence of disease severity on nitrite and cytokine production by peripheral blood mononuclear cells (PBMC) from patients with pulmonary tuberculosis (TB) Clin Exp Immunol. 2000;122(3):343–349. doi: 10.1046/j.1365-2249.2000.01394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lamsal M, et al. Evaluation of lipid peroxidation product, nitrite and antioxidant levels in newly diagnosed and two months follow-up patients with pulmonary tuberculosis. Southeast Asian J Trop Med Public Health. 2007;38(4):695–703. [PubMed] [Google Scholar]
- 55.Tsai MC, et al. Characterization of the tuberculous granuloma in murine and human lungs: Cellular composition and relative tissue oxygen tension. Cell Microbiol. 2006;8(2):218–232. doi: 10.1111/j.1462-5822.2005.00612.x. [DOI] [PubMed] [Google Scholar]
- 56.Shi L, et al. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci USA. 2005;102(43):15629–15634. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6(12):3051–3064. [PubMed] [Google Scholar]
- 58.Ding AH, Nathan CF, Stuehr DJ. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J Immunol. 1988;141(7):2407–2412. [PubMed] [Google Scholar]
- 59.Stermann M, Bohrssen A, Diephaus C, Maass S, Bange F-C. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J Clin Microbiol. 2003;41(7):3252–3259. doi: 10.1128/JCM.41.7.3252-3259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One. 2008;3(1):e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rand L, et al. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol Microbiol. 2003;50(3):1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 64.Beste DJV, et al. Transcriptomic analysis identifies growth rate modulation as a component of the adaptation of mycobacteria to survival inside the macrophage. J Bacteriol. 2007;189(11):3969–3976. doi: 10.1128/JB.01787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.