Abstract
The diversity of protistan assemblages has traditionally been studied using microscopy and morphological characterization, but these methods are often inadequate for ecological studies of these communities because most small protists inherently lack adequate taxonomic characters to facilitate their identification at the species level and many protistan species also do not preserve well. We have therefore used a culture-independent approach (denaturing gradient gel electrophoresis [DGGE]) to obtain an assessment of the genetic composition and distribution of protists within different microhabitats (seawater, meltwater or slush on sea-ice floes, and ice) of the Ross Sea, Antarctica. Samples of the same type (e.g., water) shared more of the same bands than samples of different types (e.g., ice versus water), despite being collected from different sites. These findings imply that samples from the same environment have a similar protistan species composition and that the type of microenvironment significantly influences the protistan species composition of these Antarctic assemblages. It should be noted that a large number of bands among the samples within each microhabitat were distinct, indicating the potential presence of significant genetic diversity within each microenvironment. Sequence analysis of selected DGGE bands revealed sequences that represent diatoms, dinoflagellates, ciliates, flagellates, and several unidentified eukaryotes.
Phototrophic and heterotrophic protists are ubiquitous in extreme cold-water environments, where they are central to the production and utilization of energy and the cycling of elements. Understanding the structure and diversity of these communities is of fundamental importance to biological oceanography and to understanding the activities and evolution of life on our planet. Traditional microscopic approaches for documenting the diversity and population structure of protistan assemblages in the coastal regions around Antarctica have contributed greatly to our current understanding of species biogeography, microbial food web structure, and biogeochemical processes in these waters (see reviews in references 6 and 9). This work has been most instructive for species generally greater than 20 μm in size and possessing unambiguous morphological features, such as frustules, loricae, or skeletons (e.g., diatoms, tintinnid ciliates, and choanoflagellates). Although valuable, these approaches have been unable to characterize the diversity of a significant number of species that are morphologically less distinctive, nor to determine whether genetically related organisms are present over large spatial and temporal scales. This inability is due in part to the tremendous size range and morphological diversity among protists that necessitate the use of a variety of disparate approaches to identify and count them in natural communities (4, 10) and to the fact that many species of protists preserve poorly. This situation is exacerbated for many Antarctic protists, which expire quickly when warmed even a few degrees above ambient Antarctic temperatures. The latter problem can be serious, as some of the key morphological features used for protistan species identification, such as swimming behavior, are manifested only by living cells (e.g., small flagellates).
Molecular genetic methods have become fairly well established as alternatives to morphological identification of organisms in natural microbial assemblages during the last 15 years (2, 11, 24). To date, these approaches have been developed and applied primarily for assessing bacterial and archaeal diversity, because many prokaryotic species are currently unculturable and morphological criteria for their identification are virtually lacking. More recently, genetic and immunological methods have been applied in studies of protistan communities to identify and enumerate individual species of ecological interest and to examine the diversity of these assemblages (14, 17, 19, 29, 37).
Denaturing gradient gel electrophoresis (DGGE) is a PCR-based tool that has been applied extensively and effectively for analyzing the phylotype diversity of bacterial and archaeal assemblages in different environments (for examples, see references 3, 13, and 21 to 23). Application of this method to eukaryotic microbes has been accomplished in only a few studies (5, 28, 36). Our work represents some of the first to utilize the method to analyze protistan population relationships in different microhabitats (ice, seawater, and slush) in the Antarctic marine environment.
In our analysis of the genetic diversity of Ross Sea protists, we have targeted the small subunit ribosomal DNA (srDNA) with both general eukaryote-specific primers as well as primers selective for groups within the microbial eukaryotic realm (e.g., diatoms) to generate products for DGGE analysis. Banding patterns from different samples were compared, and similarities were determined for both sets of primers. The most intense bands were isolated from the gels, sequenced, and examined for their taxonomic affinities by BLAST analysis (1). This work has shown that DGGE can be an effective alternative means of assaying the genetic structure of protistan communities in the Antarctic ecosystem. This approach permitted the efficient comparison of samples from different habitats over large spatial and temporal scales and allowed the genetic identification of members of the assemblage.
MATERIALS AND METHODS
Sample collection.
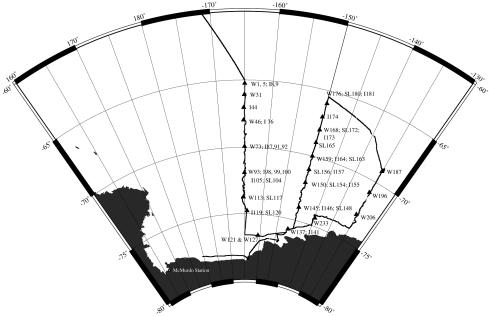
Samples of water, ice, and slush (the meltwater layer at the surface of sea ice underneath 2 to 20 cm of snow) were collected in the Ross Sea, Antarctica, during the austral summer of 1999 (1 January to 2 February 1999) onboard the RVIB Nathaniel B Palmer. Samples analyzed in this report were collected at stations each approximately 1° in latitude apart along three transects, as shown in Fig. 1. Seawater was collected in 30-liter Niskin bottles on a CTD rosette sampler at several depths between the surface and the deep chlorophyll maximum (usually between 30 and 60 m) and combined for analysis. Mixed-depth water samples were collected in triplicate to assess the consistency of extraction, amplification, and DGGE methods. Slush from areas that showed a red, brownish, or green color was collected by scooping the material with sterile containers. Ice was sampled from regions of color within ice cores collected using a Sipre corer. All ice and slush samples were brought back to the ship and thawed for 2 days at 4°C. Some ice core samples were split in half and thawed in the presence or absence of 1 liter of seawater collected from 1,000 m and 0.2-μm-pore-size filter sterilized. The sterile water was used to buffer against the destruction of cells due to extreme salinity changes during thawing but was not checked for the presence of amplifiable DNA. Water samples were prefiltered through 200-μm mesh Nitex screening to remove larger zooplankton. Protists were then collected from all samples by filtration onto 0.8-μm polycarbonate membrane filters. We filtered between 9 and 28 liters of water, 600 ml to 1.5 liters of melted ice, and 1.5 ml to 400 ml of slush. All filtrations were carried out in a walk-in cold room with the temperature at 0°C. DNA was extracted from cells collected on the filter immediately after completing filtration.
FIG. 1.
Cruise track of cruise NBP 99-01. Triangles indicate the stations sampled (see the text for samples analyzed in this report). W, water; I, ice; SL, slush. Numbers are individual sample indicators. The map was generated by Antarctic Support Associates as part of the cruise data.
DNA extraction.
Nucleic acids were extracted from cells collected on polycarbonate membrane filters using a modified combination of hot detergent lysis buffer and mechanical disruption (12). The 2× lysis buffer (100 mM Tris [pH 8.0], 40 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate) was preheated to 70°C, and 200 μl was added to the filter in 5-ml Nunc CryoTube vials (Nalge). Approximately 200 μl of zirconia-silica beads (0.5-mm diameter; BioSpec Products, Inc.) was added to the tube, and the tube was vortexed for 1 min. The sample was twice incubated at 70°C for 5 min and then vortexed for 1 min. The solution was brought to 0.7 M NaCl, followed by the addition of 10% hexadecyltrimethylammonium bromide to a final concentration of 1%, and incubated at 70°C for 15 min. An equal volume of chloroform was added, the sample was vortexed and spun, and the aqueous layer was removed to a sterile 1.5-ml microcentrifuge tube. Nucleic acids were precipitated by the addition of 0.6 volumes of isopropanol and recovered by centrifugation (10 min at 15,000 × g). The pellets were allowed to air dry and then resuspended in 50 to 100 μl of sterile Milli-Q water.
Primer design.
Primers for amplification of DGGE fragments were derived from primers that we have previously used for the amplification and sequencing of eukaryotic small subunit rRNA genes, 960F and 1200R (Table 1). A GC clamp (20) was added to the 5′ end of 960F, and the primer sequence was modified to remove the three bases at the 3′ end. This primer pair amplifies a region of the srDNA known as V7 and is just prior to the region amplified by van Hannen et al. (36).
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| 960F (eukaryote) | 5′ GGCTTAATTTGACTCAACRCG 3′ |
| 1200R (universal) | 5′ GGGCATCACAGACCTG 3′ |
| 960FbGC (GC clamped) | 5′ CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCCGGCTTAATTTGACTCTAAC 3′ |
| Biocos1R (bicosoecid) | 5′ TCTAGATGGTAAGGTTTA 3′ |
| Choan1R (choanoflagellate) | 5′ CGAWAATTACAAAGATCTA 3′ |
| ChrSyn1F (chrysophyte-synurophyte) | 5′ AATAACTTTCGGATCGAT 3′ |
| Diatom1R (diatom) | 5′ ATGCAGATTGATGATCTGCG 3′ |
| Prymn1R (prymnesiophyte) | 5′ ACATCTCTTCACGAGGAT 3′ |
In addition to the general eukaryotic primers described above, selective amplification primers were designed by aligning small subunit ribosomal sequences from organisms within particular taxonomic groups (e.g., diatoms) and generating a consensus sequence. Sequences conserved within a group, but differing from species outside the group, were identified and used to direct primer design. Potential primer sites outside of the region amplified by the DGGE primer set (960FbGC and 1200R) were chosen, and their group selectivity was assessed through BLAST (1) searches. Although the primers selected for use (Table 1) were specific for their target groups based upon BLAST results, when working with natural samples there is always the potential for nonspecific amplification. Therefore, these group-selective primers were used primarily to enrich for the desired sequences prior to amplification with DGGE primers.
PCR amplification.
PCR amplification (27) was carried out to generate products for DGGE. One microliter of extracted nucleic acid was used as the template in 50-μl (with drop of oil overlay) touchdown amplification reaction mixtures with an initial annealing temperature of 65°C for two cycles (45 s at 95°C, 45 s at 65°C, and 45 s at 72°C), followed by a decrease in annealing temperature of 2°C every two cycles down to 55°C. Twenty-five cycles were then carried out at the 55°C annealing temperature (45 s at 95°C, 45 s at 55°C, and 45 s at 72°C). All amplification reactions were carried out using a Perkin-Elmer model 480 thermal cycler. Triplicate DGGE PCRs were carried out, visualized on 1% agarose gels stained with ethidium bromide, and then combined and ethanol precipitated for analysis. The precipitated products were resuspended in 5 μl of sterile distilled water and 5 μl of DGGE loading dye (40% Ficoll 400, 10 mM Tris [pH 7.8], 1 mM EDTA, 0.1% bromphenol blue), and 3 to 5 μl was loaded per lane on the gel.
PCR amplifications using group-selective primers were carried out in standard 50-μl reaction mixtures using 1 μl of extracted nucleic acid as template and 35 cycles of 45 s at 95°C, 45 s at 42°C, and 3 min at 72°C. Each group-selective primer was used in conjunction with a general eukaryotic small subunit ribosomal gene end primer (18). Primer A was paired with reverse group-selective primers, and primer B was paired with forward group-selective primers (Table 1). One microliter of this reaction mixture was used as the template for amplification with DGGE primers as described previously.
DGGE standard markers.
Standard markers to enable the comparison of different DGGE gels to each other were generated from protistan cells that were micropipetted from seawater or slush samples. These cells included an oligotrich ciliate, two different tintinnid ciliates, a copepod, the foraminifer Neogloboquadrina pachyderma, and solitary and colonial forms of Phaeocystis. Each of the cell nucleic acid extracts for these species yielded a single band by amplification and in DGGE analysis, and each of the bands had a different position or melting point in the gel. The DGGE amplification products were cloned, and plasmid preps were generated as stocks for future amplification of markers. To generate marker mixes, each plasmid was amplified separately, products were precipitated separately, and the presence of a single DGGE band was confirmed. All seven products were then combined in approximately equal proportions.
DGGE gels and analysis.
Our DGGE gels were run with a denaturing gradient of 35 to 85% urea at 60°C at 95 V overnight (16 h) using the CBS Scientific model DGGE-2000 gel apparatus. Band patterns were analyzed within and between gels using the BioImage whole-band analyzer software (Millipore Corporation). Bands were automatically detected by the software, but final confirmation of their presence (or absence) was accomplished manually. We added bands that we believed were overlooked, or we deleted ones that appeared to be miscalled by the program. The use of markers permitted the standardization of gels and allowed us to compare lanes within and between gels. Bands were identified as being the same using a 1% deviation in band position to account for slight differences in gel runs. Density differences between bands were not established, as the amount of PCR product loaded for each sample was not quantified. Dendrograms based upon the presence or absence of bands at each position were generated using the unweighted pair group method with averages (UPGMA) cluster analysis parameters in the BioImage software.
Sequence analysis of isolated DGGE bands.
Individual bands were cut from the DGGE gel using new razor blades, placed in 25 μl of sterile Milli-Q water, and allowed to elute overnight at 4°C. Isolated bands were identified by a gel identification number and a band number (e.g., in Fig. 2A below, band 1 is number 12500.1). The water containing the eluted DGGE band was recovered and placed in a new sterile microcentrifuge tube. Two or three microliters of the eluted band was reamplified with non-GC-clamped 960F and 1200R primers. These products were ethanol precipitated to remove excess primers and then resuspended in 10 μl of sterile Milli-Q water. Five microliters of product was sequenced directly using an ABI Prism Big Dye Terminator cycle sequencing ready reaction kit and the 960F or 1200R primer. Sequencing reactions were run on an ABI 377 apparatus (PE Applied Biosystems) and analyzed using Sequencher version 4.1 (Gene Codes Corporation). BLAST (1) searches were accomplished to determine relative taxonomic affinities of the DGGE band sequences. Bands that gave what appeared to be mixed sequences were cloned into the pGEM-T Easy vector system I (Promega). Four clones were picked for each band, and individual clones were sequenced as described previously.
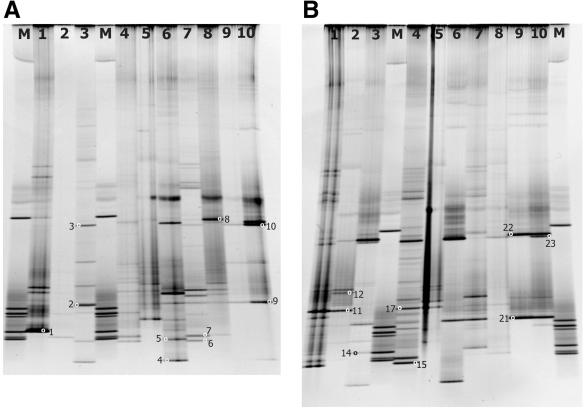
FIG. 2.
Denaturing gradient gels for samples collected from the middle transect line of the cruise (Fig. 1). (A) Gel number 12500. Lane 1, W126; lane 2, W137; lane 3, I141; lane 4, W145; lane 5, SL148; lane 6, I 146; lane 7, W150; lane 8, SL154; lane 9, I155a; lane 10, I155b. (B) Gel number 12500. Lane 1, W159; lane 2, SL163; lane 3, I164; lane 4, W168; lane 5, SL172; lane 6, I 173; lane 7, W176; lane 8, SL180; lane 9, I181a; lane 10, I181b. Small circles with numbers indicate bands recovered and sequenced. a and b designate different melting methods for duplicate ice samples.
RESULTS
DGGE analysis.
We established parameters for running our DGGE gels using PCR products generated from two clonal cultures of dinoflagellates maintained in our laboratory, Gymnodinium beii and Scrippsiella nutricula. A single band was obtained from each culture when analyzed by DGGE. Perpendicular gels were run to assess the melting characteristics of the fragment, which was a smooth disassociation profile that started at approximately 47% denaturant and ended at approximately 60% denaturant. Therefore, we chose to use a 35 to 85% urea gradient for the analysis of our environmental samples. A 16-h run time and relatively low voltage (95 V) were empirically determined to be better than shorter times at a higher voltage (200 V) by comparing the clarity of bands and lack of smearing. Additionally, reduced mobility of the bands was not observed in time travel loadings that ran for a total of 10 h, so a longer run time was used to ensure adequate band separation. The markers were also used as indicators of quality control for DGGE runs. If clear separation of the seven bands was not observed, the gel was not analyzed. DGGE profiles were examined for a limited number of our triplicate mixed-depth water samples and showed almost identical band profiles (data not shown). The difference of a single faint band was noted in some cases, and we believe that this could have been due as much to PCR amplification variability as to sample differences. Due to the overall high similarity between replicates, we chose to analyze only one mixed-depth water sample from each station.
The greatest yield in number of bands from a single sample was nearly 30, but most samples yielded between 10 and 20 bands (Fig. 2). The gels in Fig. 2 are examples of several that were run and analyzed for this project. Some lanes had failed or faint reactions (e.g., Fig. 2A, lane 2), and these were reamplified and run on another gel for analysis. When samples were compared, usually more than half of the bands appearing on a gel were unique for each sample, with relatively few bands shared between samples. Ice samples shared the most bands when analyzed by DGGE, and these samples were also generally observed to have the lowest total number of bands. In the instances where we melted duplicate ice samples in 1 liter of sterile seawater or without seawater, the same major bands were usually present (Fig. 2A and B, lanes 9 and 10). Some minor bands differed between these replicates, indicating that the melting conditions may affect recovery of rare or delicate organisms. However, the dominant bands were still present, and thus the overall genetic pattern being detected often did not change appreciably. We did not assess whether the filtered sterile seawater had amplifiable DNA present, because we believed that any potentially contaminating DNA would have been in very low abundance compared to the target organisms present in colored ice or slush. This view was supported by the fact that we saw very little difference between duplicate ice sample DGGE patterns that were melted with or without the sterile seawater.
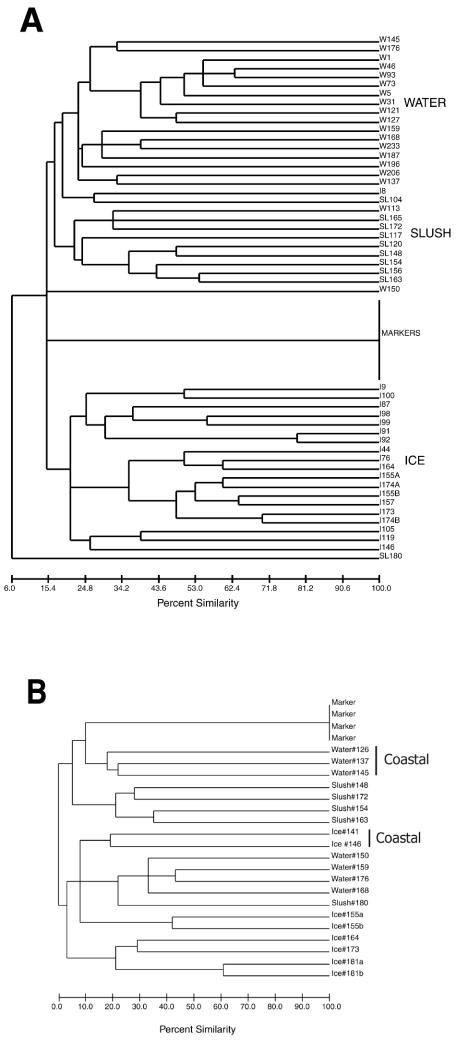
Dendrograms generated by comparisons of the banding patterns from all samples indicated that patterns obtained from the same microhabitat (i.e., water, ice, or slush) generally shared more bands than patterns from different microhabitat types (i.e., water versus ice, ice versus slush, or water versus slush), even when comparing samples collected from different locations (Fig. 3). Furthermore, assemblages collected from coastal samples of water and ice tended to group separately from those collected from the pack ice region of the Ross Sea when a single transect was examined (Fig. 3B). It should be noted that even when sample types were more similar to each other, they usually did not share a large number of bands (Fig. 3 [percent similarity scale]).
FIG. 3.
Dendrogram generated by a UPGMA cluster analysis comparison of DGGE patterns from direct amplification of ice, water, and slush samples collected in the Ross Sea, Antarctica. (A) All samples; (B) samples from the second transect only. Water samples 126, 137, and 148 and ice samples 141 and 146 were all coastal samples. a and b designate different melting methods for the same ice sample.
Slush and water samples tended to be more similar to each other than either were to the ice samples, although there were occasional exceptions (Fig. 3). The SL180 sample was very different in color (bright green instead of brownish or reddish) from all other slush samples, a macroscopic difference that was reflected in the position of the DGGE patterns in both dendrograms. Ice core sample I8 fell within the water-slush group (Fig. 3A). This ice sample was identified as a band of slight greenish color in porous ice about 21 cm from the top of an ice core. The color of this band may have indicated a history that was unique from that of the other ice samples, perhaps a community that was recently introduced into the porous matrix. The bottom part of the same ice core yielded the brown ice sample I9, whose banding pattern fell within the cluster of samples from other ice microhabitats (Fig. 3A). All other ice samples were collected from brownish bands near, or at, the bottom of ice cores and displayed DGGE patterns that were more similar to one another than to samples from either the water or slush microhabitats. Our results imply the development of unique protistan communities within these different environments.
Sequences of recovered DGGE bands.
Partial nuclear srDNA sequences for microalgae (phototrophic protists), protozoa (heterotrophic protists), copepods, and metazoa were identified from the recovered and reamplified DGGE bands (Tables 2 and 3). It was feasible to obtain sequence information directly from some of the reamplified bands, but more often than not it was difficult to obtain unambiguous sequences. It seemed likely that more than one band was being recovered, either due to the presence of multiple bands that were not separated on the gels or because the method of cutting out bands did not resolve very closely spaced bands. Therefore, DNA from bands that did not give clear sequencing results were cloned. We rarely recovered more than one sequence type when analyzing cloned bands. This result could have been due to either the low number of clones that we sampled and sequenced (usually four) or to the fact that only the dominant sequence type was recovered. A small amount of contaminating sequence may have caused a problem for direct sequence analysis but yet not have contributed significantly in the cloning process.
TABLE 2.
Primary BLAST matches of sequences obtained from DGGE bands from direct amplification products
| Band no. | BLAST identification | No. of bases in BLAST match/no. in band (% similarity with identified sequence) |
|---|---|---|
| 12500.1 (W126) | Neocalanus cristatus (copepod) | 188/201 (93) |
| 12500.2C (I141) | Thalassiosira antaractica (diatom) | 217/258 (84) |
| 12500.3 (I141) | Uncultured turbellarian (Pelophila) | 104/125 (83) |
| 12500.4 (I146) | Cancrincola plumipes (copepod) | 168/178 (94) |
| 12500.5A (I146) | Uncultured copepod AT2-8 | 105/155 (68) |
| 12500.5C (I146) | Uncultured stramenopile CCW50 (diatom) | 194/231 (83) |
| 12500.6 (W150) | Uncultured marine eukaryote S721 (copepod) | 164/170 (91) |
| 12500.7B (W150) | Phaeocystis antarctica (prymnesiophyte) | 165/231 (71) |
| 12500.8A and 8B (SL154) | Chaetoceros sp. (diatom) | 215/258 (83) |
| 219/238 (92) | ||
| 12500.9 (I155B) | Thalassiosira antarctica (diatom) | 197/211 (93) |
| 12500.10 (I155B) | Chaetoceros rostratus (diatom) | 145/175 (83) |
| 12500.11C (SL163) | Uncultured eukaryote E163 (dinoflagellate) | 207/247 (83) |
| 12500.12A (SL163) | Uncultured eukaryote E163 (dinoflagellate) | 205/256 (79) |
| 12500.14B (I164) | Uncultured eukaryote S721 (copepod) | 163/239 (67) |
| 12500.15B (W168) | Brachiopod | ∼110/196 (56) |
| 12500.15C and D (W168) | Uncultured eukaryote S721 (copepod) | 237/241 (61) |
| 12500.17 (W168) | Ichthyosporean (Sphaerothecum destruens) | 158/166 (96) |
| 12500.21 (I181A) | Thalassiosira antarctica (diatom) | 164/173 (94) |
| 12500.22 (I181A) | Uncultured turbellarian (Pelophila) | 152/211 (72) |
| 12500.23 (I181B) | Chaetoceros rostratus (diatom) | 222/224 (99) |
TABLE 3.
Sequences of DGGE bands from group-selective amplification products
| Band no. | BLAST identification | No. of bases in BLAST match/no. in band (% similarity with identified sequence) | Group-selective amplification |
|---|---|---|---|
| 120600.1 (W145) | Noctiluca scintillans (dinoflagellate) | 168/195 (86) | Choanoflagellate |
| 120600.4 (W145) | Strombidium (ciliate) | 217/231 (94) | Bicosoecid |
| 120600.6 (W145) | Uncultured chlorophyte CCW13 | 189/205 (92) | Diatom |
| 120600.7 (W145) | Thalassiosira antarctica (diatom) | 222/231 (96) | Diatom |
| 120600.10 (W145) | Uncultured diatom CCW27 | 152/224 (68) | Diatom |
| 120600.11 (W145) | Neocalanus cristatus (copepod) | 170/226 (75) | Diatom |
| 120600.13 (W145) | Neocalanus cristatus (copepod) | 170/208 (82) | Prymnesiophyte |
| 120600.14 (W145) | Phaeocystis antarctica (prymnesiophyte) | 229/233 (98) | Chrysophyte |
| 120600.15 (W145) | Paraphysomonas foraminifera (chrysophyte) | 220/221 (99) | Chrysophyte |
| 120600.25 (W150) | Unidentified dinoflagellate | 200/217 (92) | Choanoflagellate |
| 120600.27 (W150) | Ciliate | 189/222 (85) | Choanoflagellate |
| 120600.28 (W150) | Ciliate | 139/209 (67) | Bicosoecid |
| 120600.34 (W150) | Unidentified dinoflagellate | 220/242 (90) | General eukaryote |
| 120600.37 (W150) | Uncultured eukaryote S721 (copepod) | 227/243 (93) | General eukaryote |
| 120600.39 (W150) | Unidentified dinoflagellate | 220/234 (94) | Prymnesiophyte |
| 120600.42 (W150) | Phaeocystis jahnii (not 14; prymnesiophyte) | 222/237 (94) | Prymnesiophyte |
| 120600.43 (W150) | Phaeocystis jahnii (not 14; prymnesiophyte) | 222/237 (94) | Chrysophyte |
BLAST (1) analysis of DGGE fragment sequences enabled identification of their general taxonomic affinities to organisms with ribosomal sequences available in the database. In order to calculate the percent similarity between a band and its match, the number of bases matched by the search were divided by the total length of the DGGE fragment. None of our fragments was identical to any sequences in the database, and only three bands had similarities indicative of reliable identification (>98%): 12500.23 (99% Chaetoceros), 120600.14 (98% Phaeocystis), and 120600.15 (99% Paraphysomonas). Most of the bands had similarities that ranged between approximately 60 and 90%, and several were most similar to uncultured eukaryote sequences obtained directly from environmental samples. This result seems to support the presence of a considerable number of uncultured, and potentially undescribed, protistan taxa in marine ecosystems (15). Interestingly, the DGGE bands recovered by direct amplification seemed to share taxonomic affinities with sequences obtained directly from environmental samples from other studies more often than bands recovered through group-selective amplification conducted in the present study (9 out of 19 in Table 2, versus 3 out of 14 in Table 3).
Bands isolated from similar positions in the denaturing gradient (but different samples) generally yield the same sequence, but in several instances bands from different positions yielded similar sequences. For example, bands 12500.2, 12500.9, and 12500.21 (Fig. 2; Table 1) share the same position and the same diatom sequence affinity (Thalassiosira antarctica), while band 120600.7 shares the same sequence affinity but not the same gel position. The unidentified dinoflagellate bands 120600.34 and 120600.39 occurred at the same gel position, but 120600.25 did not. These similarities in taxonomic affinity, but differences in gel position, indicate that DGGE was able to detect genetic differences that may relate to species- or strain-level variation that would normally be impossible to identify using traditional methods. It is possible that in our evaluation of the DGGE sequences we have missed some of the sequence information that could account for the difference in gel position, because the length of sequence that we were able to recover from the bands varied (from 105 to 237 bases out of approximately 250 total bases) and even a single base difference can result in the distinct separation of two bands by DGGE (30).
Some of the bands recovered from the direct DGGE amplification of seawater samples yielded copepod sequences despite our prefiltration through 200-μm Nitex screening (5 out of 22 bands sequenced). Juvenile specimens, and/or appendages lost from adult copepods, may have gone through the screening, with the multicellular structures contributing significantly in the amplification of eukaryotic ribosomal genes in some of our samples. Copepod sequences were also recovered from ice samples, but these samples weren't prefiltered prior to extraction. It is of interest that the copepod sequences yielded the same type of BLAST search results that our protistan sequences did—they were similar but generally not identical to the sequences in the database.
Group-selective amplification.
The recovery of sequences from some protistan populations that were visually observed to be abundant in the samples (e.g., small flagellates) was less than anticipated (Table 2). This result may have been due to the inherent bias of PCR to amplify the most abundant sequences present in the sample, inefficient extraction (or damage) of nucleic acids from some protistan species, and our inability to recover all bands from the gel. These issues led to our development and application of group-selective amplification primers. Group-selective oligonucleotides were utilized in a primary amplification from three water DNA samples. DGGE primers were then used to reamplify the group-selective products, and these fragments were separated by DGGE.
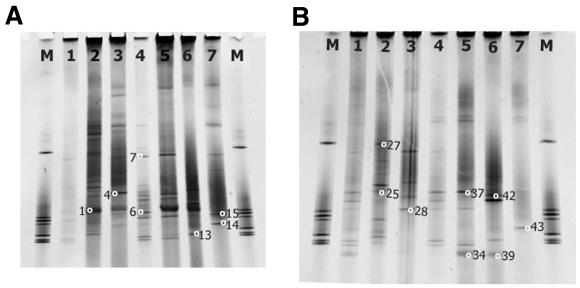
Using group-selective primers for the primary amplification, it was possible to acquire bands not detected in the original amplification (Fig. 4). The band profiles shown for each of the group-selective amplifications contained many bands not represented in either the original direct amplification (compare lane 1 to the others) or in the other selective amplifications from the same sample. In Fig. 4A, 29 of the bands observed using the various selective amplifications were not found in the direct amplification. In Fig. 4B, 22 of the bands were not observed in the direct amplification. This result indicates the efficacy of the group-selective primers to recover previously undetected sequences and illustrates that direct amplification DGGE examines only a portion of what appears to be a highly diverse community.
FIG. 4.
Denaturing gradient gels for group-selective amplifications of W145 (gel ID number 120600) (A) and W150 (gel ID number 120600) (B). Lanes correspond to different amplification primers. Lane 1, direct DGGE amplification; lane 2, choanoflagellate-selective primer amplification; lane 3, bicosecid-selective primer amplification; lane 4, diatom-selective primer amplification; lane 5, A/B primer amplification; lane 6, prymnesiophyte-selective primer amplification; lane 7, chrysophyte/synurophyte-selective primer amplification. M, marker. Small circles with numbers indicate bands recovered and sequenced.
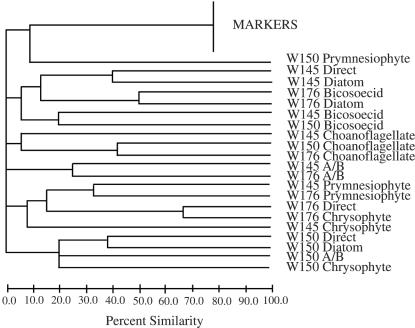
Dendrograms created for the selective amplification of DGGE gels indicated that the band patterns from amplifications by the same selective primers tended to group together, although this was not always the case (Fig. 5). This result implies the presence of some of the same organisms in different samples. It is also of interest to note that the diatom-selective amplifications were generally most similar to the original direct amplification, an expectation given the dominance of these algae in the Antarctic planktonic environment.
FIG. 5.
Dendrogram generated by UPGMA cluster analysis comparison of DGGE patterns from group-selective amplifications of three water samples. Labels indicate the sample number (W145, W150, and W176) followed by the group-selective amplification primer used.
The sequences of bands recovered from amplifications using group-selective primers indicated that the selective primers were not necessarily specific for the group for which they were designed and/or that the target sequences were very rare (Table 2). This result was not entirely unexpected, because the selective PCR amplifications were performed using a low annealing temperature (42°C). Furthermore, nonspecific amplification can occur when the appropriate templates are present at low abundances in a background of nontarget DNA (16, 35). The selective nature of the primers did reduce the recovery of copepod bands and has the potential to expand our ability to analyze and compare the diversity of the protistan assemblage, because the bands recovered in these amplifications were different than those from the direct amplifications using general eukaryote primers.
General eukaryotic primers A and B (18) were also used to amplify full-length nuclear srDNA, the products of which were reamplified with DGGE primers. After examining the gels in Fig. 4, it was apparent that some of the change in band patterns was likely due to the change in template abundances as a result of the reamplification, although several of the bands that were detected in the original gels were also present. We view this general approach as being useful for expanding the amount of genetic information recovered from a sample and providing a more complete taxonomic profile of the community.
DISCUSSION
DGGE has been used to conduct a comparison of the general protistan diversity in seawater, ice, and ice floe meltwater (slush) samples from the Ross Sea, Antarctica. The dendrograms in Fig. 3 revealed that only about half of our samples shared even 50% of their bands when samples were amplified directly with general eukaryotic primers. This finding would appear to indicate a high degree of uniqueness (endemism) among the protistan assemblages within the individual microhabitats sampled. It is important to recognize, however, that this approach is not yet capable of fully characterizing the complex protistan assemblages present in each sample, but it likely identifies the most common phylotypes present in these communities.
Phylotypes that we recovered from within ice cores were typically quite different than those recovered from seawater. This finding is contrary to studies that have suggested that species found commonly in the plankton are also common in sea ice (8, 9, 25). The uniqueness of the banding patterns from ice and seawater samples does not necessarily mean that the same species were not present in both microhabitats, but rather it implies that the dominant species in the two types of microhabitats differed markedly (or that morphologically similar but genetically distinct strains were present).
Despite the low degree of commonality among all samples in this study, samples from the same microhabitat but from different sampling sites showed more similarity than samples from the same sampling site but different microhabitats. This finding implies that environmental conditions within each of the microhabitat types enrich the abundances of habitat-specific types of protists from within the complex mixture of species present in the Ross Sea ecosystem. Some degree of similarity among the microflora and microfauna is not surprising given the results of previous microscopic studies, but the horizontal extent of this trend is striking given the temporal and spatial sampling scales involved in this study. Each station on a transect is 1° in latitude away from its closest neighbor, which is 60 nautical miles. Samples along the same transect were collected within 7 days of each other, whereas almost a month elapsed between the start of the first transect and the end of the third. It is highly noteworthy that we have been able to discern a general pattern of genetic similarity in different microhabitats despite the significantly large scales involved.
Analyses of samples from a single transect enabled discrimination between the samples that we considered coastal (open water within the polynya on the southern side of the pack ice and next to the continent, including land-fast ice) versus those that were collected from within the pack ice (Fig. 3B). Coastal water samples and ice samples formed two separate groups within the pack ice water, slush, and ice samples. These groupings indicate subtle differences in the protistan assemblages of the coastal and pack ice environments. Land-fast ice (I141 and I146) may contain multiyear growth of microbial communities and thus might be expected to contain different assemblages than ice in the pack ice region. Similarly, succession in coastal water within the polynya could explain the divergence of these planktonic communities from the water within the pack ice. The latter is more ephemeral temporally and may differ significantly from water in the polynya in physical features and nutrient chemistry. Trends related to coastal versus pack ice noted along individual transects were not evident when all samples were considered together (Fig. 3A). This inability to distinguish clearly between coastal and pack ice samples when all three transects were combined was likely due to the smaller number of bands actually shared between the samples as the communities changed over the month-long sampling scale of the project.
In DGGE analyses, the total number of bands in a lane is often taken to be a measurement of the diversity of that sample, but it is perhaps more appropriate to view the number of bands as representing the composition of the dominant populations in the sample (7). This view is supported by both the gel results (Fig. 2 and 4) and the sequence results (Tables 2 and 3). On our DGGE gels, we were able to observe up to 27 bands in a single sample by the direct amplification of that sample (Fig. 2B, lane 5). When we used the group-selective amplification strategy, we increased the total number of bands observed per sample by almost an equal amount (20 to 30). This finding indicates a high degree of diversity in the protistan assemblages from the Ross Sea, implying that our direct amplification results potentially represent only the more abundant organisms in the samples. This finding also implies that further work and new priming strategies will be necessary to fully characterize the diversity of these assemblages using this molecular approach.
Our sequencing results for DGGE bands further indicated that we had characterized only some of the protistan diversity in our samples. We were successful in recovering sequences from some taxa that have been described as conspicuous components of Antarctic protistan assemblages, such as Phaeocystis and dinoflagellates (Tables 2 and 3), but we had only limited success for others, particularly diatoms. Numerous diatom species have been reported in samples from the Antarctic environment, yet our results here indicated only three. We speculate that nucleic acid extraction efficiencies were low for some protistan species in our samples and, although the combination of bead beating and hot detergent was fairly rigorous, we may have had limited success at breaking open some cells.
A practical consideration explaining the relatively low number of bands recovered and sequenced in this study was that we were not able to exhaustively sample all of our DGGE bands. Some bands were simply too faint to be isolated reliably from the gels, and so we were constrained to analyzing the brightest and most intense bands. The brightness of a band is indicative of its concentration in the total PCR product, and that can reflect the abundance of the original template, although there are other factors that can affect this as well (26, 34). By examining the brighter bands, we were presumably observing a limited population of the most dominant or abundant organisms present in the original sample. Better methods for band isolation and removal and cloning of reamplified products will likely help to resolve this issue, although some bands will remain difficult to recover.
Morphology-based studies of Antarctic protistan diversity have described the presence of diverse and dynamic assemblages of protists in the water surrounding the continent (as examples, see references 9, 25, and 31 to 33). Our molecular approach to community composition generally is in agreement with those published reports in the sense that we have shown that the ice, slush, and seawater assemblages are genetically quite diverse and harbor protistan assemblages that can be quite heterogeneous on both the macroscale (Ross Sea) and microscale (ice and slush). In particular, our genetic results compare very well with the morphological analyses of Garrison and Buck (8). As in that study, we were able to document the distinct assemblages of organisms formed by ice and water samples, along with spatial heterogeneity among the water, ice, and slush samples. Interestingly, our DGGE analysis indicated that there was little genetic overlap between different sample types. Even samples that were considered similar still often shared fewer than 50% of their bands. Our finding of high diversity (low similarity) among samples in the same group is in contrast to the 54% similarity of the organisms from different water samples in Garrison and Buck's morphological analysis (8). This difference may be due to the fact that we examined the total microbial eukaryotic community rather than restricting our analysis to the phytoplankton. Nevertheless, the qualitative similarities between morphology-based and culture-independent approaches for assessing protistan diversity are encouraging. Given the relatively youthful state of development of DGGE and the improvements in resolution and speed of analysis expected in the future, this molecular approach should prove highly useful for ecological studies of these species. Continued development should provide insightful, culture-independent techniques for assessing the diversity of protistan assemblages in Antarctic microhabitats.
Acknowledgments
This work was supported by National Science Foundation grants OPP-9714299 and OPP-0125437.
We thank everyone involved in making the NBP99-01 cruise a success, especially the captain and crew of the RVIB Nathaniel B Palmer and Martin Jeffries (chief scientist). Special thanks are given to Alison Murray for her assistance with the DGGE methods. We also thank Peter Countway, Dawn Moran, David Beaudoin, and three anonymous reviewers for their insightful and productive comments regarding the manuscript.
Footnotes
WHOI contribution number 11028.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bano, N., and J. T. Hollibaugh. 2002. Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caron, D. A., R. J. Gast, E. L. Lim, and M. R. Dennett. 1999. Protistan community structure: molecular approaches for answering ecological questions. Hydrobiologia 401:215-227. [Google Scholar]
- 5.Diez, B., C. Pedros-Alio, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Sayed, S. Z., and G. A. Fryxell. 1993. Phytoplankton, p. 65-122. In E. I. Friedmann (ed.), Antarctic microbiology. Wiley-Liss, Inc., New York, N.Y.
- 7.Fromin, N., J. Hamelin, S. Tarnawaski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 8.Garrison, D. L., and K. R. Buck. 1985. Sea-ice algal communities in the Weddell Sea: species composition in ice and plankton assemblages, p. 103-122. In J. S. Gray and M. E. Christiansen (ed.), Marine biology of polar regions and effects of stress on marine organisms. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 9.Garrison, D. L., and M. M. Gowing. 1993. Protozooplankton, p. 123-165. In E. I. Friedmann (ed.), Antarctic microbiology. Wiley-Liss, Inc., New York, N.Y.
- 10.Gifford, D. J., and D. A. Caron. 1999. Sampling, preservation, enumeration and biomass of marine protozooplankton, p. 193-221. In R. Harris, P. H. Wiebe, J. Lenz, H. R. Skjoldal, and M. Huntley (ed.), ICES zooplankton methodology manual. Academic Press, London, England.
- 11.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuske, C. R., K. L. Banton, D. L. Adorada, P. C. Stark, K. K. Hill, and P. J. Jackson. 1998. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen, A., T. Castberg, R. A. Sandaa, C. P. D. Brussaard, J. Egge, M. Heldal, A. Paulino, R. Thyrhaug, E. J. Van Hannen, and G. Bratbak. 2001. Population dynamics and diversity of phytoplankton, bacteria and viruses in a seawater enclosure. Mar. Ecol. Prog. Ser. 221:47-57. [Google Scholar]
- 14.Lim, E. L., D. A. Caron, and M. R. Dennett. 1999. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment culture. Limnol. Oceanogr. 44:37-51. [Google Scholar]
- 15.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea plankton. Nature 409:603-606. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig, W., and K. H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 17.Massana, R., L. Guillou, B. Diez, and C. Pedros-Alio. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 19.Moon-Van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 20.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray, A. E., C. M. Preston, R. Massana, L. T. Taylor, A. Blakis, K. Wu, and E. F. DeLong. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64:2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 23.Ogino, A., H. Koshikawa, T. Nakahara, and H. Uchiyama. 2001. Succession of microbial communities during a biostimulation process as evaluated by DGGE and clone library analyses. J. Appl. Microbiol. 91:625-635. [DOI] [PubMed] [Google Scholar]
- 24.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 25.Palmisano, A. C., and D. L. Garrison. 1993. Microorganisms in Antarctic sea ice, p. 167-218. In E. I. Friedmann (ed.), Antarctic microbiology. Wiley-Liss, Inc., New York, N.Y.
- 26.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saiki, R., P. S. Walsh, C. Levenson, and H. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-494. [DOI] [PubMed] [Google Scholar]
- 28.Schabereiter-Gurtner, C., G. Pinar, W. Lubitz, and S. Rolleke. 2001. Analysis of fungal communities on historical church window glass by denaturing gradient gel electrophoresis and phylogenetic 18S rDNA sequence analysis. J. Microbiol. Methods 47:345-354. [DOI] [PubMed] [Google Scholar]
- 29.Scholin, C. A., K. R. Buck, T. Britschgi, G. Cangelosi, and F. P. Chavez. 1996. Identification of Pseudo-nitzchia australis (Bacillariophyceae) using rRNA-targeted probes in whole cell and sandwich hybridization formats. Phycologia 35:190-197. [Google Scholar]
- 30.Smith-Sørensen, B., M. C. Gebhardt, P. Kloen, J. McIntyre, F. Aguilar, P. Ceratti, and A. L. Borresen. 1993. Screening for TP53 mutations in osteosarcomas using constant denaturant gel electrophoresis (CDGE). Hum. Mutat. 2:274-285. [DOI] [PubMed] [Google Scholar]
- 31.Stoecker, D. K., K. R. Buck, and M. Putt. 1993. Changes in the sea-ice brine community during the spring-summer transition, McMurdo Sound, Antarctica. 2. Phagotrophic protists. Mar. Ecol. Prog. Ser. 95:103-113. [Google Scholar]
- 32.Stoecker, D. K., K. R. Buck, and M. Putt. 1992. Changes in the sea-ice brine community during the spring-summer transtition, McMurdo Sound, Antarctica. 1. Photosynthetic protists. Mar. Ecol. Prog. Ser. 84:265-278. [Google Scholar]
- 33.Stoecker, D. K., M. Putt, and T. Moisan. 1995. Nano- and microplankton dynamics during the spring Phaeocystis sp. bloom in McMurdo Sound, Antarctica. J. Mar. Biol. Assoc. UK 75:815-832. [Google Scholar]
- 34.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by templated annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teo, I. A., J. W. Choi, J. Morlese, G. Taylor, and S. Shaurak. 2002. LightCycler qPCR optimisation for low copy number target DNA. J. Immunol. Methods 270:119-133. [DOI] [PubMed] [Google Scholar]
- 36.Van Hannen, E. J., M. P. Van Agterveld, H. J. Gons, and H. J. Laanbroek. 1998. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J. Phycol. 34:206-213. [Google Scholar]
- 37.Vrieling, E. G., and D. M. Anderson. 1996. Immunofluorescence in phytoplankton research: applications and potential. J. Phycol. 32:1-16. [Google Scholar]