Abstract
A 492- to 495-bp fragment of the gene coding for the large subunit of the form I ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) (rbcL) was amplified by PCR from facultatively lithotrophic aerobic CO-oxidizing bacteria, colorless and purple sulfide-oxidizing microbial mats, and genomic DNA extracts from tephra and ash deposits from Kilauea volcano, for which atmospheric CO and hydrogen have been previously documented as important substrates. PCR products from the mats and volcanic sites were used to construct rbcL clone libraries. Phylogenetic analyses showed that the rbcL sequences from all isolates clustered with form IC rbcL sequences derived from facultative lithotrophs. In contrast, the microbial mat clone sequences clustered with sequences from obligate lithotrophs representative of form IA rbcL. Clone sequences from volcanic sites fell within the form IC clade, suggesting that these sites were dominated by facultative lithotrophs, an observation consistent with biogeochemical patterns at the sites. Based on phylogenetic and statistical analyses, clone libraries differed significantly among volcanic sites, indicating that they support distinct lithotrophic assemblages. Although some of the clone sequences were similar to known rbcL sequences, most were novel. Based on nucleotide diversity and average pairwise difference, a forested site and an 1894 lava flow were found to support the most diverse and least diverse lithotrophic populations, respectively. These indices of diversity were not correlated with rates of atmospheric CO and hydrogen uptake but were correlated with estimates of respiration and microbial biomass.
Several distinct functional groups comprise the bacterial lithotrophs. Obligate lithotrophs include sulfide-, sulfur-, metal-, ammonium-, and nitrite-oxidizing bacteria, many of which have been described in detail previously (2-5, 14-16, 28, 29, 35, 36, 39). In contrast, facultative lithotrophs include aerobic hydrogen- and CO-oxidizing bacteria, few of which have been described (22-24). Most facultative lithotrophs are heterotrophs that preferentially oxidize organic substrates or that grow mixotrophically with CO (or hydrogen) and organics (22, 23). However, in spite of their physiological, ecological, and phylogenetic diversity, all lithotrophs are united by their ability to incorporate CO2 for cell carbon through the activity of ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO).
RubisCO, the most abundant protein on Earth (33), has been thoroughly characterized (8, 10, 13, 17, 20, 30, 32, 33, 37, 40). Based on various biochemical analyses, two forms have been documented. All known lithotrophic bacteria contain form I, while a relatively small number of bacteria, e.g., Hydrogenovibrio marinus, Rhodobacter capsulatus, Rhodobacter sphaeroides, and several Thiobacillus species, also contain form II (33). Sequence analyses of the gene coding for the large subunit of RubisCO (rbcL) have revealed two major phylogenetically distinct form I groups, a green alga-like group and a red alga-like group, each with two subgroups, IA and IB and IC and ID, respectively. Obligate lithotrophs among the α-, β-, and γ-Proteobacteria dominate the form IA subgroup, while facultative lithotrophs among the α- and β-Proteobacteria dominate the form IC subgroup (32, 33, 40). The exceptions include Hydrogenophaga pseudoflava, a CO and hydrogen oxidizer in the form IA subgroup, and a number of ammonia-oxidizing Nitrosospira strains in the form IC subgroup (33, 38).
In addition to elucidating evolutionary relationships among lithotrophs, rbcL sequence analyses have also facilitated primer development for PCR applications (44). rbcL primers have been applied successfully in analyses of phototroph diversity and population structure (for examples, see references 42 and 43) and more recently in analyses of bacterial lithotrophs in groundwater communities (1). In the latter case, primers were developed for bacteria in the form IA and IB subgroups. Clone libraries derived from PCRs performed with these primers contained sequences most closely related to obligate lithotrophs, but their ecological roles in the environments sampled were uncertain (1).
We report here the development and application of rbcL primers for analysis of obligately and facultatively lithotrophic bacteria containing form IA and IC rbcL genes, respectively. Our primers target different sites than those described by Alfreider et al. (1). Primer efficacy was evaluated with a set of proteobacterial, facultatively lithotrophic, CO-oxidizing isolates obtained from soil, plant roots, and marine macroalgae (19), several isolates known to contain rbcL (Bradyrhizobium japonicum, Methylococcus capsulatus Bath, and Oligotropha carboxidovorans), and isolates not known to oxidize CO or contain rbcL (Cycloclasticus spirillensus and Lutibacterium anuloederans [6]). We also used DNA extracts from salt marsh sulfide-oxidizing microbial mats as PCR templates and analyzed clone libraries constructed from the products to assess the ability of the primers to amplify rbcL genes in obligate lithotrophs.
After establishing primer efficacy, we analyzed clone libraries prepared from DNA extracts from volcanic deposits at four sites in or near Kilauea Caldera, Hawaii. Atmospheric CO and hydrogen uptake at three of these sites have been previously shown to contribute significantly to respiratory metabolism, and all sites actively consume both gases (18). Clones from each of the libraries yielded rbcL sequences that were similar to form IC but not IA. Phylogenetic analysis of these sequences indicated that they were most similar to those of facultative lithotrophs. In addition, statistical analyses indicated that the diversity of rbcL sequences differed significantly among clone libraries from each of the sites. Results from the volcanic deposits were consistent with observed patterns with respect to the diversity of large-subunit CO dehydrogenase genes (coxL) (K. E. Dunfield and G. M. King, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. N-250, 2003) and agreed with biogeochemical observations that have documented CO and H2 uptake but not ammonium oxidation (18). Initial applications of the rbcL primers were reported by King and Crosby (G. M. King and H. Crosby, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. I-4, 2002).
MATERIALS AND METHODS
CO-oxidizing isolates.
Isolates enriched as CO oxidizers (see reference 19) or obtained from other investigators were used for the PCR amplification of rbcL. The isolates included Aminobacter strain COX; Bradyrhizobium strain CPP (Urbana Labs, Urbana, Ill.); Bradyrhizobium japonicum USDA 6 (from P. van Berkum, U.S. Department of Agriculture, Beltsville, Md.); Burkholderia strain JS150 (J. Spain, Tyndall Air Force Base, Fla.); Burkholderia strain LUP; Burkholderia fungorum LB400 (J. Haddock, Southern Illinois University); Cycloclasticus spirillensus (6); Lutibacterium anuloederans (6); Mesorhizobium strain NMB1; Methylococcus capsulatus Bath (R. Hanson, University of Minnesota); Oligotropha carboxydovorans (American Type Culture Collection); Rhizobium leguminosarum and Sinorhizobium fredi (P. van Berkum); Stappia aggregata, Stappia stellulata, Stappia strain CV812-530, and Stappia strain CV902-700 (all from K. Boettecher, University of Maine); Stappia sp. strain M4; Stappia sp. strain M8; Stenotrophomonas strain LUP; and Xanthobacter strain COX. All strains were grown in pyruvate-yeast extract-mineral salts medium (12), harvested by centrifugation, and processed for DNA as described below.
Site description and sampling. (i) Microbial mats.
Microbial mats containing colorless and purple photosynthetic sulfide oxidizers were sampled during November 2002 from shallow depressions in the Cod Cove salt marsh, which borders the Sheepscot River near Wiscassett, Maine. The mats were transferred to sterile Whirlpak bags and transported to the Darling Marine Center for further processing. Based on phase-contrast microscopy, the colorless mats were dominated by rod-shaped cells with occasional Beggiatoa-like filaments and purple photosynthetic sulfide oxidizers. Chromatium-like cells containing sulfur granules dominated the purple microbial mats.
(ii) Volcanic deposits.
Four sites in or near Kilauea Caldera were sampled during April 2002. The site ages, moisture regimes, and successional development differed substantially (18). Halema'uma'u (ID) and Caldera Rim (IE) tended to be drier than the other sites based on water contents. Both sites supported limited, patchy growth of pioneering ferns, shrubs, and Ohia (Metrosideros polymorpha) on weathered lava flows with overlying ash and tephra deposits. Pu'u Puai (IIA) supported patches of Ohia and Myrica faya, which grow on the coarse tephra and cinders deposited in 1959. A forested site (IIC) supports a closed canopy dominated by Metrosideros polymorpha and Myrica faya growing on weathered ash overlying lava with a litter layer typically <1 cm thick; no organic horizon was evident (7). Material from a depth of 0 to 2 cm was collected at all sites with a bleach-sterilized trowel and transferred to sterile Whirlpak bags. Within 1 to 2 h after collection, samples were frozen and held at −20°C until being transported to Maine at −80°C. Samples were held at −20°C until further processing (about 5 months).
DNA extraction and PCR amplification of rbcL.
DNA was extracted from CO-oxidizing isolates by use of three freeze-thaw cycles (−80 and 65°C, respectively) for cell suspensions followed by a bead-beating method (MoBio Labs Inc., Carlsbad, Calif.). DNA was extracted similarly from triplicate 0.5-g (fresh weight) samples of colorless and purple sulfide-oxidizing microbial mats. Triplicate 10-g (fresh weight) samples of material from each of the Kilauea sites were extracted by a bead-beating method with an Ultraclean Mega Soil DNA extraction kit (MoBio Labs Inc.) preceded by three freeze-thaw cycles at −80°C and 65°C, respectively.
A 492- to 495-bp fragment of the large-subunit gene of RubisCO, rbcL, was amplified using primers K2f and V2r, which were modified from those used by Xu and Tabita (44) and which target motifs TT(I)KPKLG and V(A)VGKLEG, respectively. Primer K2f is 5′-ACCA[C/T]CAAGCC[G/C]AAGCT[C/G]GG-3′, and primer V2r is 5′-GCCTTC[C/G]AGCTTGCC[C/G]ACC[G/A]C-3′.
PCR mixtures totaled 50 μl and contained the recommended concentrations of buffers, deoxynucleoside triphosphates, magnesium ions, and 1.25 U of MasterTaq DNA polymerase (Brinkmann Inc., Westbury, N.Y.). Primer concentrations were 1 μM. PCR was performed with an Eppendorf Mastercycler thermocycler (Brinkmann Inc.) with an initial denaturation step of 3 min at 94°C and a hot start at 80°C. The reaction continued with 30 cycles of 45 s at 94°C, 60 s at 62°C, and 90 s at 72°C, with a final extension of 20 min at 72°C. The presence and size of PCR products were determined by electrophoresis in 1% agarose with ethidium bromide staining. PCR products were stored at −20°C overnight or used within a few hours for cloning.
rbcL clone libraries and sequence analysis.
Triplicate PCR products from each site and from the colorless and purple sulfide-oxidizing microbial mats were combined and cloned into Escherichia coli by use of TOPO TA cloning kits for sequencing according to the manufacturer's instructions (Invitrogen Life Technologies, Carlsbad, Calif.). Transformed colonies were arbitrarily selected from each library and grown overnight in Luria-Bertani broth prior to plasmid extraction with a PerfectPrep Plasmid 96 Spin Direct kit, performed according to the manufacturer's instructions (Brinkmann). Plasmids were screened for the correct insert size using 1% agarose gel electrophoresis and a supercoiled DNA marker (2 to 10 kb; Promega Corporation, Madison, Wis.). Inserts in suitably sized plasmids were sequenced by the University of Maine Sequencing Facility using vector primer T3 and an ABI model 377 sequencer (Applied Biosystems, Foster City, Calif.).
Clone sequences were subjected to BLAST screening to determine their similarity to known rbcL sequences. Sequences for which rbcL was the most similar match were submitted to ExPASy (http://us.expasy.org/tools) to obtain the inferred amino acid sequences. The correct reading frames were determined from the presence of diagnostic motifs, including forward and reverse primer sites. Inferred amino acid sequences were aligned with the corresponding rbcL sequences from known obligately and facultatively lithotrophic bacteria with ClustalX software version 1.0.1, and manual adjustments were made as necessary.
Phylogenetic analyses were performed with PAUP version 4.0b (Sinauer Associates, Inc., Sunderland, Mass.) using a neighbor-joining algorithm (1,000 bootstrap replicates) and a Jukes-Cantor correction after eliminating gapped positions. The rbcL sequences from Prochlorococcus and Synechococcus were used as outgroups to root trees.
The computer program LIBSHUFF (34) was used to estimate homologous and heterologous coverages of clone libraries as a function of evolutionary distance for pairwise reciprocal comparisons (library A compared with library B and vice versa). The significance of differences in coverage versus evolutionary distance between libraries was estimated by a bootstrap algorithm. Differences in coverage were considered significant at P values of ≤0.05.
Analysis of molecular variance (AMOVA) as implemented with Arlequin (31) was used to estimate the significance of differences in population pairwise fixation indices (FST values) among volcanic deposit libraries. Arlequin was also used to estimate nucleotide diversity, average pairwise differences, and mismatch distributions for each library (see reference 31 for details). For a given set of aligned sequences within a library, nucleotide diversity is a measure of the probability that two randomly chosen homologous nucleotides will differ; average pairwise difference is a measure of the number of nucleotide differences observed when each clone sequence is compared with all other clone sequences (31).
A phylogeny test, or P test, was used to determine the significance of covariation between the distribution of unique sequences within libraries and phylogeny (21). One thousand random trees were generated by PAUP version 4.0b from the combined clone libraries. The 95% lower confidence limit of the random tree lengths was compared to the minimum length of the trees obtained using maximum parsimony with a heuristic search algorithm. The P test was considered significant if maximum parsimony tree lengths were less than the 95% lower confidence limit of the random tree lengths.
Nucleotide sequence accession numbers.
rbcL isolates have been deposited in GenBank under accession numbers AY422046 through AY422057, microbial mat sequences have been deposited under AY422058 through AY422067, and volcanic deposit sequences have been deposited under AY422838 through AY422948.
RESULTS
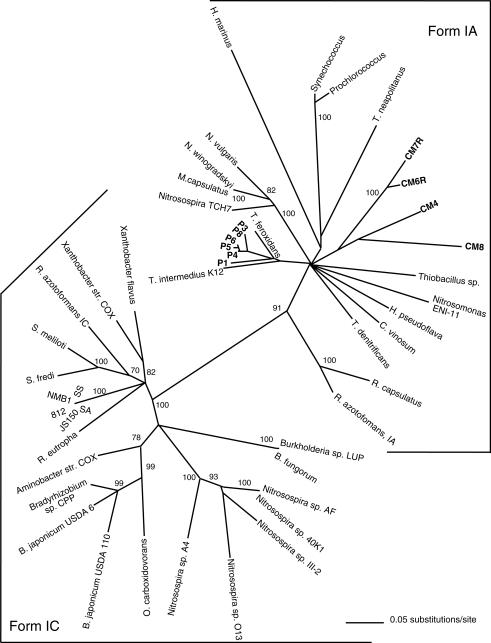
PCR products of the expected size were obtained from 14 of 17 CO or hydrogen oxidizers and a methanotroph known to contain rbcL. PCR products were not obtained from three CO oxidizers (Rhizobium leguminosarum and marine isolates Stappia sp. strain CV902-700 and Stappia sp. strain M8) or from two isolates that do not oxidize CO and are not otherwise known to contain rbcL (Cycloclasticus spirillensus and Lutibacterium anuloederans). BLAST analyses indicated that the sequences of all the PCR products were most similar to those of rbcL. Phylogenetic analysis indicated that the sequences from CO oxidizers belonged to the rbcL form IC clade (Fig. 1). rbcL from the genome sequences of several known and newly recognized CO oxidizers (e.g., Bradyrhizobium japonicum, Bradyrhizobium fungorum LB400, and Sinorhizobium meliloti) also fell within this clade (Fig. 1). All the sequences derived from CO oxidizers were distinct from those of the Nitrosospira strains that formed a coherent group within form IC (Fig. 1). All the form IC rbcL sequences were 492 bp in length, while those of form IA were 495 bp in length. The different lengths were due to the presence of an additional glycine codon 100 residues downstream from the forward primer in the form IA sequences.
FIG. 1.
Neighbor-joining analysis of 401 bp of bacterial rbcL sequences by use of 1,000 bootstrap replicates. Bootstrap values of ≥70% are indicated at the nodes. Colorless (CM) and purple (P) sulfide-oxidizing mat clone sequences are indicated in bold. GenBank accession numbers for isolates: Aminobacter strain COX, AY422046; Bradyrhizobium japonicum USDA 110, AF041820; Bradyrhizobium japonicum USDA 6, AY422048; Bradyrhizobium strain CPP, AY422047; Burkholderia strain JS150, AY422049; Burkholderia strain LUP, AY422050; Hydrogenophaga pseudoflava, U55037; Hydrogenovibrio marinus, D43622; Mesorhizobium strain NMB1, AY422051; Methylococcus capsulatus, AF447860; L32182Nitrosomonas europea, AF426427; Nitrobacter vulgarus, L22885; Nitrobacter winogradskyi strain IFO14297, AF109915; Nitrosomonas sp. strain ENI-11, AB061373; Nitrosopira sp. strain O13, AF426422; Nitrosospira sp. strain 40K1, AF426428; Nitrosospira sp. strain III2, AF426416; Nitrosospira sp. strain AF, AF426415; Nitrosospira sp. strain TCH716, AF459718; Oligotropha carboxidovorans, AY422052; Prochlorococcus marinus, AE017162; Ralstonia eutropha, U20585; Rhodobacter sphaeroides, M64624; Rhodobacter azotoformans, AB062779 and AB062780; Rhodobacter capsulatus, L82000; Sinorhizobium meliloti, AF211846; Sinorhizobium fredi USDA 205, AY422053; Stappia stellulata, AY422054; Stappia aggregata, AY422055; Stappia strain CV812-530, AY422056; Synechococcus sp. strain CcmK, U46156; Thiobacillus denitrificans, L42940; Thiobacillus ferroxidans, X70355; Thiobacillus intermedius K12, AF046933; Thiobacillus neapolitanus, AF038430; Thiobacillus sp., M34536; Xanthobacter flavus, Z22705; Xanthobacter strain COX, AY422057. Accession numbers for the mat clone sequences are AY422058 through AY422067.
The clones prepared with rbcL amplified from colorless and purple sulfide-oxidizing microbial mats yielded sequences that fell within form IA (Fig. 1); no form IC sequences were observed. Although only a small number of clones were sequenced, the results indicated that the two mat communities were dominated by different lithotrophic populations. However, the clone sequences from the purple sulfide-oxidizing mat were most closely related to the sequences of two thiobacilli and distinct from the sequence for a purple sulfide oxidizer, Chromatium vinosum (Fig. 1). In contrast, the clone sequences from the colorless sulfide-oxidizing mat were not clearly associated with known thiobacilli or other obligate lithotrophs (Fig. 1).
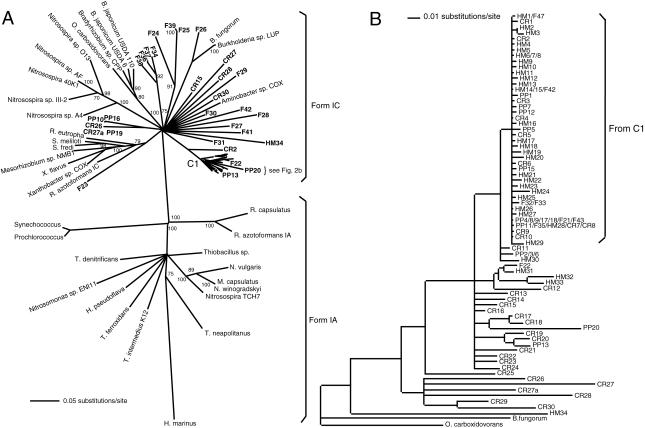
The clone sequences derived from volcanic-deposit DNA clustered exclusively with form IC rbcL (Fig. 2). Each of the 138 sequences was 492 bp in length and lacked the glycine codon found in form IA sequences (see above). The clone sequences from Forest and Pu'u Puai were distributed throughout the form IC clade but were not closely associated with the sequences from known CO oxidizers (Fig. 2A). The Halema'uma'u and Caldera Rim clone sequences also differed from the known CO oxidizer rbcL sequences but formed a distinct cluster of closely related sequences (Fig. 2B).
FIG. 2.
(A) Neighbor-joining analysis of 400 bp of rbcL clone sequences by use of 1,000 bootstrap replicates. Bootstrap values of >70% are indicated at the nodes. Clone sequences are indicated in bold. Abbreviations for clone sequences: CR, Caldera Rim; F, Forest; HM, Halema'uma'u; PP, Pu'u Puai. Accession numbers for the isolate sequences are provided in the legend to Fig. 1. Clone sequence accession numbers are AY422838 through AY422948. (B) Expanded view of clone sequences shown in panel A. Clone relationships derived from a neighbor-joining analysis are presented as a phylogram with sequences from Burkholderia fungorum and Oligotropha carboxydovorans used to root the tree. Abbreviations and accession numbers are as defined for panel A. Sequences with multiple designations occurred multiple times in one or more of the libraries.
Statistical comparisons of clone sequences performed with Arlequin revealed significant differences in diversity within and among libraries (Tables 1 and 2). Nucleotide diversity was highest for the Forest and lowest for the Halema'uma'u clone libraries, while the values for Pu'u Puai and Caldera Rim were essentially identical (Table 1). A similar trend was observed for average pairwise differences [θ(π)]. Comparisons of clones within and among libraries by AMOVA (Table 2) revealed significant differences (P < 0.05) for all average pairwise differences and for pairwise fixation indices, FST (Table 2).
TABLE 1.
Characteristics of rbcL clone libraries sequences for genomic extracts from recent volcanic depositsa
| Site | Age (yr) | No. of clones | No. of VP | ND | θ(π) |
|---|---|---|---|---|---|
| PP | 43 | 23 | 169 | 0.086 (0.043) | 42.4 (21.3) |
| Forest | ∼300 | 20 | 255 | 0.22 (0.11) | 108.1 (48.5) |
| HM | 108 | 54 | 163 | 0.029 (0.015) | 14.3 (7.2) |
| CR | 212 | 41 | 192 | 0.089 (0.043) | 43.7 (21.5) |
The number of clones analyzed by Arlequin is indicated for each site. Variable positions (VP) represent the number of variable sites (out of 492 total) within each library. Nucleotide diversity (ND) and average pairwise difference [θ(π)] are expressed as means (±1 standard deviation) for each site. Site abbreviations: PP, Pu'u Puai; HM, Halema'uma'u; CR, Caldera Rim.
TABLE 2.
Corrected average pairwise differences and pairwise fixation indicesa
| Site | θ[π] or FST for site:
|
|||
|---|---|---|---|---|
| PP | Forest | HM | CR | |
| PP | 105.4 | 30.8 | 46.1 | |
| Forest | 0.294 | 103.4 | 108.8 | |
| HM | 0.111 | 0.528 | 34.9 | |
| CR | 0.065 | 0.344 | 0.181 | |
Corrected average, pairwise differences (θ[π]) are shown above diagonal; pairwise fixation indices (FST) are shown below diagonal. All values are significant at a P of ≤ 0.05. Site abbreviations: PP, Pu'u Puai; HM, Halema'uma'u; CR, Caldera Rim.
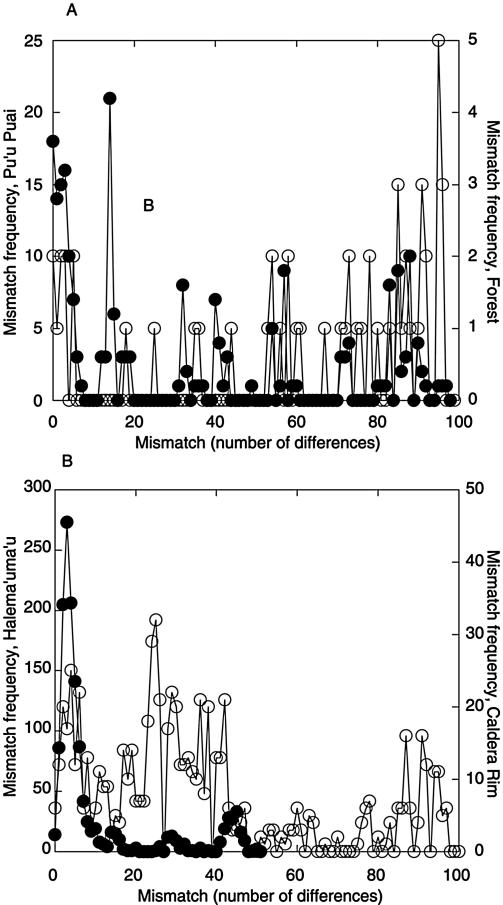
The occurrence of mismatches obtained from pairwise clone comparisons was plotted as a function of the mismatch number for each library (Fig. 3). The distributions for Pu'u Puai, Forest, and Caldera Rim were multimodal or ragged, but the Forest distribution was substantially damped relative to that of other sites (Fig. 3). In contrast, the Halema'uma'u distribution was very strongly skewed, with a prominent peak representing one to six mismatches (Fig. 3B).
FIG. 3.
(A) Frequency distribution of mismatches in Forest (○) and Pu'u Puai (•) clone libraries; (B) frequency distribution of mismatches in Caldera Rim (○) and Halema'uma'u (•) clone libraries. Note differences in frequency scales.
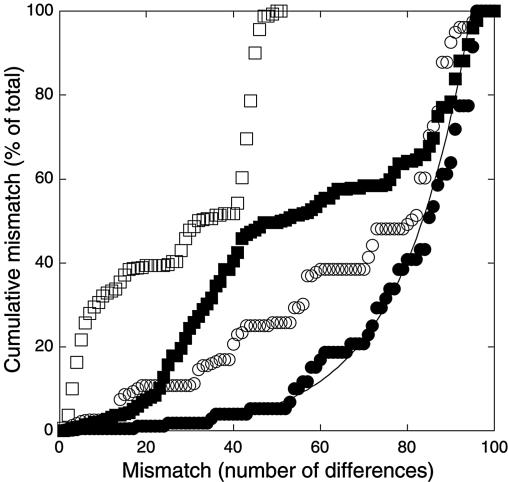
Plots of the cumulative percentage of total mismatches revealed distinct patterns for the four libraries (Fig. 4). A relatively large fraction of closely related Halema'uma'u clone sequences (≤10 mismatches or ≥98% similarity) accounted for about 30% of total mismatches, while a smaller fraction of more distantly related clones (40 to 50 mismatches) accounted for about 50% of the total. For the Forest library, the cumulative percentage of total mismatches increased exponentially with the mismatch level (Fig. 4). The patterns for Pu'u Puai and Caldera Rim were intermediate between those for Halema'uma'u and Forest.
FIG. 4.
Cumulative mismatch as a percentage of total mismatch within clone libraries versus mismatch level for Forest (•), Pu'u Puai (○), Caldera Rim (□), and Halema'uma'u (▪) clone libraries.
The “evaluate random trees” algorithm of PAUP version 4.0b was used to create 1,000 random trees from pooled volcanic-deposit clone libraries with parsimony as an optimality criterion. The 95% lower confidence limits of the random tree lengths were compared to the lengths of trees resulting from heuristic searches using parsimony. The latter lengths were always substantially lower than the 95% lower confidence limits of the random trees (P < 0.05).
LIBSHUFF analysis of the clone libraries indicated that homologous coverage, a measure of the extent of sequence similarity within libraries, ranged from 20 to 39% (Table 3). Heterologous coverage, which measured the representation of a given library in another and vice versa, ranged from 6 to 35% and in general was lower than homologous coverage. Pairwise reciprocal library comparisons (Table 3) indicated that the Forest and Caldera Rim libraries were significantly different from all others (P < 0.05), that Halema'uma'u differed marginally from Pu’u Puai (P = 0.053) and Caldera Rim (P = 0.064), and that Pu’u Puai differed from Halema’uma’u (P = 0.001).
TABLE 3.
LIBSHUFF comparison of clone librariesa
| Clone library | Covhom (%) | Covhet (%) | P |
|---|---|---|---|
| Forest | 20.0 | 10.0 | 0.001 |
| Halema'uma'u | 25.9 | 5.6 | 0.068 |
| Forest | 10.0 | 0.001 | |
| Caldera Rim | 22.0 | 12.2 | 0.008 |
| Forest | 10.0 | 0.001 | |
| Pu'u Puai | 39.1 | 30.4 | 0.081 |
| Halema'uma'u | 9.3 | 0.064 | |
| Caldera Rim | 17.1 | 0.001 | |
| Halema'uma'u | 5.6 | 0.301 | |
| Pu'u Puai | 30.4 | 0.001 | |
| Caldera Rim | 14.6 | 0.005 | |
| Pu'u Puai | 34.8 | 0.947 |
Homologous (Covhom) and heterologous (Covhet) coverages of libraries are given. Probability values (P) for the significance of differences between homologous and heterologous coverages in reciprocal comparisons as a function of evolutionary distance are also given. See the work of Singleton et al. (34) for details.
DISCUSSION
The gene that codes for the large subunit of RubisCO, rbcL, has been targeted for molecular assays of phytoplankton population structure, diversity, and dynamics (25-27, 41-44). Several primers have proven suitable for form I genes in marine and freshwater phototrophs. Xu and Tabita (44) have examined rbcL expression and phytoplankton diversity at several sites and depths in Lake Erie. Pichard et al. (27) and Paul et al. (25, 26) have documented spatial diversity for form IA, IB, and ID rbcL sequences in the Gulf of Mexico. Wyman et al. (42, 43) have used group-specific primers to determine diatom and prymnesiophyte dynamics during nutrient-stimulated blooms. Wawrik et al. (41) have also used form ID-specific primers for real-time PCR assays of rbcL expression in cultures and natural phytoplankton assemblages.
In contrast, little is known about bacterial rbcL distribution and diversity. Elsaied and Naganuma (9) have reported sequences from unknown form IA and IB and form II lineages from various deep-sea samples. They have documented several Thiobacillus-like sequences but have observed no form IC sequences (9). Alfrieder et al. (1) have examined various groundwater systems by use of a different set of form IA and IB primers. Like Elsaied and Naganuma (9), Alfrieder et al. (1) have reported sequences from unknown lineages and a few sequences similar to those of sulfide oxidizers. Several form IA sequences similar to rbcL from a known aerobic CO and H2 oxidizer (Hydrogenophaga pseudoflava) were observed, but again form IC sequences were absent (1). However, the latter observation may reflect primer specificity rather than the relative contributions of the two groups to groundwater lithotrophic communities.
Although the primers designed by Xu and Tabita (44) were based on highly conserved regions of phototroph rbcL sequences, they appear unsuitable for bacterial rbcL assays due to significant nucleotide mismatches in the targeted regions (King and Crosby, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol.). However, the modifications described here have facilitated rbcL amplification from both facultative and obligate bacterial lithotrophs (Fig. 1). The former includes various CO- and hydrogen-oxidizing Proteobacteria, with the latter represented by Thiobacillus-like sequences obtained from purple and colorless sulfide-oxidizing microbial mats (Fig. 1).
The results presented here indicate that the primers used in this study amplify a broad spectrum of form IC rbcL genes. The results from analyses of microbial mats and volcanic materials, and from an examination of primer binding sites in diverse form IA and IC sequences from GenBank, support the general utility of the primers. Similar levels of nucleotide identity within primer binding sites were found for sequences representative of each form (data not shown), which suggests that bias among the forms may be limited.
In contrast to the results from mat (Fig. 1) and phytoplankton surveys (25-27, 41-44), only form IC clone sequences have been obtained from volcanic deposits (Fig. 2). As with other studies (1, 9), few of the clone sequences are sufficiently similar to published rbcL sequences to allow unambiguous inferences about taxonomic or phylogenetic affinities. Indeed, most of the sequences appear to have been derived from undescribed organisms or organisms not known to contain rbcL.
Phylogenetic analyses of clone sequences from volcanic deposits suggest that the four sites examined harbor distinct lithotrophic populations (Fig. 2). Statistical analyses support this proposition (Fig. 3 and 4; Tables 1 to 3). Both nucleotide diversity and average pairwise difference within library sequences are about eightfold higher for Forest than for Halema'uma'u (Table 1), which indicates that the oldest, most biologically complex site, Forest, supports the most divergent sequences, while the least divergent sequences are found in the least developed site, Halema'uma'u.
Based on AMOVA, the average pairwise differences derived from paired library comparisons are all statistically significant (P < 0.05) (Table 2) and are consistently higher for comparisons with the data for the Forest site, which agrees with the phylogenetic results (Fig. 2) and buttresses the notion that the Forest site contains the most divergent lineages. Average pairwise differences from paired libraries also provide further evidence for the presence of distinct populations among sites.
FST, or fixation index, also derived from AMOVA, measures genetic diversity within a population compared to the total diversity of the pooled populations (21, 31). Low values indicate similar levels of total and within-population diversity, while high values indicate that total diversity exceeds that within populations. Accordingly, the greatest differentiation among sites occurs between Forest and Halema'uma'u (Table 2), which represent two extremes with respect to ecosystem development. The least differentiation based on FST occurs between Caldera Rim and Pu'u Puai (Table 2), which is consistent with phylogenetic analyses that indicate that some of the clones from Pu'u Puai tend to be distributed among those from Caldera Rim (Fig. 2). This may reflect the impact on microbial communities of similar levels of plant community development at the two sites.
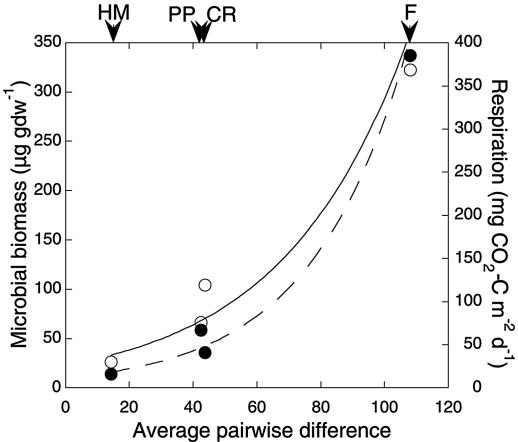
Mismatch distribution plots also reveal distinct differences among the sites (Fig. 3). For Forest, the results indicate that populations consist of an approximately even distribution of closely related (few mismatches) and distantly related (many mismatches) taxa (Fig. 3A), which likely accounts for the exponential increase in the cumulative fraction of total mismatches as a function of observed mismatches (Fig. 4). In contrast, the data from Halema'uma'u suggest that relatively closely related taxa with few divergent groups dominate lithotroph populations. One interpretation of these patterns is that they reflect an increase in phyletic diversity with increasing ecosystem complexity. Such a relationship may also account for the positive correlations of respiration and microbial biomass with average pairwise difference (Fig. 5) and nucleotide diversity (data not shown).
FIG. 5.
Exponential relationship for microbial biomass (r = 0.99) (○) and respiration (r = 0.99) (•) with average pairwise difference within clone libraries (Table 1); library abbreviations are as defined in the legend to Fig. 2. Biomass and respiration data are taken from a report by King (18). Arrows indicate average pairwise differences for each library.
Two additional statistical analyses corroborate population differentiation among sites. A P test (21) shows that sequence distribution within libraries covaries significantly with phylogenetic position. A significant P test along with significant FST values strongly suggests that differences in site age, plant development, and various physical and chemical parameters (18) affect the composition of lithotrophic communities.
The results from LIBSHUFF analyses also confirm population differentiation but provide somewhat different insights. Paired reciprocal library comparisons show that the Forest library differs significantly from all others up to evolutionary distances of 0.2 or more but the Pu’u Puai and Halema’uma’u libraries do not differ from that of the Forest site (Table 3). One explanation for this pattern is that the diversity of the Forest library sequences encompasses and largely describes sequences in the other libraries, while they account for only a portion of the Forest diversity (12, 37). This explanation is consistent with the phylogenetic distribution of sequences (Fig. 2). By analogy, the Caldera Rim diversity encompasses that of Halema'uma'u and Pu’u Puai, but the reverse is not true, and the Pu’u Puai diversity largely encompasses that of Halema’uma’u, while the reverse does not hold true.
Collectively, LIBSHUFF and other analyses suggest that although the populations among the four sites differ significantly, they may share a core set of phylogenetically similar taxa that is retained throughout successional development. Additional, more divergent taxa recruit over time as a function of local characteristics, resulting in different community structures. Populations at Halema'uma'u, the least developed site, may be dominated by examples of core populations. In contrast, the Forest appears to provide examples of populations common in more complex and mature systems.
At present, it is not possible to relate physiological or ecological function definitively to phylogenetic position within the form IC clade. Although facultative lithotrophs such as CO- and hydrogen-oxidizing bacteria dominate the clade, several obligately lithotrophic, ammonium-oxidizing Nitrosospira strains form a distinct IC cluster (Fig. 1) (38). This cluster indicates that multiple functional groups contain form IC rbcL. The same is true for form IA rbcL, which occurs in sulfide and ammonium oxidizers, including Nitrosospira species (33, 38), and at least one oxidizer of CO and hydrogen (8, 33).
Nonetheless, several lines of evidence suggest that most of the clone sequences likely represent CO and hydrogen oxidizers. First, all of the clone sequences are phylogenetically distinct from Nitrosospira sequences with a high level of bootstrap support (Fig. 2). Second, ammonium oxidation has been detected consistently only at the Forest site, and the rates were relatively low (18). Third, efforts to amplify ammonia oxygenase genes have proven successful only for Forest samples (V. Gomez-Alvarez, G. M. King, and K. Nüsslein, unpublished data). In contrast, all of the sites consume CO and hydrogen (18) and contain CO dehydrogenase genes (Dunfield and King, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol.).
Even though CO and H2 oxidizers are distributed ubiquitously and may account for a large fraction of observed rbcL sequences, the indices of form IC diversity (Table 1) do not appear to vary significantly with CO and hydrogen oxidation rates (r < 0.6). The limited correlation between these variables may reflect the limitations of the data set rather than the principles governing relationships between diversity and metabolic activity. In particular, only four sites have been assessed thus far, and with the exception of the Forest site, CO and hydrogen uptake rates do not differ substantially among the sites (18).
Regardless, two related measures of diversity within clone libraries, nucleotide diversity and average pairwise difference (Table 2), correlate positively (Fig. 5) with respiration (r = 0.99) and microbial biomass (r = 0.99). A positive correlation indicates that increases in facultative lithotroph diversity are associated with system-level changes that elevate microbial biomass and total heterotrophic activity measured as respiration. Changes in the latter parameters reflect increases in organic matter that accompany successional development from initially barren volcanic deposits to a closed-canopy forest (18). Increased facultative lithotroph diversity is potentially consistent with increasing heterotrophic substrate availability, since facultative lithotrophs may grow mixotrophically in situ. In addition, changes in biotic and physical complexity (e.g., development of plant communities and soil) accompanying ecosystem development may contribute to patterns of microbial diversity.
In conclusion, rbcL sequence data reveal distinct populations of largely undescribed facultative lithotrophs among recent volcanic deposits differing in depositional age and successional state. These populations appear to dominate the bacterial autotroph communities at the sites, which is consistent with published biogeochemical measurements (18). Indices of rbcL diversity increase with estimates of respiration and biomass, a trend not previously reported for other functional genes. The ability of the primers used in this study to amplify rbcL from obligate and facultative lithotrophs suggests that analysis of RubisCO genes may provide valuable insights into the structure and dynamics of diverse microbial communities.
Acknowledgments
This work was supported in part by the National Science Foundation (BIO-0085495).
We thank K. Roache for technical assistance and W. Gilmartin for use of Hale Mahana.
Footnotes
Contribution 391 from the Darling Marine Center.
REFERENCES
- 1.Alfreider, A., C. Vogt, D. Hoffman, and W. Babel. 2003. Diversity of ribulose 1,5-bisphosphate carboxylase/oxygenase large-subunit genes from groundwater and aquifer microorganisms. Microb. Ecol. 45:317-328. [DOI] [PubMed] [Google Scholar]
- 2.Arp, D. J., L. A. Sayavedra-Soto, and N. G. Hommes. 2002. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178:250-255. [DOI] [PubMed] [Google Scholar]
- 3.Bock, E., and M. Wagner. 2001. Oxidation of inorganic nitrogen compounds as an energy source. In M. Dworkin et al. (ed.). The prokaryotes: an evolving electronic resource for the microbiological community. Springer-Verlag. [Online.] http://link.springer-ny.com/link/service/books/10125/.
- 4.Cannon, G. C., S. H. Baker, F. Soyer, D. R. Johnson, C. E. Bradburne, J. L. Mehlman, P. S. Davies, Q. L. Jiang, S. Heinhorst, and J. M. Shively. 2003. Organization of carboxysome genes in the thiobacilli. Curr. Microbiol. 46:115-119. [DOI] [PubMed] [Google Scholar]
- 5.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung, W. K., and G. M. King. 2001. Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl. Environ. Microbiol. 67:5585-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crews, T. E., K. Kitayama, J. H. Fownes, R. H. Riley, D. A. Herbert, D. Mueller-Dombois, and P. M. Vitousek. 1995. Changes in soil phosphorous fractions and ecosystem dynamics across a long chronosequence in Hawaii. Ecology 76:1407-1424. [Google Scholar]
- 8.Delwiche, C. F., and J. D. Palmer. 1996. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13:873-882. [DOI] [PubMed] [Google Scholar]
- 9.Elsaied, H., and T. Naganuma. 2001. Phylogenetic diversity of ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes from deep-sea microorganisms. Appl. Environ. Microbiol. 67:1751-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn, M. W., and F. R. Tabita. 2003. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furlong, M. A., D. R. Singleton, D. C. Coleman, and W. B. Whitman. 2002. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl. Environ. Microbiol. 68:1265-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, K. R., and G. M. King. 2001. Enrichment of high-affinity CO oxidizers in Maine forest soil. Appl. Environ. Microbiol. 67:3671-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartman, F. C., and M. R. Harpel. 1994. Structure, function, regulation and assembly of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 63:197-234. [DOI] [PubMed] [Google Scholar]
- 14.Heinhorst, S., S. H. Baker, D. R. Johnson, P. S. Davies, G. C. Cannon, and J. M. Shively. 2002. Two copies of form I RuBisCO genes in Acidithiobacillus ferroxidans ATCC 23270. Curr. Microbiol. 45:115-117. [DOI] [PubMed] [Google Scholar]
- 15.Karavaiko, G. I., T. P. Turova, T. F. Kondrat'eva, A. M. Lysenko, T. V. Kolganova, S. N. Ageeva, L. N. Muntyan, and T. A. Pivovarova. 2003. Phylogenetic heterogeneity of the species Acidithiobacillus ferrooxidans. Int. J. Syst. Evol. Microbiol. 53:113-119. [DOI] [PubMed] [Google Scholar]
- 16.Kelly, D. P., and A. P. Wood. 2000. Reclassification of some species of Thiobacillus to the newly designed genera Acidithiobacillus gen. nov., Halothiobacillus gen. nov. and Thermithiobacillus gen. nov. Int. J. Syst. Evol. Microbiol. 50:511-516. [DOI] [PubMed] [Google Scholar]
- 17.Kim, E. Y., Y. T. Ro, and Y. M. Kim. 1997. Purification and some properties of ribulose 1,5-bisphosphate carboxylase/oxygenases from Acinetobacter sp. strain JC1 and sp. strain JC1 and Hydrogenophaga pseudoflava. Mol. Cells. 7:380-388. [PubMed] [Google Scholar]
- 18.King, G. M. 2003. Contributions of atmospheric CO and hydrogen uptake to microbial dynamics on recent Hawaiian volcanic deposits. Appl. Environ. Microbiol. 69:4067-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, G. M. 2003. Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusian, B., and B. Bowien. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135-155. [DOI] [PubMed] [Google Scholar]
- 21.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer, O., and H. G. Schlegel. 1983. Biology of aerobic carbon monoxide-oxidizing bacteria. Annu. Rev. Microbiol. 37:277-310. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, O., K. Frunzke, D. Gadkari, S. Jacobitz, I. Hugendieck, and M. Kraut. 1990. Utilization of carbon monoxide by aerobes: recent advances. FEMS Microbiol. Rev. 87:253-260. [Google Scholar]
- 24.Mörsdorf, G., K. Frunzke, D. Gadkari, and O. Meyer. 1992. Microbial growth on carbon monoxide. Biodegradation 3:61-82. [Google Scholar]
- 25.Paul, J. H., A. Alfreider, and B. Wawrik. 2000. Micro- and macrodiversity in rbcL sequences in ambient phytoplankton populations from the southeastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 198:9-18. [Google Scholar]
- 26.Paul, J. H., A. Alfreider, J. B. Kang, R. A. Stokes, D. Griffin, L. Campbell, and E. Ornolisdottir. 2000. Form IA rbcL transcripts associated with a low salinity/high chlorophyll plume (′Green River') in the eastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 198:1-8. [Google Scholar]
- 27.Pichard, S. L., L. Campbell, and J. H. Paul. 1997. Diversity of the ribulose bisphosphate carboxylase/oxygenase form I gene (rbcL) in natural phytoplankton communities. Appl. Environ. Microbiol. 63:3600-3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H.-P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 30.Robinson, J. J., J. L. Stein, and C. M. Cavanaugh. 1998. Cloning and sequencing of a form II ribulose-1,5-bisphosphate carboxylase/oxygenase from the bacterial symbiont of the hydrothermal vent tubeworm Riftia pachyptila. J. Bacteriol. 180:1596-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider, S., D. Roesseli, and L. Excoffier. 2000. Arlequin version 2.000, a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
- 32.Shively, J. M., W. Devore, L. Stratford, L. Porter, L. Medlin, and S. E. Stevens, Jr. 1986. Molecular evolution of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase (rubisCO). FEMS Microbiol. Lett. 37:251-257. [Google Scholar]
- 33.Shively, J. M., G. van Keulen, and M. G. Meijer. 1998. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu. Rev. Microbiol. 52:191-230. [DOI] [PubMed] [Google Scholar]
- 34.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, R., M. Ohishi, M. Ohshima, M. Saitoh, K. Omata, and T. Tokuyama. Characteristics of an ammonia-oxidizing bacterium with a plasmid isolated from alkaline soils and its phylogenetic relationship. J. Biosci. Bioeng. 92:232-236. [DOI] [PubMed]
- 36.Taylor, G. T., M. Iabichella, T.-Y. Ho, M. I. Scranton, R. C. Thunell, F. Mueller-Karger, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 37.Uchino, Y., and A. Yokota. 2003. “Green-like” and “red-like” rubisCO cbbL genes in Rhodobacter azotoformans. Mol. Biol. Evol. 20:821-830. [DOI] [PubMed] [Google Scholar]
- 38.Utåker, J. B., K. Andersen, Å. Aakra, B. Moen, and I. F. Nes. 2002. Phylogeny and functional expression of ribulose 1,5-bisphosphate carboxylase/oxygenase from the autotrophic ammonia-oxidizing bacterium Nitrosospira sp. isolate 40KI. J. Bacteriol. 184:468-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakaao, N., K. Hanada, A. Takahashi, Y. Sakurai, and H. Shojota. 1991. Morphological, physiological, and chemotaxonomical characteristics of iron and sulfur oxidizing bacteria isolated from acid mine drainage waters. J. Gen. Appl. Microbiol. 7:35-48. [Google Scholar]
- 40.Watson, G. M. F., and F. R. Tabita. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Microbiol. Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]
- 41.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wyman, M. 1999. Diel rhythms in ribulose-1,5-bisphosphate carboxylase/oxygenase and glutamine synthetase gene expression in a natural population of marine picoplanktonic cyanobacteria (Synechococcus spp.). Appl. Environ. Microbiol. 65:3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wyman, M., J. T. Davies, D. W. Crawford, and D. A. Purdie. 2000. Molecular and physiological responses of two classes of marine chromophytic phytoplankton (diatoms and prymnesiophytes) during the development of nutrient-stimulated blooms. Appl. Environ. Microbiol. 66:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, H. H., and F. R. Tabita. 1996. Ribulose-1,5-bisphosphate carboxylase/oxygenase gene expression and diversity of Lake Erie planktonic microorganisms. Appl. Environ. Microbiol. 62:1913-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]