Abstract
BACKGROUND:
Aurora-A/STK15 is a serine/threonine kinase critical for regulated chromosome segregation and cytokinesis. We investigated the association between 2 nonsynonymous single nucleotide polymorphisms in the coding region of STK15, T91A (Phe31Ile) and G169A (Val57Ile), and clinical outcome of esophageal cancer treated with preoperative chemoradiation.
METHODS:
Genotypes at Phe31Ile and Val57Ile were assessed from peripheral blood lymphocytes of 190 esophageal cancer patients and were correlated to response to treatment, recurrence rate, risk of death, disease-free survival (DFS) and median survival time (MTS).
RESULTS:
All patients had resectable esophageal or gastroesophageal junction cancer and received preoperative chemoradiation followed by esophagectomy. The heterozygous variant Phe31/Ile variant was significantly associated with tumor recurrence (odds ratio [OR] = 4.39; 95% confidence interval [CI], 2.12-8.94; P < .001), shorter DFS (P = .0001), and shorter MTS (P = .012). For patients receiving cisplatin-based therapy, only the variant Phe31/Ile had an adverse effect on response (OR = 2.8; 95% CI, 1.01-5.17; P = .048) and MTS (P = .026). The variant 91A-169G haplotype carried a significant risk for lack of complete response (OR = 2.54; 95% CI, 1.15-5.54) and higher rate of recurrence (OR = 2.73; 95%CI, 1.00-7.29). The presence of at least 1 variant allele at each locus further increased the risk of recurrence (adjusted OR = 6.21; 95% CI, 2.28-17.11; P = <.001), and was associated significantly shorter DFS (P = .003).
CONCLUSIONS:
Our study shows that functional SNPs in the STK15 gene are associated with higher rate of recurrence, higher likelihood of chemoratiotherapy-resistance, shorter DFS, and shorter MTS. Confirmation of our data and understanding the mechanisms through which STK15 functional SNPs mediate resistance to chemoradiotherapy are warranted.
Keywords: Aurora-A, polymorphism, chemoradiotherapy, esophageal cancer, outcome
INTRODUCTION
Carcinoma of the esophagus or and gastroesophageal junction (E/GEJC) remains a highly virulent malignancy, with approximately 16,980 new cases and 14,710 deaths expected in the United states in 2011.1 E/GEJC is 1 of the few cancers that have exhibited a dramatic increase in incidence with more than 400% increase in its rate in 25 years accompanied by 8% in the death rate in 13 years 1990 to 2003.2 Despite aggressive therapeutic multidisciplinary approaches, the 5-year survival rate of E/GEJC is <20%.3
For patients with localized E/GEJC, representing about 50% of all patients at diagnosis, a commonly recommended approach is preoperative chemoradiotherapy (CTXRT) followed by surgical resection, although primary resection remains an option.4-11
The current preoperative treatment regimens are associated with substantial morbidity and benefit only a limited group of patients. Level 1 evidence for preoperative CTXRT has not been published because of equivocal results from several randomized trials. It is, however, clear that patients who have a highly CTXRT sensitive cancer (eg, when no cancer cells or only a few cancer cells are identified in the resected specimen) tend to survive better than those who have relatively resistant cancer.12-16
Unfortunately, the majority of E/GEJC are resistant to currently utilized CTXRT. Clearly, it would be important to identifying factors that mediate chemoradiation resistance and metastatic progression. Unfortunately, pre-treatment clinical parameters, such as age, sex, histology, or location of the primary cancer, clinical stage (whether II, III, or IVA), imaging studies, do not predict the outcome and the identification of genetic and molecular biomarkers to optimize therapy is warranted.
Aurora A, also known as STK15, BTAK, AIKI, and AURKA in humans, belongs to a multigenic family of mitotic serine/threonine kinases including 2 other members, Aurora-B and Aurora-C.17 STK15 is an oncogene located on chromosome 20q13, a region frequently amplified in a variety of human neoplasia, including esophageal cancers.17 In physiological conditions, the Aurora-A protein is expressed and active at the highest level during the G2-M phase of the cell cycle and falls again during the exit from mitosis into the next G1.18-20 Aurora-A localizes at the centrosomes of interphase cells, and becomes phophorylated and activated in the centrosomes late in the G2 phase.20 Aurora-A is necessary for the recruitment of proteins required for spindle formation and centrosome maturation process, such as TACC, Kinesin 5, and γ-tubulins.17 In addition, Aurora-A also plays key roles in the organization and alignment of chromosomes, as well as the exit from mitosis and proper cytokinesis. Aurora-A activation is inhibited by the late G2 DNA damage checkpoint, although its sustained expression abrogate the G2-M DNA damage checkpoint resulting in the inappropriate commitment to mitosis, and further promoting genomic instability.21 Aurora-A elevated expression, with or without gene amplification, has been reported in a variety of human cancers, and has been correlated with chromosomal instability and clinical aggressiveness.17
Two coding single nucleotide polymorphisms (SNPs) in the STK15 gene have been reported to fall within in the NH-2 terminal region of the Aurora-A protein and have been associated with functional consequences.22-27 The T91A SNP, which causes an amino acid change at codon 31 from phenylalanine to an isoleucin, was initially associated with enhanced cell transformation in vitro and significantly increased chromosomal instability in human colon cancers.23 Several studies, including a meta-analysis, suggested that the T91A polymorphism is associated with low penetrance cancer susceptibility affecting multiple cancers types, including esophageal cancer;27 it can lead to earlier ages of diagnosis in pancreatic cancer, and may have synergistic effects with other genes.22 However, the 91A allele has also been shown to yield protective effects, such as in patients with hereditary nonpolyposis colorectal cancer (HNPCC).22 The second functional SNP, the G169A at codon 57, causes an amino acid change from valine to an isoleucine, and reduces Aurora-A kinase activity.23 The T91A and G169A haplotype combinations (91A-169A; T91-169A) have been shown to be significantly associated with esophageal cancer risk.
Regulated Aurora-A is critical for maintenance of genomic integrity after DNA damage and the STK15 SNPs disrupt some physiologic functions of Aurora-A; therefore, we hypothesized that the STK T91A and G169A SNPs would mediate CTXRT resistance and lead to adverse outcomes in patients with E/GEJC. We discuss our results in 190 patients in whom we studied STK15 T91A and G169A.
MATERIALS AND METHODS
Patients
This study included 190 histologically confirmed esophageal cancer patients who consented for research blood collection and were recruited between 1985 and 2003 at the University of Texas MD Anderson Cancer Center (Houston, Tx). These patients were diagnosed had potentially resectable adenocarcinoma or squamous cell carcinoma of the esophagus or gastroesophageal junction and had received chemoradiotherapy followed by surgery or induction chemotherapy followed by chemoradiotherapy and surgery.
Clinical Data Collection
Patients were evaluated by computed tomography, barium-swallow esophagogram, endoscopic ultrasound, and positron emission tomography (when feasible). Chemotherapeutic agents used were: platinum analogs, fluoropyrimidines, and taxanes. The recommended dose of radiotherapy was 50.4 Gy in 28 fractions. Approximately 5 to 6 weeks after the completion of chemoradiotherapy, patients underwent surgical resection. The resected specimens were reviewed systematically to assess the degree of residual cancer and lymph nodal involvement.15 All participants voluntarily donated blood for research after an informed consent originating from a protocol approved by The University of Texas MD Anderson institutional review board.
STK15 Genotyping
Genomic DNA extracted using the PicoPure DNA extraction kit (Arcturus Bioscience, Mountain View, Calif). Genotyping was performed using the Taqman SNP assay. Probes were labeled fluorescently with either 6-FAM or VIC on the 5′ end and a nonfluorescent minor groove binder (MGB) quencher on the 3′ end (Applied Biosystems, Bedford, Mass). The primer and probe sequences used were as follows: for Phe31Ile, 5′-CTG GCC ACT ATT TAC AGG TAA TGG A (forward primer), 5′-TGG AGG TCC AAA ACG TGT TCT C (reverse primer), VIC-ACT CAG CAA TTT CCT T-MGB (T allele for Phe), 6-FAM-CTC AGC AAA TTC CTT-MGB (A allele for Ile); for Val57Ile, 5′-CGG CTT GTG ACT GGA GAC A (forward primer), 5′-GGG TCT TGT GTC CTT CAA ATT CTT C (reverse primer), 6-FAM-CAG CGC GTT CCT T-MGB (G allele for Val), VIC-CAG CGC ATT CCT T- MGB (A allele for Ile). Typical amplification mixes (5 μL) contained sample DNA (5 ng), 1 × TaqMan PCR buffer A, 200 μM dNTPs, 5 mM MgCl2, 0.65 units of AmpliTaq Gold, 900 nM each primer and 200 mM each probe. Reactions were carried out on a dual-384-well GeneAmp PCR system 9700, with thermal conditions being 95°C for 10 minutes, followed by 50 cycles of 95°C for 15 seconds and 62°C (for Phe31Ile) or 60°C (for Val57Ile) for 1 minute. Reaction plates were then read with the ABI Prism 7900HT Sequence Detection System, with analyzed fluorescence results auto-called into genotypes using SDS version 2.1 software (Applied Biosystems). Water control, previously genotyped samples, and internal controls of 5% of the samples were randomly selected and run in duplicates with 100% concordance.
Statistical Analysis
The Fisher and chi-square tests were used to assess the association between recurrence/survival and demographics, clinical variables, and treatments. We set the study endpoint event as the date of the patient’s first recurrence or overall death and applied the Cox proportional hazard model to estimate the hazard ratios (HRs) associated with the Phe31Ile and Val57Ile polymorphisms. The outcome variables for patients lost to follow-up or those who experienced recurrence or death after the end of the study were flagged as censored.
HRs were estimated by fitting the Cox model while adjusting for age, gender, radiation dosage, CTXRT sequence, clinical stage, chemoregimens, histology, tumor location, pathology, and histological viability. Kaplan-Meier survival function and log-rank tests were used to assess clinical outcomes in relation to the polymorphisms. The above statistical analyses were completed with the STATA software (Version 8, College Station, Tx).
RESULTS
Patient Characteristics
The median age of the 190 patients was 61 years (range, 32-78 years). There were 165 men (86.8%) and 25 women (13.2%). All patients were Caucasians. Histology distribution included 85.3% adenocarcinoma (162 patients) and 14.7% squamous cell carcinoma (28 patients). Clinical stage distribution was as follows: stage IIA or IIB in 57.4%, stage III in 37.4%, and stage IVA in 5.2%. Induction chemotherapy followed by CTXRT was given to 81 patients (42%), with the remaining 109 patients (58%) receiving only preoperative CTXRT. The distribution for chemotherapeutic treatments was: cisplatin in 136 (71.6%) patients, fluoropyrimidine in 181 (95.3%) patients, and taxane in 97 (51.0%) patients.
Pathologic complete response (absence of residual cancer cells, pathCR) was observed in 64 (33.7%) patients, with the remaining 126 (66.3%) had less than a pathCR. At the median follow-up time of 18.7 months (range, 0.23-178.43 months), 60 (31.6%) of the 190 patients had local-regional or distant relapse. The overall survival rates at 3 and 5 years were 47.9% and 34.4%, respectively, with a median overall survival at 33.6 months (95% CI: 22.9,47.3). There was no association was found between preoperative clinical variables (ie, age, gender, histology, tumor location, clinical stage, radiation dosage, chemoradiotherapy sequence, chemoregimens) and patient outcomes (median survival, disease-free survival [DFS], rate of relapse, overall survival, or response to CTXRT). These results were similar to our previous published clinical studies.14,15
Effect of Preoperative CTXRT on Response and Survival
The schedule of preoperative treatment (eg, induction chemotherapy [CT] followed by concomitant CTXRT vs CTXRT alone) nor the class of chemotherapeutic agents was associated with pathological response or survivals. However, a difference in clinical outcome was associated with total dose of radiotherapy received by the patients. Seventy-three (57%) of the 128 patients who received >45 Gy were alive compared with 22 (37.2%) of the 59 patients who received <45 Gy (P = .012). Similarly, local-regional relapse was reduced when the recommended dose of radiation could be administered (P = .025).
STK15 Genotype Characteristics
The frequencies for the Phe31Ile (T91A) SNP were 71.8% for the Phe allele and 28.2% for Ile allele. For the Val57Ile SNP, frequencies for the Val and Ile alleles were 86.4% and 13.6%, respectively.
Effects of Individual STK15 Polymorphisms on Response and Clinical Outcomes
Table 1a illustrates the association between STK15-T91A SNP and response to CTXRT, recurrence rate, and survivals. The T91A SNP was not associated with response to CTXRT. Compared with patients carrying the wild-type TT genotypes (Phe/Phe), patients with the heterozygous variant AT genotype (Phe31/Ile) were at significantly increased risk of tumor relapse with an adjusted odds ratio (OR) of 4.39 (95% confidence interval [CI], 2.12-8.94) (P < .001). There was no association for the homozygous variant AA genotype (Ile/Ile) with risk of relapse; however, there was a significant association of combined variant alleles (AT + AA) with risk of relapse with an adjusted OR of 3.73 (95%CI, 2.12-8.94) (P = .016). Kaplan-Meier estimates demonstrated that both variant allele and combined variant alleles were associated with significantly shorter DFS compared with the wild-type allele (AT: 12.2 months, AT + AA: 12.63 months vs TT: 42.17 months; P = .0001 and 0.0006, respectively). Although the STK15-T91A SNP was not significantly associated with risk of death, the median survival time (MTS) of patients carrying 1 copy of the Phe31Ile allele was significantly shorter than in patients with the wild-type genotype (MTS, TA: 22.5 months vs TT: 51.3 months; P = .012).
Table 1a.
Risk Estimates for the Phe31Ile SNP in Esophageal Cancer Patients Treated by Cisplatin-Based Preoperative CTXRT
| Genotypes | Clinical | Outcome | Adjusted ORa
(95% CI) |
|---|---|---|---|
| <Path CR | Path CR | ||
| TT | 58 | 37 | Reference |
| AT | 57 | 23 | 1.62 (0.82-3.18) |
| AA | 9 | 4 | 1.43 (0.37-5.56) |
| AT + AA | 1.59 (0.83-3.05) | ||
| Recurrence | No recurrence | ||
| TT | 20 | 75 | Reference |
| AT | 38 | 42 | 4.39 (2.16-8.94) |
| AA | 2 | 11 | 0.99 (0.20-4.81) |
| AT + AA | 3.73 (1.85-7.52) | ||
| Dead | Not dead | ||
| TT | 41 | 54 | Reference |
| AT | 48 | 32 | 1.34 0.85-2.13 |
| AA | 3 | 10 | 0.47 0.13-1.64 |
| AT + AA | 1.23 0.78-1.94 | ||
| Dead or recurrence |
Not dead or recurrence |
||
| TT | 43 | 52 | Reference |
| AT | 52 | 28 | 1.59 1.01-2.50 |
| AA | 3 | 10 | 0.44 0.13-1.52 |
| AT + AA | 1.41 0.90-2.21 |
Abbreviations: CI, confidence interval; CTXRT, chemoradiotherapy; OR, odds ratio; SNP, single nucleotide polymorphism.
Adjusted by age, gender, tumor type, tumor location, histology, clinical stage, radiation dose, chemoradiation sequence, and chemotherapy regimens.
The STK15 Val57Ile polymorphism did not have an effect on clinical outcome (Table 1b).
Table 1b.
Risk Estimates for the Val57Ile SNP in Esophageal Cancer Patients Treated by Preoperative CTXRT
| Genotypes | Clinical | Outcome | Adjusted ORa
(95% CI) |
|---|---|---|---|
| <Path CR | Path CR | ||
| GG | 92 | 48 | Reference |
| AG | 30 | 15 | 1.05 0.49-2.24 |
| AA | 2 | 1 | 0.59 0.03-10.39 |
| AG + AA | 1.02 0.49-2.16 | ||
| Recurrence | No recurrence | ||
| GG | 43 | 97 | Reference |
| AG | 16 | 29 | 1.39 0.70-2.76 |
| AA | 1 | 2 | 2.62 0.17-39.78 |
| AG + AA | 1.43 0.73-2.80 | ||
| Dead | Not dead | ||
| GG | 68 | 72 | Reference |
| AG | 23 | 22 | 1.26 0.74-2.15 |
| AA | 1 | 2 | 2.96 0.37-23.54 |
| AG + AA | 1.29 0.76-2.20 | ||
| Dead or recurrence |
Not dead or recurrence |
||
| GG | 72 | 68 | Reference |
| AG | 25 | 20 | 1.16 0.69-1.95 |
| AA | 1 | 2 | 1.65 0.16-17.21 |
| AG + AA | 1.18 0.71-1.95 |
Abbreviations: CI, confidence interval; CTXRT, chemoradiotherapy; OR, odds ratio; SNR single nucleotide polymorphism.
Adjusted by age, gender, tumor type, tumor location, histology, clinical stage, radiation dose, chemoradiation sequence, and chemotherapy regimens.
Effects of Individual STK15 Polymorphisms on Clinical Outcomes in Cisplatin-5 Fluorouracil-Treated Patients
When analyzing the effects of the STK15 SNP only within the 134 patients treated with cisplatin-based chemotherapy, the T91A variant Phe31/Ile was significantly associated with lack of response to CTXRT (Table 2a). Patients with the heterozygous variant phenotype
Table 2a.
Risk Estimates for the Phe31Ile SNP in Esophageal Cancer Patients Treated by Cisplatin-Based Preoperative CTXRT
| Genotypes | Clinical | Outcome | Adjusted OR* (95% CI) |
|---|---|---|---|
| <Path CR | Path CR | ||
| TT | 39 | 30 | Reference |
| AT | 43 | 15 | 2.28 1.01-5.17 |
| AA | 5 | 2 | 1.30 0.19-8.80 |
| AT + AA | 2.15 0.98-4.75 | ||
| Recurrence | No recurrence | ||
| TT | 16 | 53 | |
| AT | 29 | 29 | 3.76 1.62-8.70 |
| AA | 1 | 6 | 0.97 0.11-8.59 |
| AT + AA | 3.43 1.48-7.96 | ||
| Dead | Not dead | ||
| TT | 33 | 36 | Reference |
| AT | 38 | 20 | 1.20 0.70-2.08 |
| AA | 1 | 6 | 0.19 0.02-1.58 |
| AT + AA | 1.09 0.64-1.88 | ||
| Dead or recurrence |
Not dead or recurrence |
||
| TT | 34 | 35 | Reference |
| AT | 40 | 18 | 1.47 0.85-2.53 |
| AA | 1 | 6 | 0.20 0.03-1.61 |
| AT + AA | 1.30 0.76-2.24 |
Abbreviations: CI, confidence interval; CTXRT, chemoradiotherapy; OR, odds ratio; SNR single nucleotide polymorphism.
Adjusted by age, gender, tumor type, tumor location, histology, clinical stage, radiation dose, chemoradiation sequence, and chemotherapy regimens.
(AT) carried a 2.28-fold (95%CI, 1.01-5.17) higher risk of lacking response than patients with homozygous wild-type genotype (P = .048). Similarly, combined variant alleles were significantly associated with lack of response to CTXRT with an adjusted OR of 2.15 (95%CI, 0.98-4.75; P = .047). In addition, heterozygous Phe31Ile variant genotype and combined variant alleles (AT + AA) were significantly associated with increased risk of relapse (AT OR = 3.76; 95% CI, 1.62-8.70; P = .002; AT + AA OR = 3.42; 95% CI, 1.48-7.96; P = .044).
Both AT genotype combined AT + AA genotypes were associated with significantly shorter DFS (AT: 12.2 months, AT + AA: 12.4 months) compared with the TT wild-type genotype group (DFS: 35.67 months; P = .003 and 0.008, respectively). Intriguingly, only the variant Phe31/Ile allele was associated with shorter median survival time (MTS: 20.67 months) compared with the wild-type Phe31/Phe31 allele (MTS: 34.8; P = .026) but the combined variant allele was not (P = .13). The Val57Ile polymorphism alone was not significantly associated with risk of response to CTXRT, relapse, or death in the cisplatin-treated group (Table 2b).
Table 2b.
Risk Estimates for the Val57Ile SNP in Esophageal Cancer Patients Treated by Cisplatin-Based Preoperative CTXRT
| Genotypes | Clinical | Outcome | Adjusted ORa
(95% CI) |
|---|---|---|---|
| <Path CR | Path CR | ||
| GG | 64 | 33 | Reference |
| AG | 21 | 13 | 0.87 0.37-2.07 |
| AA | 2 | 1 | 0.61 0.03-11.18 |
| AG + AA | 0.86 0.37-2.00 | ||
| Recurrence | No recurrence | ||
| GG | 33 | 64 | Reference |
| AG | 12 | 22 | 1.55 0.67-3.62 |
| AA | 1 | 2 | 2.45 0.10-61.22 |
| AG + AA | 1.59 0.69-3.63 | ||
| Dead | Not dead | ||
| GG | 52 | 45 | Reference |
| AG | 19 | 15 | 1.42 0.76-2.65 |
| AA | 1 | 2 | 3.67 0.45-29.85 |
| AG + AA | 1.47 0.80-2.72 | ||
| Dead or recurrence |
Not dead or recurrence |
||
| GG | 52 | 45 | Reference |
| AG | 19 | 15 | 1.42 0.76-2.65 |
| AA | 1 | 2 | 3.67 0.45-29.85 |
| AG + AA | 1.47 0.80-2.72 |
Abbreviations: CI, confidence interval; CTXRT, chemoradiotherapy; OR, odds ratio; SNR single nucleotide polymorphism.
Adjusted by age, gender, tumor type, tumor location, histology, clinical stage, radiation dose, chemoradiation sequence, and chemotherapy regimens.
STK15 Haplotypes and Clinical Outcome
There were 4 possible STK15 haplotypes derived from the 2 known genotypes (ie, 91T-169G, 91A-169G, 91T-169A, and 91A-169A). Because both 91T and 169G wild-types were favorable alleles we used the 91T-169G haplotype as the reference. As shown in Table 3, compared with the reference, only the variant 91A-169G haplotype carried a significant risk for lack of complete response (adjusted OR = 2.53; 95% CI, 1.15-5.54) and recurrence (adjusted OR = 4.39; 95%CI, 2.16-8.94). None of the other haplotypes was significantly associated with clinical outcome.
Table 3.
Risk Estimates for STK15 Phe31Ile and Val57Ile Haplotypes in Esophageal Cancer Patients Treated by Preoperative CTXRT
| Haplotype | Clinical | Outcome | Adjusted OR* (95% CI) |
|---|---|---|---|
| STK31- STK57 | <path CR | path CR | |
| wild-wild | 37 | 28 | Reference |
| variant-wild | 55 | 20 | 2.53 1.15-5.54 |
| wild-variant | 20 | 9 | 2.21 0.73-6.15 |
| variant-variant | 12 | 7 | 0.91 0.26-3.17 |
| STK31- STK57 | Recurrence | No recurrence | |
| wild-wild | 13 | 524 | Reference |
| variant-wild | 29 | 46 | 4.39 (2.16-8.94) |
| wild-variant | 7 | 22 | 0.66 0.16-2.76 |
| variant-variant | 10 | 9 | 2.93 0.68-12.68 |
| STK31- STK57 | Dead | Not dead | |
| wild-wild | 27 | 38 | Reference |
| variant-wild | 43 | 32 | 1.65 0.69-3.92 |
| wild-variant | 14 | 15 | 1.27 0.35-4.56 |
| variant-variant | 11 | 8 | 3.79 0.74-19.35 |
| STK31- STK57 | Dead or recurrence |
Not dead or recurrence |
|
| wild-wild | 29 | 36 | Reference |
| variant-wild | 45 | 30 | 1.64 0.69-3.89 |
| wild-variant | 14 | 15 | 0.85 0.25-2.91 |
| variant-variant | 13 | 6 | 3.28 0.75-14.36 |
Abbreviations: CI, confidence interval; CTXRT, chemoradiotherapy; OR, odds ratio; SNP, single nucleotide polymorphism.
Adjusted by age, gender, tumor type, tumor location, histology, clinical stage, radiation dose, chemoradiation sequence, and chemotherapy regimens.
Joint Effects of Combined STK15 Polymorphisms on Clinical Outcomes in Caucasians
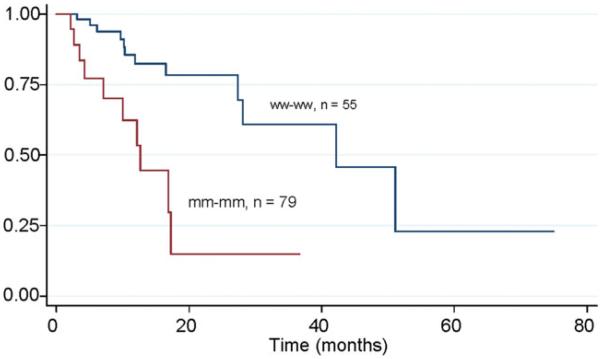
The risk estimates for response to CTXRT, relapse, death, and death/recurrence when combining the 2 STK15 polymorphisms are shown in Table 4. The reference group had the favorable genotype combination TT + GG. Variant allele at the PheIle31 locus (AT or AA) and wild-type allele at Val57 carried a significant risk of relapse with an adjusted OR of 3.49 (95%CI, 1.54-7.91; P = .03). The presence of at least 1 variant allele at each locus further increased the risk of relapse (adjusted OR = 6.21; 95% CI, 2.28-17.11; P = <.001). The Kaplan-Meier estimates demonstrated that patients with at least 1 variant allele at the Ile31 and Val57 loci had a significantly shorter disease-free survival time of 12.63 months than patients in the reference genotype group (DFS = 42.17; P = .003 (Fig. 1). None of the other groups were significantly associated with esophageal cancer outcome (Table 4).
Table 4.
Joint Effects of Risk Estimates for STK15 Phe31Ile and Val57Ile Genotypes and Esophageal Cancer Treatment Outcome
| Joint Effect | Clinical | Outcome | Adjusted OR* (95% CI) |
|---|---|---|---|
| STK31-STK57 | <path CR | path CR | |
| ww_ww | 37 | 28 | Reference |
| ww_wm+mm | 21 | 9 | 1.93 0.72-5.17 |
| wm+mm_ww | 55 | 20 | 2.27 1.06-4.85 |
| wm+mm_wm+wm | 11 | 7 | 1.06 0.33-3.42 |
| STK31-STK57 | Recurrence | No recurrence | |
| ww_ww | 13 | 52 | Reference |
| ww_wm+mm | 7 | 23 | 1.36 0.48-3.89 |
| wm+mm_ww | 30 | 45 | 3.49 1.54-7.91 |
| wm+mm_wm+wm | 10 | 8 | 6.25 2.28-17.11 |
| STK31-STK57 | Dead | Not dead | |
| ww_ww | 27 | 38 | Reference |
| ww_wm+mm | 14 | 16 | 1.43 0.72-2.84 |
| wm+mm_ww | 41 | 34 | 1.32 0.78-2.24 |
| wm+mm_wm+wm | 10 | 8 | 1.56 0.69-3.58 |
| STK31-STK57 | Dead or recurrence |
Not dead or recurrence |
|
| ww_ww | 29 | 36 | Reference |
| ww_wm+mm | 14 | 16 | 1.17 0.59-2.33 |
| wm+mm_ww | 43 | 32 | 1.40 0.83-2.37 |
| wm+mm_wm+wm | 12 | 6 | 1.76 0.83-3.72 |
Abbreviations: CI, confidence interval; OR, odds ratio.
Adjusted by age, gender, tumor type, tumor location, histology, clinical stage, radiation dose, chemoradiation sequence, and chemotherapy regimens.
Figure 1.
This shows Kaplan-Meier disease-free survival estimates by joint effects of STK15 polymorphisms at Ile31 and Val57 loci for patients with esophageal cancer treated by preoperative chemoradiation therapy.
DISCUSSION
This is the first study to investigate the association between STK15 functional polymorphisms and outcome in patients with esophageal cancer treated with preoperative CTXRT. The 3 major findings of this study are that: 1) the heterozygous variant allele of the Phe31Ile is associated with significantly higher risk of tumor relapse and shorter DFS after preoperative CTXRT, 2) the increasing number of variant alleles at both loci doubles the risk of relapse, and 3) the variant Phe31/Ile alone has an impact on the response to CTXRT, relapse rate, DFS, and median survival in patients receiving cisplatin-based CTXRT.
STK15 has been linked to tumor development and progression through it amplification and/or overexpression in a variety of human cancers, including colon, bladder, breast, ovarian, liver, pancreas, gastric and lung.17 STK15 has been reported to interact and phosphorylate several important proteins involved in stress response and cell cycle checkpoint after DNA damage.19 It has been also shown that aberrant expression of STK15 leads to deregulation of proteins mediating DNA damage response, such as BRCA1, p53, and CDC25B.19 Overexpression of STK15 leads to diminished transcriptional activity and increased degradation of p53, causing checkpoint defects and genetic instability and ultimately facilitating cancer development and progression. The Phe31Ile SNP is of special interest since Ewart-Toand et al20 showed that the variant allele (Ile) was more oncogenic than the wild-type Phe31, was preferentially amplified, and associated with aneuploidy in human cancers. In our study the variant Ile imparted the strongest risk of adverse outcome with increased recurrence rate and shortest DFS. One possible functional explanation is that esophageal cancers developing in a STK15 variant Ile background harbor enhanced genomic instability and increased clonal diversity more likely to lead to the generation of clones capable of invasion and metastasis.
Several Previous Studies of the Val5
Val57Ile
Several previous studies of the Val57Ile polymorphism and cancer risk did not find significant associations.22-27 Our data suggest that although alone the Val57Ile SNP is not associated with clinical biology, the presence of the Ile variant allele enhances the adverse effect of the adverse effect of the Phe31Ile variant. In vitro, the combination of variant Phe31Ile and Val57Ile significantly reduces Aurora-A kinase activity (to 15% compared with the wild type) and most importantly induces abnormal nuclear morphology and genomic instability in human immortalized fibroblasts. This evidence underscores the biological plausibility of our results.
The strengths of our study include the relatively large sample size considering the targeted population, and a homogenous Caucasian population, thus reducing the risk of confounding by population stratification. The limitations of our study include the hospital-based study design with which we could not exclude the possibility of selections bias. In addition, even though our study is of fairly large size considering the targeted population, it is underpowered for some stratified analyses, such as the effects of the STK15 SNPs on the clinical outcome of patients treated with taxanes-based therapy. Because of the critical role of Aurora-A in microtubules depending functions, such as centrosome maturation, spindle formation, and cytokinesis, it would be of interest to investigate the effects of thee functional SNPs on microtubules targeting drugs. Further studies are needed to confirm these observations, and to characterize the mechanisms by which these variant differentially modify esophageal cancer outcome. A better understanding of the functional value of these allelic variant may lead to the use of Aurora-A inhibitors to improve the clinical outcome in esophageal cancer.
In summary, we found that in esophageal cancer patients undergoing preoperative CTXRT, the heterozygous STK15-Ile31 variant is associated with increased rate of relapse and shorter DFS. In particular, for patients who were treated with cisplatin-based combination, it was associated with poor response to CTXRT and shorter survival. Phe31Ile and Val57Ile variant produce an allele dependent cumulative adverse effect on relapse rate and DFS. Our data suggest that these STK15 functional SNPs may modify the effects of preoperative CTXRT in esophageal cancer. Confirmation of our data in a larger cohort and understanding the molecular mechanisms through which STK15 functional SNPs mediate resistance to CTXRT are necessary.
Acknowledgments
FUNDING SUPPORT
This work was supported in part by The University of Texas MD Anderson Multidisciplinary Esophageal Research Grant, Riverkreek Foundation, Dallas, Cantu, Smith, and Park Families (to J.A.A.), National Cancer Institute Grants #CA74880, CA91846, and CA142072-0109>
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors made no disclosures
REFERENCES
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer death. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–251. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 4.Hofstetter W, Swisher SG, Correa AM, et al. Treatment outcomes of resected esophageal cancer. Ann Surg. 2002;236:376–384. doi: 10.1097/00000658-200209000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelsen DP, Ginsberg R, Pajak TF, et al. Chemotherapy followed by surgery compared with surgery alone for localized esophageal cancer. N Engl J Med. 1998;339:1979–1984. doi: 10.1056/NEJM199812313392704. [DOI] [PubMed] [Google Scholar]
- 6.Al Sarraf M, Martz K, Herskovic A, et al. Progress report of combined chemoradiotherapy versus radiotherapy alone in patients with esophageal cancer: an intergroup study. J Clin Oncol. 1997;15:277–284. doi: 10.1200/JCO.1997.15.1.277. [DOI] [PubMed] [Google Scholar]
- 7.Bosset JF, Gignoux M, Triboulet JP, et al. Chemoradiotherapy followed by surgery compared with surgery alone in squamous-cell cancer of the esophagus. N Engl J Med. 1997;337:161–337. doi: 10.1056/NEJM199707173370304. [DOI] [PubMed] [Google Scholar]
- 8.Cooper JS, Guo MD, Herskovic A, et al. Chemoradiotherapy of locally advanced esophageal cancer: long-term follow-up of a prospective randomized trial (RTOG 85-01). Radiation Therapy Oncology Group. JAMA. 1999;281:1623–1627. doi: 10.1001/jama.281.17.1623. [DOI] [PubMed] [Google Scholar]
- 9.Malthaner RA, Wong RK, Rumble RB, et al. Neoadjuvant or adjuvant therapy for respectable esophageal cancer: a systematic review and meta-analysis. BMC Med. 2004;2:35–40. doi: 10.1186/1741-7015-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahl M, Stuschke M, Lehmann N, et al. Chemoradiation with and without surgery in patients with locally advanced squamous cell carcinoma of the esophagus. J Clin Oncol. 2005;23:2310–2317. doi: 10.1200/JCO.2005.00.034. [DOI] [PubMed] [Google Scholar]
- 11.Kaklamanos IG, Walker GR, Ferry K, et al. Neoadjuvant treatment for respectable cancer of the esophagus and the gastroesophageal junction: a meta-analysis of randomized clinical trials. Ann Surg Oncol. 2003;10:754–761. doi: 10.1245/aso.2003.03.078. [DOI] [PubMed] [Google Scholar]
- 12.Berger AC, Farma J, Scott WJ, et al. Complete response to nonadjuvant chemoradiotherapy in esophageal carcinoma is associated with significantly improved survival. J Clin Oncol. 2005;23:4330–4337. doi: 10.1200/JCO.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 13.Jin J, Liao Z, Zhang Z, et al. Induction chemotherapy improved outcomes of patients with respectable esophageal cancer who received chemoradiotherapy followed by surgery. Int J Radiat Oncol Biol Phys. 2004;60:427–436. doi: 10.1016/j.ijrobp.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Rohatgi P, Swisher SG, Correa AM, et al. Characterization of pathologic complete response after preoperative chemoradiotherapy in carcinoma of the esophagus and outcome after pathologic complete response. Cancer. 2005;104:2365–2372. doi: 10.1002/cncr.21439. [DOI] [PubMed] [Google Scholar]
- 15.Chierac LR, Swisher SG, Ajani JA, et al. Posttherapy pathologic stage predicts survival in patients with esophageal carcinoma receiving preoperative chemoradiation. Cancer. 2005;103:1347–1355. doi: 10.1002/cncr.20916. [DOI] [PubMed] [Google Scholar]
- 16.Swisher SG, Hofstetter W, Wu TT, et al. Proposed revision of the esophageal staging system to accommodate pathologic response (pP) following preoperative chemoradiation (CRT) Ann Surg. 2005;241:810–817. doi: 10.1097/01.sla.0000161983.82345.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 18.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell. 2003;96:215–229. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Marumoto T, Honda S, Hara T, et al. Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem. 2003;278:51786–51795. doi: 10.1074/jbc.M306275200. [DOI] [PubMed] [Google Scholar]
- 20.Marumoto T, Hirota T, Morisaki T, et al. Roles of aurora-A kinase in mitotic entry andG2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 22.Kimura MT, Mori T, Conroy J, et al. Two functional coding single nucleotide polymorphisms in STK15 (Aurora-A) coordinately increase esophageal cancer risk. Cancer Res. 2005;65:3548–3554. doi: 10.1158/0008-5472.CAN-04-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Li D, Wei C, et al. Aurora-A and p16 polymorphisms contribute to an earlier age at diagnosis of pancreatic cancer in Caucasians. Clin Cancer Res. 2007;13:3100–3104. doi: 10.1158/1078-0432.CCR-06-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J, Sen S, Amos CI, et al. Association between Aurora-A kinase polyporphisms and age of onset of hereditary nonpolyposis colorectal cancer in a Caucasian population. Mol Carcinog. 2007;46:249–256. doi: 10.1002/mc.20283. [DOI] [PubMed] [Google Scholar]
- 25.Chen SS, Chang PC, Cheng YW, Tang FM, Lin YS. Suppression of the STK15 oncogenic activity requires a transactivation-independent p53 function. EMBO J. 2002;21:4491–4499. doi: 10.1093/emboj/cdf409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao X, Sun T, Wang Y, Zhang X, Tan W, Lin Lin. Functional STK15 Phe31Ile polymorphism is associated with the occurrence and advanced disease status of esophageal squamous cell carcinoma. Cancer Res. 2004;64:2680–2683. doi: 10.1158/0008-5472.can-04-0651. [DOI] [PubMed] [Google Scholar]
- 27.Ewart-Toland A, Dai Q, Gao YT, et al. Aurora-A/STK15 T+91A is a general low penetrance cancer susceptibility gene: a meta-analysis of multiple cancer types. Carcinogenesis. 2005;26:1368–1373. doi: 10.1093/carcin/bgi085. [DOI] [PubMed] [Google Scholar]