Abstract
A strain affiliated with the Roseobacter clade and producing a new antibiotic named tropodithietic acid (L. Liang, Ph.D. thesis, University of Göttingen, Göttingen, Germany, 2003) was isolated from the German Wadden Sea. The compound showed strong inhibiting properties with respect to marine bacteria of various taxa and marine algae. Antibiotic production was found to occur during the complete growth phase. Strain mutants without antagonistic properties appeared several times spontaneously.
Since the first antibiotic from a marine bacterium was described in 1966 (6), the number of new compounds has increased constantly during the years. Even though only a few compounds from marine organisms might be interesting for the pharmaceutical market today (7), some bacterial species are already used as biocontrols and are added to aquaculture stocks. Most secondary metabolites from marine bacteria found so far were isolated from Streptomyces and Alteromonas species (24). In recent studies wide arrays of marine bacteria were tested for bacterial antagonistic effects, and it was demonstrated that this trait appears to be a widespread feature in marine habitats and present in many bacterial groups (5, 9, 15). Production of secondary metabolites by bacteria of the Roseobacter group has been reported previously (8, 13), and some organisms of this group are thought to be either probiotic or antibiotic in different aquacultures (3, 19). Here we describe the antagonistic activity of a new strain of the Roseobacter clade against marine bacteria and algae.
A water sample was taken above an intertidal mud flat of the German Wadden Sea (53°42′20"N, 07°43′11"E) on 25 October 1999 (water temperature, 9.6°C; pH, 7.9; O2 saturation, 94%; salinity, 34‰). The sample was taken directly to the laboratory for further processing. Tubes containing 9 ml of marine broth 2216 (Difco) were inoculated with 1 ml of seawater, and the contents were vigorously mixed. Using these tubes, we prepared dilution series with 1:10 steps. The cultures were incubated in the dark at 4, 15, 20, and 28°C (with shaking) for 4 weeks. Growth was determined microscopically and by monitoring turbidity. Aliquots (100 μl) from the highest and lowest most-probable-number dilutions of each parallel experiment in which growth was obtained were spread on agar plates with marine agar 2216 (Difco). Plates were incubated at 4, 15, 20, or 28°C in the dark. Different types of colonies obtained were streaked out on fresh plates and transferred at least three times for purification. Bacterial strains were compared by colony morphology and color and by denaturing gradient gel electrophoresis as previously described (23). For a screening of the production of new antimicrobial substances, strains were selected which showed pigmentation or for which no closely related organisms were found by BLAST analysis of their 16S rRNA gene fragments.
To detect antimicrobial activity, agar diffusion tests with standard test organisms were performed using Bacillus subtilis and Staphylococcus aureus (both from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany) (DSM 10 and DSM 20231, respectively) and Escherichia coli strain K-12 (obtained from H.-P. Fiedler, University of Tübingen, Tübingen, Germany). B. subtilis was grown in medium containing the following ingredients (in grams per liter): glucose (5.0), Na citrate · 2H2O (0.5), KH2PO4 (3.0), K2HPO4 (7.0), MgSO4 · 7H2O (0.1), and (NH4)2SO4 (1.0). S. aureus and E. coli were grown in medium containing the following ingredients (in grams per liter): Bacto nutrient broth (8.0) and NaCl (5.0). For preparation of the test plates, cultures were adjusted to an optical density at 600 nm (OD600) of 1.0 for B. subtilis and 1.3 for E. coli and S. aureus. Plates were prepared with the media described above containing 15.0 g of agar/liter and were overlaid with top agar containing 1% of the cell suspensions. Strains tested for antibacterial activity were grown in marine broth 2216 at 20°C until the stationary phase was reached. Sterile antibiotic assay disks (Schleicher and Schuell, Dassel, Germany) (diameter, 6 mm) were soaked with culture broth of the strains and put on plates with B. subtilis, S. aureus, and E. coli. The plates were incubated at 37°C in the dark overnight and checked afterwards for inhibition zones. For one of our strains (T5), strong antibacterial activity was detected. Isolation and structure elucidation of the new antibiotic tropodithietic acid obtained from this strain have been described previously by Liang (14).
Subsequently, growth inhibition of marine bacteria by strain T5, by a mutant of T5 (strain T5-3) (see below), by Roseobacter gallaeciensis BS107 (from the Collection de l'Institut Pasteur, Paris, France) (CIP 105210), by Ruegeria algicola (DSM 10251), and by Ruegeria gelatinovorans (DSM 5887) was investigated by agar diffusion tests. These strains (as well as 14 other strains, all of which were obtained during this study) were also used as target organisms. These isolates were chosen for inhibition experiments because of their differences (at least on the genus level) in phylogenetic affiliations (Table 1). Strains were pregrown for 4 days at 20°C with shaking in 5 ml of marine broth 2216. A culture broth (100 μl) with an adjusted OD600 of ∼0.5 was spread on plates with marine agar 2216. For strains T4 and T15, an OD600 of only ∼0.07 was obtained because of weaker growth. Sterile antibiotic assay disks were put on these plates, to which 10 μl of culture broth (OD600, ∼0.5) of the organisms (which were tested for production of antibiotic compounds) was applied. A 10-μl portion of medium was tested as a control, and 10 μl of a pure 1 mM tropodithietic acid solution (prepared from the isolated and purified compound and dissolved with 1% dimethyl sulfoxide [DMSO]) was tested for comparison to the culture broths. The sensitivity of bacterial strains to a 1% DMSO solution (10 μl) was also tested. Penicillin G, streptomycin sulfate, and chloramphenicol (all from Sigma, Deisenhofen, Germany) (10 μl of a 1 mM solution) were used as references. All inhibition experiments were carried out at least in parallel. Plates were incubated for 2 to 4 days (depending on growth of the test strains) at 20°C.
TABLE 1.
Phylogenetic affiliations of bacterial isolates and dilution steps they were obtained from
| Strain | Dilution | Phylogenetic group (class) | Closest described relativea (accession no.) | 16S rRNA similarity (%) |
|---|---|---|---|---|
| T5 | 10−0 | α-Proteobacteria | Roseobacter gallaeciensis (Y13244) | 99 |
| T3 | 10−4 | α-Proteobacteria | Erythromicrobium ramosum (AB013355) | 96 |
| T11 | 10−0 | α-Proteobacteria | Roseovarius tolerans (Y11551) | 95 |
| TK | 10−4 | α-Proteobacteria | Mesorhizobium tianshanense (AF041447) | 97 |
| TL | 10−5 | α-Proteobacteria | Leisingera methylohalidivorans (AY005463) | 95 |
| T1 | 10−0 | γ-Proteobacteria | Shewanella colwelliana (AF170794) | 98 |
| T8 | 10−0 | γ-Proteobacteria | Pseudoalteromonas haloplanktis subsp. tetraodonis (AF214730) | 99 |
| T16 | 10−0 | γ-Proteobacteria | Vibrio furnissii (X76336) | 96 |
| T17 | 10−0 | γ-Proteobacteria | Marinomonas mediterranea (AF063027) | 95 |
| TAI | 10−0 | γ-Proteobacteria | Colwellia maris (AB002630) | 93 |
| BIA | 10−6 | Flavobacteria | Psychroserpens burtonensis (U62912) | 93 |
| T15 | 10−5 | Flavobacteria | Tenacibaculum mesophilum (AB032504) | 93 |
| TN | 10−5 | Flavobacteria | Zobellia uliginosa (M62799) | 90 |
| T2 | 10−4 | Actinobacteria | Aeromicrobium fastidiosum (Z78209) | 97 |
| T4 | 10−4 | Actinobacteria | Pseudonocardia alni (Y08535) | 99 |
Affiliations determined by BLAST analysis (http://www.ncbi.nlm.nih.gov/blast). Only validly published organisms were taken into account.
Liang (14) described inhibition of freshwater algae by tropodithietic acid. To study effects on marine algae, we performed inhibition experiments with one axenic algal culture of Skeletonema costatum (from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton) (CCMP 1332) and one nonaxenic culture of a Nitzschia sp. obtained from the Wadden Sea and provided by E. Rhiel, University of Oldenburg, Oldenburg, Germany. S. costatum was grown on sterile Guillard seawater medium (f/2) (10) prepared with filtered (2-μm-pore-size filter) seawater from the Wadden Sea. The Nitzschia sp. was grown on a medium described by Schlösser (21). The cultures were incubated at 15°C in a 12-h-12-h dark-light cycle at approximately 85 μE m−2 s−1. For inhibition experiments, the organisms were pregrown in liquid cultures and then spread on agar plates containing the same medium and 0.9% (wt/vol) agar. Inhibition experiments with culture broths of strain T5, R. gallaeciensis, R. gelatinovorans, R. algicola, tropodithietic acid (dissolved with 1% DMSO), and a 1% DMSO solution were performed as described above for bacterial isolates and were carried out in parallel.
Tropodithietic acid production by T5, R. gallaeciensis, and T5-3 strains was measured by high-pressure liquid chromatography analysis. Overnight shaken cultures of the strains (6 ml) grown with marine broth 2216 were used. For extraction of tropodithietic acid, the pH of the sample was adjusted to pH 3 and the sample was centrifuged for 5 min at 6,240 × g. A total of 5 ml of the supernatant was passed over a column with 1 ml of Amberlite-XAD-16 (Sigma-Aldrich, Deisenhofen, Germany). After being washed with 10 ml of distilled water, tropodithietic acid was eluted with 3 ml of methanol. The extract was vacuum dried with a rotary evaporator at 37°C and redissolved in 1 ml of methanol. Reversed-phase high-pressure liquid chromatography (Thermo Separation Products, San Jose, Calif.) was used for measurement of the tropodithietic acid concentration. Samples were separated with a reversed-phase column (Knauer, Berlin, Germany) (250 by 3 mm; Nucleosil C 18/5 μm; integrated precolumn; injection volume, 20 μl; flow rate, 0.5 ml min−1) and 0.1% phosphoric acid-acetonitrile as an eluent (60% isocratic; 0.1% phosphoric acid and 40% acetonitrile). Tropodithietic acid was detected after 8.4 min with a diode array detector (La Chrom L-7450; Merck-Hitachi, Darmstadt, Germany) at 303 nm.
Covariance between growth and antibiotic production was investigated in a temperature- and pH-controlled 1.5-liter batch fermenter system (FairMenTec, Göttingen, Germany). The experiment was carried out at temperature and pH optima (30°C and pH 7.5) (T. Heidorn, unpublished results). Autoclaved and filtered marine broth 2216 was used as a growth medium. The fermenter was inoculated with a 10% volume of an overnight shaken preculture. Growth was determined by measuring the OD650 offline in a photometer (LKB, Cambridge, England). Antibiotic production was determined by agar diffusion tests with Bacillus amyloliquefaciens (DSM 7) as the test organism. For this purpose, 10 μl of culture broth was pipetted every hour onto a 6-mm-diameter filter paper disk and placed on an overlaid test plate. On each plate, one disk with 5 μl of penicillin G (0.3 mM) served as a reference. The experiment was carried out in triplicate. After incubation overnight at 30°C, diameters of the inhibition zones (the distances between antibiotic assay disks and bacterial lawn of the test strain) were determined. Relative inhibition was calculated as the ratio of the sample value to the value seen with the corresponding reference preparation.
For determining a standard curve between the OD and dry weight of the bacterial biomass, a well-grown culture was diluted to OD650 values in the range of 0.2 to 0.8. Distilled water was used for an OD650 = 0.0. For measuring dry weight, different volumes (from 5 ml at OD650 0.8 to 25 ml at OD650 0.2 and 0.0) of the dilutions were filtered onto 1.0-μm-pore-size glass fiber filters (type 13440; Sartorius, Göttingen, Germany). Each filter was washed with 50 ml of distilled water, blown with the same volume of air, dried at 105°C overnight, and cooled to room temperature in an exsiccator. The filters were weighed before and after filtration, and the difference was determined as the dry weight.
PCR amplification and sequencing of the almost complete 16S rRNA genes of the isolates were performed according to the methods described by Brinkhoff and Muyzer (4). For all isolates, more than 700 bp were determined. Sequences were compared with similar sequences of reference organisms by BLAST searches (http://www.ncbi.nlm.nih.gov/blast) (1).
Phylogenetic analysis was performed with ARB software (O. Strunk, O. Gross, B. Reichel, M. May, S. Hermann, N. Stuckmann, B. Nonhoff, M. Lenke, A. Ginhart, A. Vilbig, T. Ludwig, A. Bode, K.-H. Schleifer, and W. Ludwig, ARB: a software environment for sequence data; Department of Microbiology, Technische Universität München, Munich, Germany, 1998 [http://www.arb-home.de]). For tree calculation, only sequences with more than 1,300 bp were investigated using maximum-likelihood analysis. Shorter sequences were added later (using the maximum parsimony option) to the final tree.
From the dilution cultures, 33 different isolates were obtained. In agar diffusion tests, the culture broth of one of these organisms, strain T5, showed remarkable antibacterial activity against B. subtilis (inhibition zones of >6 mm between the paper disk and bacterial lawn of the target strain). The inhibiting substance was described as tropodithietic acid (14). Thiotropocin, a tropolone derivative with antibiotic properties, is the only compound known so far which has the same molecular formula as tropodithietic acid (C8H4O3S2) and shows a similar structure. Thiotropocin has been found in bacteria of different phylogenetic lineages, i.e., a Pseudomonas sp. (12) and a Caulobacter sp. (11). Tropodithietic acid contains a carboxylic tropone skeleton connected with a four-membered disulfide ring system (14), which was not described for thiotropocin. Compounds containing a tropone skeleton were obtained not only from bacteria but also from higher organisms. Colchicine, for example, was isolated from Colchicum autumnale, the autumn crocus, and is currently used therapeutically to reduce pain and inflammation during gout attacks.
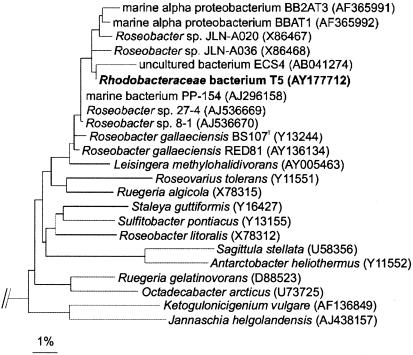
BLAST analysis of a 1,360-bp-long 16S rRNA gene fragment of strain T5 demonstrated a high level of similarity (99%) to the sequence of R. gallaeciensis BS107T and to those of seven other strains (all having sequence lengths of more than 1,300 bp and showing at least 98% similarity). Calculation of a phylogenetic tree with these sequences and those of fully characterized species of the Roseobacter clade revealed a distinct cluster of T5 and its close relatives (Fig. 1). Two shorter sequences of the uncultured bacterium ECS4 (436 bp) and of R. gallaeciensis RED81 (830 bp) falling into this cluster were also found; these sequences showed levels of similarity of 98 and 99%, respectively, to the sequence of T5. The next relative of members of this cluster is the recently described species Leisingera methylohalidivorans (20), which shows 97% sequence similarity to strain T5.
FIG. 1.
Maximum-likelihood tree showing the phylogenetic affiliations of strain T5. Selected sequences from the γ subclass of Proteobacteria strains were used to root the tree. Bar, 1% sequence divergence.
As described for R. gallaeciensis (18), colonies of strain T5 grown on marine agar are smooth, convex, and brownish with regular edges; both organisms produce a brownish diffusible pigment during growth in marine broth or on marine agar. Weak inhibition with streptomycin sulfate and strong inhibition by penicillin G and chloramphenicol were observed for T5 and R. gallaeciensis. For penicillin G, our results are in contrast to the results of Ruiz-Ponte et al. (18), who reported that cells of R. gallaeciensis are resistant to this antibiotic.
Colorless mutants of strain T5 appeared spontaneously several times on agar plates (marine agar) after aliquots from liquid cultures of strain T5 were plated without prior use of mutagenic agents or treatments. Comparison of the 16S rRNA genes of these strains and of T5 yielded identical sequences. In inhibition experiments, these mutants expressed no antibiotic activities (data not shown). No reversion to the brown phenotype was observed. One of these mutants, strain T5-3, was used for a detailed comparison to strain T5 in inhibition experiments. The highest tropodithietic acid concentration in the culture broth reached by strain T5 (and also by R. gallaeciensis) was 0.25 mM. For strain T5-3, a much lower concentration of 0.01 mM was detected.
The results of the agar diffusion tests with Wadden Sea bacteria are summarized in Table 2. While the tested Flavobacteria and Actinobacteria strains were clearly inhibited by the presence of culture broths of strain T5 and R. gallaeciensis, some α-Proteobacteria strains were resistant. No inhibition (or only reduced growth) was observed with γ-Proteobacteria strains. No effects on other isolates could be observed with the non-antibiotic-producing mutant strain T5-3, Ruegeria gelatinovorans, and Ruegeria algicola. In tests with bacterium-free culture broths of strain T5 and R. gallaeciensis, no antibiotic effect (except for that seen with two strains) was observed. For strains TL and T2, small inhibition zones of 1 to 2 mm were measured, corresponding to 25% or less of the size of the inhibition zones measured with the culture broths containing cells of strain T5 and R. gallaeciensis. Tests with pure tropodithietic acid (dissolved in 1% DMSO) showed inhibition or reduced growth of all tested strains. The organisms were not affected by the presence of a 1% DMSO solution. While culture broths showed no autotoxicity for strain T5 and R. gallaeciensis, both were inhibited by a pure 1 mM tropodithietic acid solution. Comparison of tropodithietic acid with penicillin G, streptomycin sulfate, and chloramphenicol revealed differences in the inhibition spectrum, and none of these antibiotics inhibited all of the strains which were inhibited by tropodithietic acid. The strongest effects were obtained with penicillin G, while tropodithietic acid inhibited or at least reduced the growth of all strains tested.
TABLE 2.
Antimicrobial activities of culture broths and pure antibiotics in agar diffusion tests with Wadden Sea isolates and described Roseobacter and Ruegeria spp.
| Substance or strain | Activity (mm) ofa:
|
|||||
|---|---|---|---|---|---|---|
| T5 culture broth | R. gallaeciensis culture broth | Tropodithietic acid | Penicillin G | Streptomycin sulfate | Chloramphenicol | |
| α-Proteobacteria | ||||||
| T5 | —c | — | 2 | 16 | 1 | 8 |
| T5-3 | — | — | 2 | 16 | 1 | 8 |
| R. gallaeciensis | — | — | 2 | 18 | 2 | 9 |
| R. gelatinovorans | — | — | 5 | 26 | 3 | 12 |
| R. algicola | 3 | 3 | 8 | 26 | 3 | 9 |
| T3 | 4 | 4 | 9 | — | — | 3b |
| T11 | — | — | 1 | — | 1 | 4b |
| TK | — | — | 2b | 13b | — | 2b |
| TL | 10 | 9 | 14 | 35 | 4 | 5 |
| γ-Proteobacteria | ||||||
| T1 | — | — | 3b | — | — | 4b |
| T8 | — | — | 1 | — | 1b | — |
| T16 | — | — | 1 | — | 1b | 5 |
| T17 | — | — | 1 | — | 3 | 5 |
| TAI | 3b | 2b | 2 | 4b | 2 | 4b |
| Flavobacteria | ||||||
| BIA | 13 | 12 | 13 | 13 | — | — |
| T15 | 10 | 9 | 9 | 12 | — | — |
| TN | 7 | 8 | 9 | — | — | — |
| Actinobacteria | ||||||
| T2 | 7 | 8 | 11 | 21 | 5 | 6 |
| T4 | 4 | 4 | 3 | 3b | 5 | — |
Samples (10 μl) of the different solutions were tested. The concentration of the antibiotics was 1 mM. Activity is expressed as the diameter (in millimeters) of the inhibitory zone between a paper disk and the bacterial lawn of the target strain.
No clear inhibition but areas with reduced growth were seen.
—, no growth inhibition.
Some strains used as targets allowed growth of T5 and R. gallaeciensis around the filter paper disks, while others clearly did not. No growth was observed on plates with strains T1, T8, and T5-3. Growth of strain T5 and R. gallaeciensis (visible as the brown color of the colonies of these organisms) was detected on plates with all other organisms.
Inhibition experiments with algae as target organisms revealed that none of the culture broths (with or without cells) of strain T5, R. gallaeciensis, R. gelatinovorans, and R. algicola had any effect on the algae. A pure 1 mM tropodithietic acid solution, however, inhibited growth of the tested algae. Inhibition zones (distances between alga lawns and antibiotic assay disks) had diameters of 3 to 4 mm for S. costatum and 2 mm for the nonaxenic Nitzschia species. No effect was visible with the 1% DMSO solution.
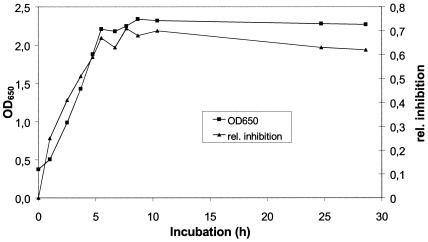
Because many bacteria exhibit production of secondary metabolites mainly in the late exponential or in the stationary phase, we investigated whether this is also the case for production of tropodithietic acid by strain T5. Inhibition zones in agar diffusion tests with Bacillus amyloliquefaciens were already observed in the early logarithmic phase and increased until the stationary phase was reached (Fig. 2). Thus, production of tropodithietic acid occurs during the complete growth phase. As OD650 and increases of biomass of T5 are highly correlated (r2 = 0.9997), growth rate and antibiotic production are also highly correlated.
FIG. 2.
Covariation between the growth of strain T5 and its production of the antibiotic. Growth is expressed as the increase in OD measured at 650 nm. Activity of the culture broth was determined using agar diffusion tests with B. amyloliquefaciens (DSM 7) as the test organism and is expressed as the inhibition of the sample relative to that of the corresponding reference (penicillin G).
Comparison of the inhibition patterns of the culture broths of strain T5 and R. gallaeciensis and of the pure antibiotic indicates that tropodithietic acid is indeed the substance causing the inhibition by the living bacteria. Target strains which were inhibited by the culture broths were also inhibited by the pure antibiotic, and organisms which were not affected by the culture broths were affected only a little by the pure antibiotic (Table 2). The inhibition efficiency of the culture broth, however, was dependent on the presence of cells of strain T5 and R. gallaeciensis. Because we observed growth of these strains around the antibiotic assay disks, we assume that this was due to ongoing antibiotic production during the tests. Production of secondary metabolites of some marine bacteria has been described as being induced by different factors (such as the presence of other bacteria) (19) or only when the bacteria are grown on a solid surface (16). Therefore, the possibility that other compounds are produced (and that other factors, e.g., competition for nutrients, causing inhibition zones are also involved) cannot be totally excluded.
Flavobacteria and Actinobacteria strains (as well as some α-Proteobacteria strains) were strongly affected by the presence of tropodithietic acid, while all γ-Proteobacteria strains tested were not influenced or showed only reduced growth. It is known that the sensitivity to antibiotics differs between different phylogenetic groups, and the observed high-level sensitivity of the Flavobacteria strains (as well as the finding that γ-Proteobacteria strains are most resilient to inhibitory substances) is in agreement with recent reports (9, 15) whereas no comparable information is so far available concerning the sensitivity of marine gram-positive bacteria. Streptomycin and chloramphenicol, however, showed no effect on the tested Flavobacteria strains. Organisms belonging to the γ subdivision of the Proteobacteria were generally the least sensitive to all tested substances. Whether this trend is generally consistent for antibiotics and what implications it may have in an ecological and also in a pharmaceutical context remains to be investigated.
Ruiz-Ponte et al. (19) showed that R. gallaeciensis BS107 (18) was antagonistic to γ-Proteobacteria strains. The authors found that cell extracts of strain BS107 significantly enhanced scallop larval survival and thus were beneficial to the rearing process. The results suggested that the inhibitory effect was displaced only in the presence of another bacterium, Vibrio anguillarum 408, or when supernatants of V. anguillarum cultures were added to pure cultures of strain BS107. The antibiotic was sensitive to trypsin (8,000 U ml−1) and stable at 100°C, and it was assumed that the substance is a peptide. In our experiments, antibacterial activity was expressed in pure cultures of strain T5 and R. gallaeciensis; since tropodithietic acid is not a peptide (14) and therefore is not sensitive to trypsin, R. gallaeciensis must be able to produce another, as-yet-undescribed antibiotic. It is known that bacteria which produce secondary metabolites often have the potential to synthesize various compounds from a single strain (2, 25). This trait seems also to be true for R. gallaeciensis.
Phylogenetic analysis of the 16S rRNA gene sequence, physiological characteristics, and similarities in the inhibition spectrum show that strain T5 is highly related to R. gallaeciensis BS107. High similarity values of at least 98% within 16S rRNA gene sequences were also found for nine other organisms (Fig. 1). For two of those strains (BBAT1 and BB2AT3 [isolated from marine aggregates]), high-level antagonistic activity was reported earlier (15); it is likely that these strains also produce tropodithietic acid. In our inhibition experiments with Ruegeria algicola and R. gelatinovorans, we observed no antagonistic activities of these bacteria even though okadaic acid production of R. algicola (formerly Roseobacter algicola) has been reported previously (13). Even Leisingera methylohalidivorans, the closest relative of members of the R. gallaeciensis cluster, does not have antagonistic properties (D. Kessler, University of Oldenburg, unpublished results); therefore, tropodithietic acid production seems to be restricted to members of the R. gallaeciensis cluster.
Organisms of the R. gallaeciensis cluster are obviously widespread in marine environments (15, 17, 18, 22), and a high level of abundance of organisms closely related to R. gallaeciensis was found by Sekiguchi et al. (22), who studied the bacterial distribution and diversity of the Changjiang estuary. Culture and molecular methods indicated that these bacteria belong to the dominant species in this area. Repeated detection of related strains in Galicia might also indicate a preferred habitat for R. gallaeciensis-like bacteria. Strain T5 was obtained from the lowest dilution of only one parallel of a most-probable-number culture series, was not detected at higher dilutions (results not shown), and is so far the only strain of the R. gallaeciensis cluster which was obtained from the Wadden Sea. Future studies have to elucidate the ecological niche of this organism and how successful it is in this habitat.
Nucleotide sequence accession number.
The sequences obtained in this study are available from GenBank under accession no. AY166703 and AY177712 to AY177725.
Acknowledgments
We thank Erhardt Rhiel for providing us with algal cultures obtained from the Wadden Sea, and we thank Axel Zeeck for helpful advice.
This work was supported by the Volkswagen Foundation within the Lower Saxonian priority program “Marine Biotechnology.”
Footnotes
We dedicate this paper to Prof. Dr. Axel Zeeck on the occasion of his 65th birthday.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode, H. B., B. Bethe, R. Höfs, and A. Zeeck. 2002. Big effects from small changes: possible ways to explore nature's chemical diversity. Chembiochem. 3:619-627. [DOI] [PubMed] [Google Scholar]
- 3.Boettcher, K. J., B. J. Barber, and J. T. Singer. 2000. Additional evidence that juvenile oyster disease is caused by a member of the Roseobacter group and colonization of nonaffected animals by Stappia stellulata-like strains. Appl. Environ. Microbiol. 66:3924-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgess, J. G., E. M. Jordan, M. Bregu, A. Mearns-Spragg, and K. G. Boyd. 1999. Microbial antagonism: a neglected avenue of natural products research. J. Biotechnol. 70:27-32. [DOI] [PubMed] [Google Scholar]
- 6.Burkholder, P. R., R. M. Pfister, and F. P. Leitz. 1966. Production of a pyrrole antibiotic by a marine bacterium. Appl. Microbiol. 14:649-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faulkner, D. J. 2000. Marine pharmacology. Antonie Leeuwenhoek 77:135-145. [DOI] [PubMed] [Google Scholar]
- 8.Gram, L., H.-P. Grossart, A. Schlingloff, and T. Kiørboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grossart, H.-P., A. Schlingloff, M. Bernhard, M. Simon, and T. Brinkhoff. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 10.Guillard, R. R. L. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 26-60. In W. L. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, N.Y.
- 11.Kawano, Y., Y. Nagawa, H. Nakanishi, H. Nakajima, M. Matsuo, and T. Higashihara. 1997. Production of thiotropocin by a marine bacterium, Caulobacter sp. and its antimicroalgal activities. J. Mar. Biotechnol. 5:225-229. [Google Scholar]
- 12.Kintaka, K., H. Ono, S. Tsubotani, S. Harada, and H. Okazaki. 1984. Thiotropocin, a new sulfur-containing 7-membered-ring antibiotic produced by a Pseudomonas sp. J. Antibiot. 37:1294-1300. [DOI] [PubMed] [Google Scholar]
- 13.Lafay, B., R. Ruimy, C. Rausch de Traubenberg, V. Breittmayer, M. J. Gauthier, and R. Christen. 1995. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45:290-296. [DOI] [PubMed] [Google Scholar]
- 14.Liang, L. 2003. Investigation of secondary metabolites of North Sea bacteria: fermentation, isolation, structure elucidation and bioactivity. Ph.D. thesis. University of Göttingen, Göttingen, Germany.
- 15.Long, R. A., and F. Azam. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long, R. A., A. Qureshi, D. J. Faulkner, and F. Azam. 2003. 2-n-Pentyl-4-quinolinol produced by a marine Alteromonas sp. and its potential ecological and biogeochemical roles. Appl. Environ. Microbiol. 69:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinhassi, J., and T. Berman. 2003. Differential growth response of colony-forming α- and γ-proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the Eastern Mediterranean Sea, and the Gulf of Eilat. Appl. Environ. Microbiol. 69:199-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Ponte, C., V. Cilia, C. Lambert, and J. L. Nicolas. 1998. Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int. J. Syst. Bacteriol. 48:537-542. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Ponte, C., J. F. Samain, J. L. Sanchez, and J. L. Nicolas. 1999. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol. 1:52-59. [DOI] [PubMed] [Google Scholar]
- 20.Schaefer, J. K., K. D. Goodwin, I. R. McDonald, J. C. Murrell, and R. S. Oremland. 2002. Leisingera methylohalidivorans gen. nov., sp. nov., a marine methylotroph that grows on methyl bromide. Int. J. Syst. Evol. Microbiol. 52:851-859. [DOI] [PubMed] [Google Scholar]
- 21.Schlösser, U. G. 1994. SAG—Sammlung von Algenkulturen at the University of Göttingen. Catalogue of strains. Bot. Acta 107:113-186. [Google Scholar]
- 22.Sekiguchi, H., H. Koshikawa, M. Hiroki, S. Murakami, K. Xu, M. Watanabe, T. Nakahara, M. Zhu, and H. Uchiyama. 2002. Bacterial distribution and phylogenetic diversity in the Changjiang estuary before the construction of the Three Gorges Dam. Microb. Ecol. 43:82-91. [DOI] [PubMed] [Google Scholar]
- 23.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner-Döbler, I., W. Beil, S. Lang, M. Meiners, and H. Laatsch. 2002. Integrated approach to explore the potential of marine microorganisms for the production of bioactive metabolites. Adv. Biochem. Eng. Biotechnol. 74:207-238. [DOI] [PubMed] [Google Scholar]
- 25.Wratten, S. J., M. S. Wolfe, R. J. Andersen, and D. J. Faulkner. 1977. Antibiotic metabolites from a marine pseudomonad. Antimicrob. Agents Chemother. 11:411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]